Abstract
Olfactory-driven behaviors are central to the lifecycle of the malaria vector mosquito Anopheles gambiae and are initiated by peripheral signaling in the antenna and other olfactory tissues. To continue gaining insight into the relationship between gene expression and olfaction, we have performed cohort comparisons of antennal transcript abundances at five time points after a blood meal, a key event in both reproduction and disease transmission cycles. We found that more than 5,000 transcripts displayed significant abundance differences, many of which were correlated by cluster analysis. Within the chemosensory gene families, we observed a general reduction in the level of chemosensory gene transcripts, although a subset of odorant receptors (AgOrs) was modestly enhanced in post–blood-fed samples. Integration of AgOr transcript abundance data with previously characterized AgOr excitatory odorant response profiles revealed potential changes in antennal odorant receptivity that coincided with the shift from host-seeking to oviposition behaviors in blood-fed female mosquitoes. Behavioral testing of ovipositing females to odorants highlighted by this synthetic analysis identified two unique, unitary oviposition cues for An. gambiae, 2-propylphenol and 4-methylcyclohexanol. We posit that modest, yet cumulative, alterations of AgOr transcript levels modulate peripheral odor coding resulting in biologically relevant behavioral effects. Moreover, these results demonstrate that highly quantitative, RNAseq transcript abundance data can be successfully integrated with functional data to generate testable hypotheses.
Keywords: chemoreceptor, bioinformatics, transcriptomics
Anautogenous female mosquitoes, including the malaria vector Anopheles gambiae, require a blood meal to complete each reproductive cycle (1). The high degree of human biting displayed by An. gambiae (2) and its competence for malaria parasite development make this species an enduring threat to human health in Africa and other parts of the world. Chemosensory inputs, most notably in the form of airborne kairomones, provide the principal sensory stimuli that drive An. gambiae blood meal host detection and selection (3, 4). Therefore, it is of great interest to further elucidate the molecular basis of olfactory-driven behaviors in disease-transmitting mosquitoes.
Host seeking in mosquitoes is episodic and activity patterns vary between species (1). Some species, such as An. gambiae, display distinctively nocturnal biting, whereas others exhibit either crepuscular or day-biting habits (5). Moreover, electrophysiological studies of both whole antennae and of individual odorant receptor neurons (ORNs) in several dipterans reveal time-of-day variability in responses to odor stimuli (6, 7). Recently, diel variation was also observed to occur at the transcript level for several An. gambiae olfactory genes, including the highly conserved An. gambiae odorant receptor coreceptor, AgamOrco (hereafter AgOrco), which displayed reduced abundance during times of day associated with behavioral inactivity (8). Importantly, An. gambiae females are refractory to host odor stimulation for a prolonged period after a blood meal (9), which correlates with reduced electrophysiological responses to some odors during the same period (10). Post-blood meal reductions in olfactory responses have also been shown in the yellow fever mosquito, Aedes aegypti, indicating that modulation of odor sensitivity may be a general characteristic of anautogenous mosquitoes (11).
In recent years, several chemosensory gene families have been identified in An. gambiae, including An. gambiae odorant (AgamOr, hereafter AgOr), An. gambiae gustatory (AgamGr, hereafter AgGr), and variant An. gambiae ionotropic glutamate (AgamIr, hereafter AgIr) receptors, as well as An. gambiae odorant binding proteins (AgamObp, hereafter AgObp) (12–15). The characterization of An. gambiae chemosensory gene expression patterns in distinct tissues along with the heterologous deorphanization of receptor sensitivities has helped refine our understanding of the links between chemosensory behavior and signaling (13, 16–22). The centrality of chemoreception to host-seeking behaviors, the shaping of the mosquito’s chemosensory receptive range by the distribution of differentially tuned chemoreceptors, and the blood meal-induced shift toward behaviors other than host seeking, suggest that expression dynamics of chemosensory genes presage overt behavioral phenotypes. Previous studies have already shown that the levels of mosquito chemosensory genes are affected by changing physiological conditions (8, 23–25). Therefore, a more exhaustive exploration of antennal transcript modulation in response to blood feeding may provide unique insights into the mechanisms of mosquito chemosensory driven behaviors central to disease transmission. To this end, we have sequenced mRNA isolated from An. gambiae antennae to compare the transcriptome profiles of non–blood-fed versus blood-fed females over the 2 d after blood feeding (Fig. S1). We have used these data, in combination with previously published AgOr response profiles, to model potential odorant sensitivity changes within the antennae subsequent to blood feeding.
Results and Discussion
Blood Feeding Globally Alters Antennal Transcript Abundance.
In 10 antennal samples, transcripts for 8,995 genes were reliably detectable above background levels. At any given time point, ∼5,000 distinct transcripts displayed significant abundance differences between paired blood-fed (Bf) and non–blood-fed (nBf) samples. In addition, 169 transcripts displayed no detectable differences in abundance between the Bf and nBf groups at any of the five time points (Dataset S1). A subset of these highly stable genes is involved in basic cellular processes (e.g., dynein, actin), and many encode protein domains associated specifically with DNA binding (e.g., WD-repeats, zinc finger, and homeobox domains). Additionally, the An. gambiae orthologs of the fruit fly circadian genes period [An. gambiae gene annotation (AGAP) 001856], timeless (AGAP008288), and cycle (AGAP005655) displayed patterns of synchronous cycling within Bf and nBf cohorts. Notably at the 24-h time point, transcripts for both period and timeless were reduced in the Bf sample (Fig. S2), a phenomenon previously documented in Bf sandflies (26, 27). These results further link physiologic state to peripheral clock gene regulation.
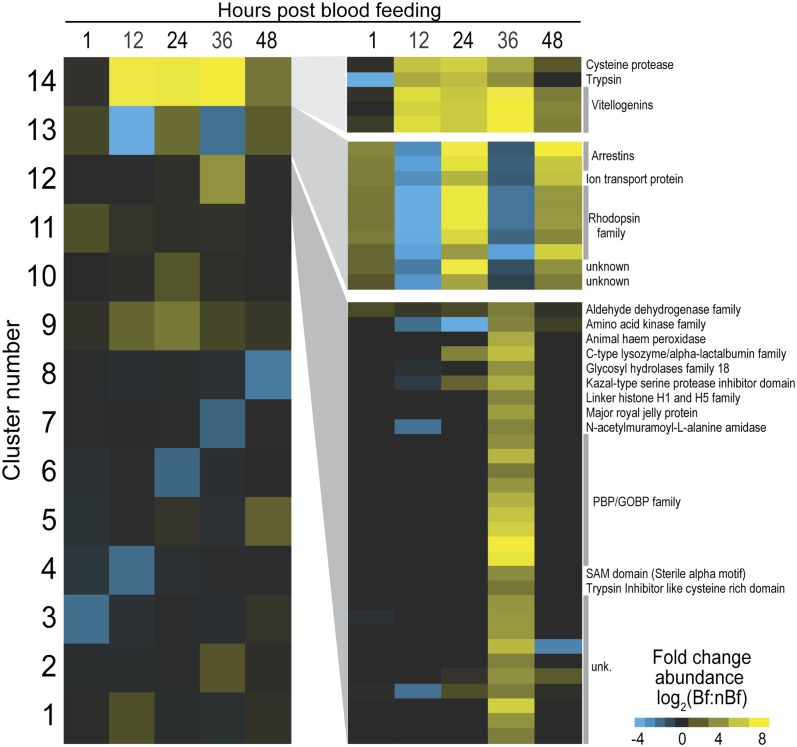
To investigate the broad patterns of antennally expressed transcripts and to further examine the integrity of our dataset, we conducted a cluster analysis of the 1,235 genes that displayed at least an absolute twofold change in abundance between Bf and nBf at one or more time points. A total of 14 clusters, each of which comprised transcripts that shared a similar differential expression profile, captured the prevalent types of variation observed in the samples (Fig. 1 and Dataset S1). Most of the clusters included transcripts that varied significantly between Bf and nBf at only a single time point, although several clusters revealed marked transcript variations across multiple time points. For example, cluster 1 comprised 712 transcripts that were greatly enhanced in the 12-h Bf antennae alone, whereas cluster 9 comprised 55 transcripts displaying marked enhancement in Bf samples across all five time points.
Fig. 1.
Cluster analysis of differential transcript abundances. Fourteen groups of genes displayed similar patterns of significant degrees of fold change in transcript abundance between blood-fed (Bf) and non–blood-fed (nBf) samples over all five time points. Log2 fold change scale indicates transcript abundances that were significantly higher (yellow) or significantly lower (blue) in Bf samples. (Left) Patterns of differential transcript abundances within each of the 14 clusters is shown in columns for 1, 12, 24, 36, and 48 h post-blood feeding. (Right) Differential expression of individual genes within cluster number 14 (Top), 13 (Middle), and 12 (Bottom). Log2 scale at Bottom Right indicates magnitude by with transcript abundances were significantly higher (yellow) or significantly lower (blue) in post-Bf samples relative to nBf samples.
Cluster 14 contained the fewest number of transcripts (5), all of which displayed very strong and sustained enhancements in the Bf samples yet were undetectable in their nBf counterparts. Interestingly, three of these transcripts encoded vitellogenin precursor proteins that are normally involved in oogenesis and expressed by fat bodies (28). This cluster also included the transcripts for the trypsin and cysteine protease genes that are typically associated with blood digestion and are enriched in An. gambiae following a blood meal (23, 24). Similarly, cluster 12 encompassed significant enrichment in transcripts whose function is not usually associated with olfaction and included transcripts for major royal jelly protein (AGAP005958), heme peroxidase (AGAP004038), and a homolog to the Drosophila gene Dmel\CG9629 (AGAP000881), all of which have been associated with dipteran ovaries or embryos (29, 30). This expression in the antenna may reflect induction by circulating signaling factors such as juvenile hormone or 20-hydroxyecdysone, which can activate transcription in tissues where they would seemingly have no obvious function (28).
The largest group of olfactory-associated transcripts also appeared in cluster 12 and included a subset of oxidases/dehydrogenases that could serve as odor-degrading enzymes (31) and nine AgObps that were only found above the threshold of detection in the 36-h Bf sample (Fig. 1). Although the exclusive expression of these AgObps in the later stages of the gonotrophic cycle may reflect their requirement for the onset of olfactory-driven oviposition behaviors, it is notable that seven of these AgObps belong to the “atypical” class of two-domain Obps (14) and were the only atypical Obps detectable within the antenna. Consequently, their dissimilarity to “classical” Obps coupled with their co-occurrence with transcripts not normally associated with olfaction suggests that they might also be subject to global regulation and play their primary roles outside the antennae.
A final cluster of particular interest was the set of 10 transcripts in cluster 13 that displayed a strong, diel oscillatory abundance pattern that is phase shifted subsequent to blood feeding; this phase shift results in the differential expression pattern displayed by this cluster. The rhythmicity seen in these transcripts suggests that peripheral circadian clock genes may be involved in their regulation. It is well established that autonomous clocks operate in a number of peripheral insect tissues, including the antennae of Drosophila melanogaster (32, 33) where odor sensitivity rhythms are affected by the circadian oscillator (34, 35). Although differential expression analysis would not detect genes that cycle synchronously between Bf and nBf, the rhythmic pattern seen in cluster 13 is the result of a decoupling of diel rhythmicity between the Bf and nBf groups following blood feeding, possibly a result of the near doubling in the abundance of the clock ortholog (AGAP005711) within the Bf cohort (Fig. S2).
The functional implications of this cluster are further suggested by the presence of five opsin G protein-coupled receptors (GPCRs) and two arrestins. The presence of opsin transcripts in An. gambiae antennae was reported by our group (36), and three of the opsins in cluster 13 (AGAPs 13149, 12985, and 12982) have high sequence similarity to D. melanogaster opsins, Rh6 and ninaE. Both genes have been associated with nonvisual sensory modalities in D. melanogaster, Rh6 with audition (37), and ninaE with 18–24 °C temperature discrimination in larvae (38). Although it is unclear what role audition may play in post–blood-feeding behaviors, the differential abundance of opsins may be indicative of shifts in the thermal preferences of An. gambiae females that tend to rest in cool spaces following blood feeding (1).
More provocatively, these GPCRs may be involved in olfactory signal transduction within ORNs. Light-dark behavior cycles in adult mosquitoes are likely tied to circadian oscillators that, as discussed above, show strong and consistent variations in light-dark expression patterns across all time points. Moreover, in all samples, the abundance pattern of Gαq followed that of the transcripts in cluster 13 (Dataset S1). Isoforms of Gαq subunits have been immunolocalized within the antennal sensilla of An. gambiae (39) and have also been shown to strongly affect the electrophysiogical responses of D. melanogaster antenna to a variety of odors (40). In as much as only the clock ortholog shows a strong change in transcript abundance in response to blood feeding (Fig. S2), the expression pattern of this gene cluster may be the result of a simple regulatory mechanism that modulates the overall responsiveness of the antenna and gives rise to the diel- and blood feeding-dependent patterns of olfactory behavior and physiology.
Chemosensory Genes Show Subtle Alterations.
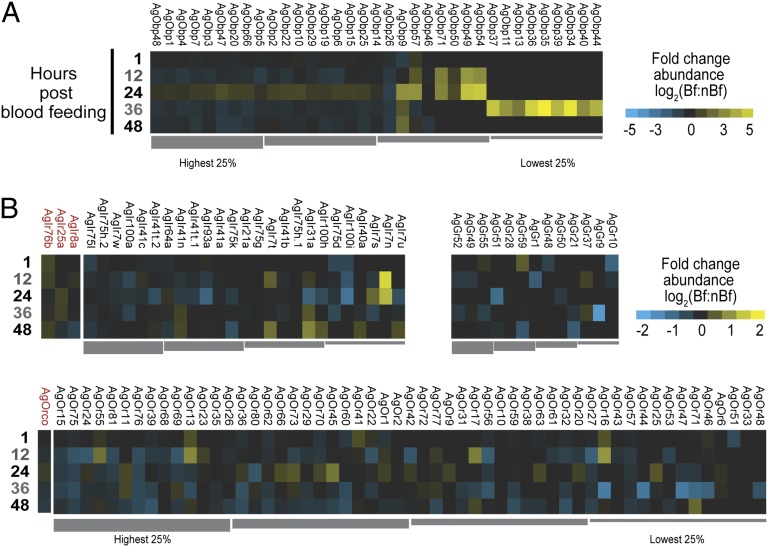
Not surprisingly, only a portion of the AgGrs, AgIrs, AgObps, and AgOrs annotated in the An. gambiae genome were detectable in the adult female antenna (Fig. 2). Although this incomplete representation may, in part, reflect limited expression in diverse subpopulations of antennal cells, some of those undetected chemosensory genes have been associated with chemosensory tissues other than the antenna (e.g., palp, labellum), whereas others are known to be exclusive to the An. gambiae larval life stage (17–20). The general absence of most of the annotated AgObps is also consistent with our previous study that showed many AgObps to be highly enriched in the adult mosquito body rather than in chemosensory tissues (36).
Fig. 2.
Antennal chemosensory differential transcript abundances following a bloodmeal. Chemosensory transcripts that were represented at significantly higher (yellow) or lower (blue) values in post–blood-fed samples; nondifferentially expressed chemosensory transcripts are denoted as zeros (black). Genes within each chemosensory gene family are arrayed left to right from most abundant to least based upon abundance levels seen within the 1-h Bf cohort. (A) Odorant binding protein family (AgObp). (B) Chemoreceptor families. (B, Upper Left) Variant ionotropic receptor family (AgIr) with the three AgIr coreceptors on the left (red); (B, Upper Right) Gustatory receptors (AgGr). (B, Lower) Odorant receptor family (AgOr) with the AgOr coreceptor (AgOrco) on the left (red). All transcripts were ordered left to right from highest to lowest RPKM values (quartile bars above or below each image). Log2 scales indicate transcript abundances that were significantly higher (yellow) or significantly lower (blue) in post-blood–fed samples. Note different scales for panel A and panel B.
As revealed by the cluster analysis above, the AgObps displayed the greatest variation among the chemosensory genes during the 2 d after blood feeding. Thirty-five AgObps were detectable in the female antenna, and most were expressed at very high levels, ranking these AgObps among the most highly expressed genes in the antenna. Following blood feeding, the most abundant 50 percent of AgObps displayed pervasive deenrichment across every time point except for 24 h, when the transcripts for nearly every AgObp showed enrichment in the blood fed cohort (Fig. 2A), suggestive of the recovery of the molecular apparatus of olfaction.
The other notable enrichment among AgObps occurred at 36 h in the Bf cohort when nine otherwise undetectable AgObps all spiked in abundance. Given the close physical proximity of these AgObps on either chromosome 2R (AgObps 13, 39, and 40) or the X chromosome (AgObps 34, 35, 36, and 37), it is likely that these AgObps share common regulatory elements. This observation is entirely consistent with previous observations of high levels of these aytpical AgObps in whole mosquitoes following blood feeding (23). Inasmuch as those AgObps that have been characterized as playing a functional role in olfaction (41) show continually high abundance in the antenna throughout our analyses, the transitory appearance of transcript for these nine AgObps is likely the result of a more global, organismal-level regulation of transcription.
In contrast to the AgObps, transcripts for the antennal chemoreceptors did not display dramatic changes in their levels between the Bf and nBf cohorts. Consistent with their primary role in contact chemosensation, AgGrs were the least abundant chemoreceptor class in the antenna and they showed the least variation in response to blood feeding (42, 43). Similarly invariant transcript levels were displayed by the AgIr and AgOr coreceptors (AgIr8a, AgIr25a, AgIr76b, and AgOrco), implying that on the whole levels, AgIr and AgOr transcripts remained relatively stable between the Bf and nBf samples (Fig. 2B). However, transcripts for individual tuning AgIrs and AgOrs showed a pattern of depletion in the Bf cohort (Fig. S3) interspersed with select instances of enrichment (Fig. 2B), suggesting that the combinations of AgIrs and AgOrs represented in the antenna changed after a blood meal. Moreover, the overall rank order of the level of abundance of individual AgIrs and AgOrs shifted following the blood meal, indicating a temporary reshuffling in the relative abundances of chemoreceptors over the days following a blood meal (Fig. S4).
Taken together, these data suggest that blood feeding may have complex consequences upon peripheral chemoreception. In Drosophila, individual ORNs are known to express multiple Irs (42) and Ors (44), and the shift in rank order observed here could reflect fluctuating receptor levels within individual ORNs, perhaps effecting subtle shifts in their responsive range following a blood meal. Indeed, a certain level of dynamism within the chemosensory periphery of An. gambiae has already been suggested by studies that show that some antennal sensilla temporarily change in their odor response profiles subsequent to a blood meal (45). Therefore, although the overall density of chemoreceptors in the antenna appears to be a constant, the composition of the chemoreceptor population may be altered in response to blood feeding, such that subtle changes in abundance levels of a subset of chemoreceptors may transiently modulate antennal sensitivity to select semiochemicals (Fig. 2B).
Integration of Physiologic Data Reveals Post-Blood Feeding Shift in Antennal Odor Receptivity.
Given that the responsiveness of mosquito chemosensory sensilla varies concordantly with even small changes in the transcript abundance of the responding odorant receptor (46), and that significant numbers of AgOrs have been functionally characterized against a representative panel of odorant molecules (21, 22), our present dataset affords a powerful opportunity to contextualize the response profiles of those deorphanized AgOrs within the mosquito antenna. By weighting the odor-response data with our measured transcript abundance levels, we calculated the relative modulation of the cumulative receptor responses to each odorant, resulting in a predictive heat map that depicts how fluctuations in AgOr expression levels might affect the odorant response spectrum of the adult female antenna following blood feeding (Fig. 3).
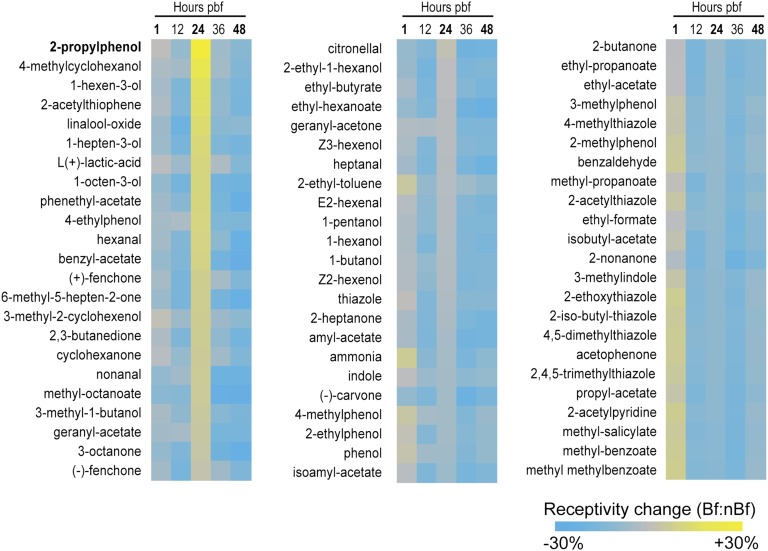
Fig. 3.
Calculated changes in AgOr-mediated odorant receptivity following a bloodmeal. Displayed are the conceptualized differences in antennal receptivity for a panel of 69 odors in post–blood-fed (Bf) vs. non–blood-fed females (nBf). Response characteristics to each chemical were based on known AgOr responses in heterologous expression systems and were then weighted by the relative antennal expression levels for each responding receptor. Results are sorted from high to low, based on the receptivity displayed by the 24-h post–blood-feeding time point. Scale bar shows calculated increases (yellow) and decreases (blue) in odor receptivity.
This analysis suggested that at 1 h after blood meal, the weighted odor response profile remained very similar between the Bf and the nBf mosquitoes, and at 12 h after blood meal, the Bf cohort displayed an overall decrease in calculated odor responsiveness to all 69 odorants (Fig. 3). This trend was seen again at both the 36-h and 48-h time points. For example, at 12 h after blood feeding, the largest sensitivity depletions to odorant occurred with respect to linalool-oxide and, second, to 1-octen-3-ol. Linalool oxide is a flower-associated aromatic and is likely involved in the flower feeding behavior of all adult mosquitoes; importantly, it along with 1-octen-3-ol are also characterized components of human skin emanations (47, 48). Moreover, 1-octen-3-ol has been widely characterized as a host-associated semiochemical for both Culcidae in general and Anophelinae in particular (49, 50). Both 36-h and 48-h time points also showed a generalized diminution in weighted odor receptivity in the blood-fed group. This general shift away from most odorants including some host cues would be in keeping with the observed refractory period in host seeking following a blood meal. Moreover, given the relative stability of AgOrco abundance, this reduction in receptivity to these odorants would also accommodate an enhancement in receptivity to a select group of other semiochemicals.
Indeed, at 24 h after blood feeding, the calculated responsiveness to a discrete set of odorants appeared to increase in the Bf sample (Fig. 3). A 10- to 20-percent increase in enhanced receptivity is observed for the compounds 2-iso-butyl-thiazole, 1-hexen-3-ol, 4-methylcyclohexanol, and 2-propylphenol (Fig. 3). Of the odors to which blood-fed females appear to have increased their receptivity, at least 10 have been classified as attractive semiochemicals, half of which are general oviposition attractants, and four of which have been characterized oviposition attractants in mosquitoes specifically (21, 51, 52). In contrast, longer chain esters have been implicated as oviposition repellants in some mosquito species (53) and the only relative receptive increase to esters in this analysis occurs with methyl octanoate, the longest chain ester assayed. This analysis shows a focused enhancement in the receptivity of the antenna that is initially reflected in the transcriptome profile 24 h after taking a blood meal. Because several of the odorants to which the antenna becomes more attuned have been previously implicated in mosquito oviposition behavior, this analytical approach suggests that the act of blood feeding may up-regulate a select subset of AgOrs in anticipation of the gravid female’s need to choose an oviposition site.
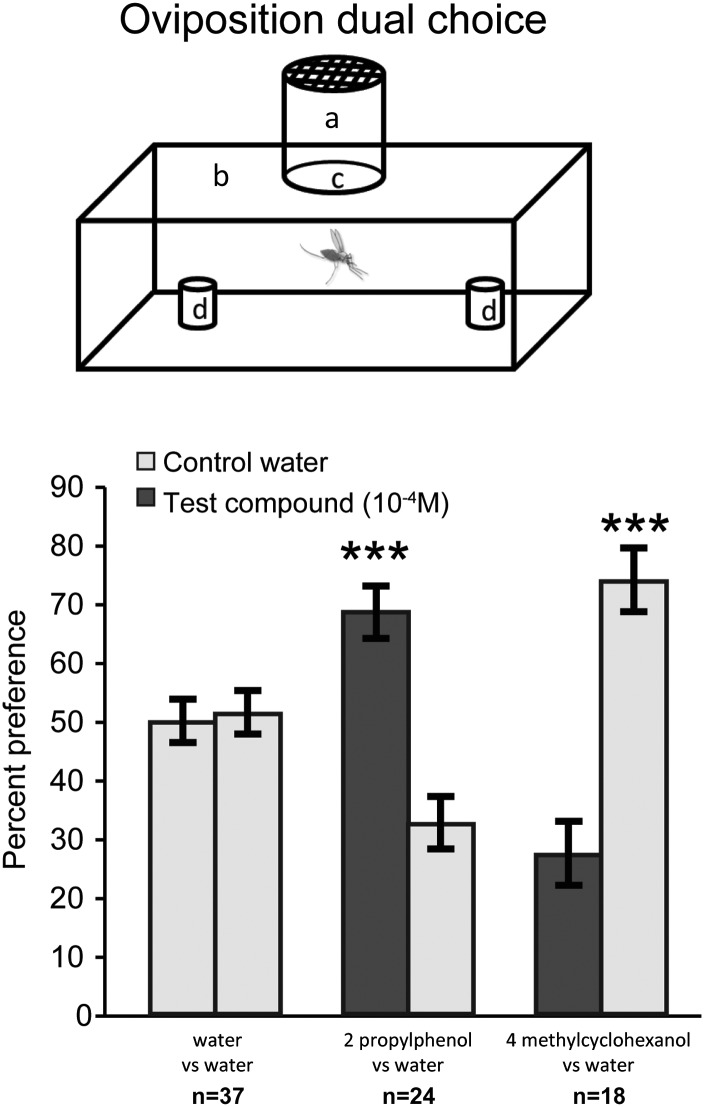
To test the hypothesis that the odors displaying increased receptivity are related to oviposition, we conducted dual choice oviposition assays by using the top two compounds displaying enhanced receptivity in the Bf cohort at the 24 h time point (2-propylphenol and 4-methylcyclohexanol). In dual-choice oviposition assays, An. gambiae females displayed a robust preference for 2-propylphenol and a significant aversion to 4-methylcyclohexanol (Fig. 4). Whereas 4-methylcyclohexanol has been shown to act as an oviposition cue for several species of Toxorhynchites (51), 2-propylphenol not been previously characterized either within the context of insect behavior in general or Anophelene oviposition behavior in particular. These observations not only demonstrate that these unitary odorants elicit robust responses from gravid, ovipositing An. gambiae females, they serve to support the implementation and interpretation of the unique receptivity analysis carried out here.
Fig. 4.
Dual choice oviposition assay. (Upper) Schematic diagram of oviposition preference bioassay cage for dual choice test. Gravid females are held in releasing chamber (a) and allowed to enter assay cage (b) after the dark cycle begins by opening a pathway (c) connecting the releasing chamber to the assay cage. Females are allowed to choose between two oviposition cups (d) containing either test water or control water. Ten females are released per cage and represent a single assay. (Lower) Gravid female responses to a choice of either water vs. water (Left), water vs. 2-propyl phenol (Center) or water vs. 4 methylcyclohexanol (Right). y axis reports percent of total eggs oviposited in either untreated water (light gray bar) or treated water (dark gray bar). All compounds were tested at a concentration of 10−4 M. The number of dual choice assays is indicated along the x axis. Error bars show SEM. ***P < 0.001, t test.
Extending on these results, we would suggest that AgOrs that were significantly enriched in the Bf cohort at the 24-h time point (e.g., AgOrs45, AgOr73, AgOr66, AgOr11) may be centrally involved in olfactory-mediated behaviors in An. gambiae post-blood feeding and oviposition site selection behaviors in particular. However, it should be noted that because individual AgOrs respond to multiple odorant ligands that can, in turn, activate multiple AgOrs (21, 22), examining any given AgOr without regard to the full diversity of expressed chemoreceptors is unlikely to provide a complete picture of peripheral chemical receptivity. Importantly, the receptivity analysis presented here takes both considerations into account and suggests that even small changes in the chemosensory transcriptome profile can produce additive and biologically significant effects when analyzed within the context of odorant receptivity.
Although this analysis does not account for the important consequences of downstream processing and integration of peripheral responses that occur in the antennal lobe and other components of the central nervous system, it has revealed that a pattern of small changes in the levels of multiple peripheral chemoreceptors may act combinatorially to transiently shift the receptive profile of the An. gambiae antenna. The timing of this shift is coincident with the female mosquito’s transition from host seeking to oviposition behaviors, and our analysis enabled us to subsequently identify two compounds that alter oviposition behavior in gravid females. More broadly, the work presented here suggests that high-resolution RNAseq data may be integrated with functional information (e.g., transcriptional cascades, cell-cell signaling pathways, or metabolic pathways) to produce informative, synthetic analyses.
Methods
Mosquito Rearing and Blood Feeding.
An. gambiae sensu stricto (SUA 2La/2La), an M-form isolate originating from Suakoko, Liberia (9), were reared in the Vanderbilt Insectary Facility as described (36).
RNA Isolation and RNA Sequencing.
Over the 2 d following blood feeding, ∼200 female mosquitoes were collected from each cohort at each of the five sequential, post–blood-feeding time points (1, 12, 24, 36, and 48 h) and antennae were resected for RNA isolation. mRNA was sequenced in 100-bp, paired-end fashion, on a single lane of an Illumina HiSeq2000.
Odorant Receptivity Changes.
Relative differences in odorant receptivity between the Bf and the nBf cohorts was calculated from physiologic, odorant-response data from previously published functional deorphinization of An. gambiae odorant receptors (21). The response of each AgOr to each odorant (in spikes per second) was weighted by the transcript abundance of that AgOr. Odorant responses in weighted spikes per second were then summed for each odorant. The post–blood-feeding “receptivity change” of the antenna to each odorant was calculated per odorant by dividing the summed weighted spikes per second for each chemical in the Bf group by the summed weighted spikes per second for each chemical in the nBf group.
Supplementary Material
Acknowledgments
We thank Travis Clark, Chelsea Baker, and the Vanderbilt Technologies for Advanced Genomics for RNA sample preparation and Illumina sequencing; and Dr. John Carlson, Dr. John Gibbons, and members of the A.R. and L.J.Z. laboratories for helpful discussions and critical readings. This work was conducted in part by using the resources of the Advanced Computing Center for Research and Education at Vanderbilt University and was supported by funds provided by the Searle Scholars Program and National Science Foundation Grant DEB-0844968 (to A.R.), grants from the Innovation and Discovery in Engineering and Science program of Vanderbilt University, and National Institute of Allergy and Infectious Diseases, National Institutes of Health Grant AI056402 (to L.J.Z.). D.C.R. was supported by the National Institute on Deafness and Other Communication Disorders through National Research Service Award F31 DC012991.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302562110/-/DCSupplemental.
References
- 1.Clements AN. The Biology of Mosquitoes. New York: CUNY Anim Behav Initiat; 1999. [Google Scholar]
- 2.Scott TW, Takken W. Feeding strategies of anthropophilic mosquitoes result in increased risk of pathogen transmission. Trends Parasitol. 2012;28(3):114–121. doi: 10.1016/j.pt.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Zwiebel LJ, Takken W. Olfactory regulation of mosquito-host interactions. Insect Biochem Mol Biol. 2004;34(7):645–652. doi: 10.1016/j.ibmb.2004.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takken W, Knols BG. Odor-mediated behavior of Afrotropical malaria mosquitoes. Annu Rev Entomol. 1999;44:131–157. doi: 10.1146/annurev.ento.44.1.131. [DOI] [PubMed] [Google Scholar]
- 5.Klowden MJ, Briegel H. Mosquito gonotrophic cycle and multiple feeding potential: Contrasts between Anopheles and Aedes (Diptera: Culicidae) J Med Entomol. 1994;31(4):618–622. doi: 10.1093/jmedent/31.4.618. [DOI] [PubMed] [Google Scholar]
- 6.Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400(6742):375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- 7.Van der Goes van Naters WM, Den Otter CJ, Maes FW. Olfactory sensitivity in tsetse flies: A daily rhythm. Chem Senses. 1998;23(3):351–357. doi: 10.1093/chemse/23.3.351. [DOI] [PubMed] [Google Scholar]
- 8.Rund SSC, Hou TY, Ward SM, Collins FH, Duffield GE. Genome-wide profiling of diel and circadian gene expression in the malaria vector Anopheles gambiae. Proc Natil Acad Sci USA. 2011;108(32):E421–E430. doi: 10.1073/pnas.1100584108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takken W, Adam W, van Loon JJ. Inhibition of host-seeking response and olfactory responsiveness in Anopheles gambiae following blood feeding. J Insect Physiol. 2001;47(3):303–310. doi: 10.1016/s0022-1910(00)00107-4. [DOI] [PubMed] [Google Scholar]
- 10.Qiu YT, et al. Behavioural and electrophysiological responses of the malaria mosquito Anopheles gambiae Giles sensu stricto (Diptera: Culicidae) to human skin emanations. Med Vet Entomol. 2004;18(4):429–438. doi: 10.1111/j.0269-283X.2004.00534.x. [DOI] [PubMed] [Google Scholar]
- 11.Siju KP, Hill SR, Hansson BS, Ignell R. Influence of blood meal on the responsiveness of olfactory receptor neurons in antennal sensilla trichodea of the yellow fever mosquito, Aedes aegypti. J Insect Physiol. 2010;56(6):659–665. doi: 10.1016/j.jinsphys.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Croset V, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6(8):e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill CA, et al. G protein-coupled receptors in Anopheles gambiae. Science. 2002;298(5591):176–178. doi: 10.1126/science.1076196. [DOI] [PubMed] [Google Scholar]
- 14.Xu PX, Zwiebel LJ, Smith DP. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol Biol. 2003;12(6):549–560. doi: 10.1046/j.1365-2583.2003.00440.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu C, et al. Distinct olfactory signaling mechanisms in the malaria vector mosquito Anopheles gambiae. PLoS Biol. 2010;8(8):e1000467. doi: 10.1371/journal.pbio.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox AN, Pitts RJ, Robertson HM, Carlson JR, Zwiebel LJ. Candidate odorant receptors from the malaria vector mosquito Anopheles gambiae and evidence of down-regulation in response to blood feeding. Proc Natl Acad Sci USA. 2001;98(25):14693–14697. doi: 10.1073/pnas.261432998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pitts RJ, Fox AN, Zwiebel LJ. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae. Proc Natl Acad Sci USA. 2004;101(14):5058–5063. doi: 10.1073/pnas.0308146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon H-W, Lu T, Rützler M, Zwiebel LJ. Olfactory responses in a gustatory organ of the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci USA. 2006;103(36):13526–13531. doi: 10.1073/pnas.0601107103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu T, et al. Odor coding in the maxillary palp of the malaria vector mosquito Anopheles gambiae. Current Biol. 2007;17(18):1533–1544. doi: 10.1016/j.cub.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia Y, et al. The molecular and cellular basis of olfactory-driven behavior in Anopheles gambiae larvae. Proc Natl Acad Sci USA. 2008;105(17):6433–6438. doi: 10.1073/pnas.0801007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carey AF, Wang G, Su C-Y, Zwiebel LJ, Carlson JR. Odorant reception in the malaria mosquito Anopheles gambiae. Nature. 2010;464(7285):66–71. doi: 10.1038/nature08834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang G, Carey AF, Carlson JR, Zwiebel LJ. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae. Proc Natl Acad Sci USA. 2010;107(9):4418–4423. doi: 10.1073/pnas.0913392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marinotti O, Nguyen QK, Calvo E, James AA, Ribeiro JM. Microarray analysis of genes showing variable expression following a blood meal in Anopheles gambiae. Insect Mol Biol. 2005;14(4):365–373. doi: 10.1111/j.1365-2583.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- 24.Marinotti O, et al. Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol Biol. 2006;15(1):1–12. doi: 10.1111/j.1365-2583.2006.00610.x. [DOI] [PubMed] [Google Scholar]
- 25.Sim CK, Perry S, Tharadra SK, Lipsick JS, Ray A. Epigenetic regulation of olfactory receptor gene expression by the Myb-MuvB/dREAM complex. Genes Dev. 2012;26(22):2483–2498. doi: 10.1101/gad.201665.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meireles-Filho ACA, Amoretty PR, Souza NA, Kyriacou CP, Peixoto AA. Rhythmic expression of the cycle gene in a hematophagous insect vector. BMC Mol Biol. 2006;7:38. doi: 10.1186/1471-2199-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meireles-Filho ACA, et al. The biological clock of an hematophagous insect: Locomotor activity rhythms, circadian expression and downregulation after a blood meal. FEBS Lett. 2006;580(1):2–8. doi: 10.1016/j.febslet.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 28.Attardo GM, Hansen IA, Raikhel AS. Nutritional regulation of vitellogenesis in mosquitoes: Implications for anautogeny. Insect Biochem Mol Biol. 2005;35(7):661–675. doi: 10.1016/j.ibmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Fakhouri M, et al. Minor proteins and enzymes of the Drosophila eggshell matrix. Dev Biol. 2006;293(1):127–141. doi: 10.1016/j.ydbio.2006.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guedes SdeM, et al. Drosophila melanogaster larval hemolymph protein mapping. Biochem Biophys Res Commun. 2003;312(3):545–554. doi: 10.1016/j.bbrc.2003.10.156. [DOI] [PubMed] [Google Scholar]
- 31.Leal WS. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58(1):373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- 32.Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278(5343):1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 33.Ivanchenko M, Stanewsky R, Giebultowicz JM. Circadian photoreception in Drosophila: Functions of cryptochrome in peripheral and central clocks. J Biol Rhythms. 2001;16(3):205–215. doi: 10.1177/074873040101600303. [DOI] [PubMed] [Google Scholar]
- 34.Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biology. 2004;14(8):638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Krishnan P, Chatterjee A, Tanoue S, Hardin PE. Spike amplitude of single-unit responses in antennal sensillae is controlled by the Drosophila circadian clock. Curr Biol. 2008;18(11):803–807. doi: 10.1016/j.cub.2008.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pitts RJ, Rinker DC, Jones PL, Rokas A, Zwiebel LJ. Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics. 2011;12:271. doi: 10.1186/1471-2164-12-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senthilan PR, et al. Drosophila auditory organ genes and genetic hearing defects. Cell. 2012;150(5):1042–1054. doi: 10.1016/j.cell.2012.06.043. [DOI] [PubMed] [Google Scholar]
- 38.Shen WL, et al. Function of rhodopsin in temperature discrimination in Drosophila. Science. 2011;331(6022):1333–1336. doi: 10.1126/science.1198904. [DOI] [PubMed] [Google Scholar]
- 39.Rützler M, Zwiebel LJ. Molecular biology of insect olfaction: Recent progress and conceptual models. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2005;191(9):777–790. doi: 10.1007/s00359-005-0044-y. [DOI] [PubMed] [Google Scholar]
- 40.Kain P, et al. Reduced odor responses from antennal neurons of Gq{alpha}, phospholipase C{beta}, and rdgA mutants in drosophila support a role for a phospholipid intermediate in insect olfactory transduction. J Neurosci. 2008;28(18):4745–4755. doi: 10.1523/JNEUROSCI.5306-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelletier J, Guidolin A, Syed Z, Cornel AJ, Leal WS. Knockdown of a mosquito odorant-binding protein involved in the sensitive detection of oviposition attractants. J Chem Ecol. 2010;36(3):245–248. doi: 10.1007/s10886-010-9762-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136(1):149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montell C. A taste of the Drosophila gustatory receptors. Curr Opin Neurobiol. 2009;19(4):345–353. doi: 10.1016/j.conb.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldman AL, Van der Goes van Naters W, Lessing D, Warr CG, Carlson JR. Coexpression of two functional odor receptors in one neuron. Neuron. 2005;45(5):661–666. doi: 10.1016/j.neuron.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 45.Qiu YT, van Loon JJA, Takken W, Meijerink J, Smid HM. Olfactory coding in antennal neurons of the malaria mosquito, Anopheles gambiae. Chem Senses. 2006;31(9):845–863. doi: 10.1093/chemse/bjl027. [DOI] [PubMed] [Google Scholar]
- 46.Bohbot JD, Durand NF, Vinyard BT, Dickens JC. March 7, 2013. Functional development of the octenol response in Aedes aegypti. Front Physiol 4:39 10.3389/fphys.2013.00039. [DOI] [PMC free article] [PubMed]
- 47.Cork A, Park KC. Identification of electrophysiologically-active compounds for the malaria mosquito, Anopheles gambiae, in human sweat extracts. Med Vet Entomol. 1996;10(3):269–276. doi: 10.1111/j.1365-2915.1996.tb00742.x. [DOI] [PubMed] [Google Scholar]
- 48.Bernier UR, et al. Synergistic attraction of Aedes aegypti (L.) to binary blends of L-lactic acid and acetone, dichloromethane, or dimethyl disulfide. J Med Entomol. 2003;40(5):653–656. doi: 10.1603/0022-2585-40.5.653. [DOI] [PubMed] [Google Scholar]
- 49.Takken W, Kline DL. Carbon dioxide and 1-octen-3-ol as mosquito attractants. J Am Mosq Control Assoc. 1989;5(3):311–316. [PubMed] [Google Scholar]
- 50.Takken W, Knols BGJ, Otten H. Interactions between physical and olfactory cues in the host seeking behaviour of mosquitoes: The role of relative humidity. Ann Trop Med Parasitol. 1997;91(2):119–120. [Google Scholar]
- 51.Collins LE, Blackwell A. Electroantennogram studies of potential oviposition attractants for Toxorhynchites moctezuma and T. amboinensis mosquitoes. Physiol Entomol. 1998;23(3):214–219. [Google Scholar]
- 52.Leal WS, et al. Reverse and conventional chemical ecology approaches for the development of oviposition attractants for Culex mosquitoes. PLoS ONE. 2008;3(8):e3045. doi: 10.1371/journal.pone.0003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ganesan K, Mendki MJ, Suryanarayana MVS, Prakash S, Malhotra RC. Studies of Aedes aegypti (Diptera: Culicidae) ovipositional responses to newly identified semiochemicals from conspecific eggs. Aust J Entomol. 2006;45(1):75–80. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.