Abstract
DNA methylation is an important epigenetic mark in many eukaryotic organisms. De novo DNA methylation in plants can be achieved by the RNA-directed DNA methylation (RdDM) pathway, where the plant-specific DNA-dependent RNA polymerase IV (Pol IV) transcribes target sequences to initiate 24-nt siRNA production and action. The putative DNA binding protein DTF1/SHH1 of Arabidopsis has been shown to associate with Pol IV and is required for 24-nt siRNA accumulation and transcriptional silencing at several RdDM target loci. However, the extent and mechanism of DTF1 function in RdDM is unclear. We show here that DTF1 is necessary for the accumulation of the majority of Pol IV-dependent 24-nt siRNAs. It is also required for a large proportion of Pol IV-dependent de novo DNA methylation. Interestingly, there is a group of RdDM target loci where 24-nt siRNA accumulation but not DNA methylation is dependent on DTF1. DTF1 interacts directly with the chromatin remodeling protein CLASSY 1 (CLSY1), and both DTF1 and CLSY1 are associated in vivo with Pol IV but not Pol V, which functions downstream in the RdDM effector complex. DTF1 and DTF2 (a DTF1-like protein) contain a SAWADEE domain, which was found to bind specifically to histone H3 containing H3K9 methylation. Taken together, our results show that DTF1 is a core component of the RdDM pathway, and suggest that DTF1 interacts with CLSY1 to assist in the recruitment of Pol IV to RdDM target loci where H3K9 methylation may be an important feature. Our results also suggest the involvement of DTF1 in an important negative feedback mechanism for DNA methylation at some RdDM target loci.
Keywords: histone modifications, small RNA, gene silencing, transposon
DNA cytosine methylation is a conserved epigenetic mark that plays important roles in maintaining genome stability, transcriptional gene silencing, and developmental regulation (1, 2). In plants, DNA methylation occurs in three sequence contexts: CG, CHG, and CHH (H = A, C, T). CG and CHG methylation are symmetric in sequence and are maintained through a semiconservative mechanism that requires the DNA methyltransferases (METHYLTRANSFERASE 1) (MET1) and CHROMOMETHYLASE 3 (CMT3), respectively. In contrast, the asymmetric CHH methylation needs to be established during each cell cycle (1, 2). In plants a 24-nt small interfering RNA (siRNA)-dependent DNA methylation pathway is involved in recruiting the de novo DNA methyltransferase DOMAINS REARRANGED METHYLASE 2 (DRM2) and is responsible for DNA methylation at many transposable elements and repetitive sequences (3, 4).
Two plant-specific homologs of RNA polymerase II play important roles in the RNA-directed DNA methylation (RdDM) pathway (5). RNA polymerase IV presumably initiates 24-nt siRNA biogenesis by specifically transcribing RdDM target loci to produce single-stranded RNA (ssRNA) transcripts, which serve as templates for RNA-dependent RNA polymerase 2 (RDR2) to generate double-stranded RNAs (dsRNAs). CLASSY 1 (CLSY1), a putative ATP-dependent chromatin remodeling protein, is proposed to be involved in one or more steps of dsRNA generation. The resulting dsRNAs are then processed by Dicer-like 3 (DCL3) to generate 24-nt siRNAs, which are loaded onto ARGONAUTE (AGO)4 or AGO6. RNA polymerase V produces scaffold RNAs that help recruit 24-nt siRNA-bound AGO4, which together with several other proteins, including RDM1, forms the guiding complex to recruit DRM2 (4). Whereas Pol V transcripts have been recently identified (6), the mechanism of Pol IV function remains to be elucidated. It is unclear what features are recognized by Pol IV to initiate RdDM at specific loci.
In addition to siRNAs, specific histone modifications can also regulate DNA methylation (7). An example of cross-talk between histone H3 lysine 9 dimethylation (H3K9me2) and CHG methylation is implemented by a positive feedback loop involving the DNA methyltransferase CMT3 and histone methyltransferase KYP (KRYPTONITE). CMT3 contains bromo adjacent homology and chromo domains that bind to H3K9me2 and catalyzes CHG methylation (8), whereas KYP harbors SET- and RING-associated domains that bind to CHG and CHH methylation and methylates histone H3K9 (9). Several histone-modifying enzymes including the histone deacetylase HDA6 (10), the putative histone demethylase JMJ14 (JUMONJI 14) (11, 12) and histone H2B deubiquitination enzyme UBP26/SUP32 (UBIQUITIN PROTEASE 26, also known as SUP32) (13) have been shown to affect DNA methylation and/or transcriptional silencing of RdDM target loci. However, direct links between specific histone modifications and the initiation of RdDM are still missing.
We previously identified an RdDM component named DNA-BINDING TRANSCRIPTION FACTOR 1 (DTF1) through a REPRESSOR OF SILENCING 1-1 (ros1-1) suppressor screen (14). Similar to other RdDM mutants that suppress ros1, dtf1 ros1 exhibits reduced DNA methylation and derepression of the RD29A-LUC transgene. DTF1/SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1) was also identified by Law et al. (15) as a Pol IV-associated protein through immunoaffinity purification of the largest Pol IV subunit NUCLEAR RNA POLYMERASE D 1 (NRPD1) (15). However, these studies did not reveal to what extent DTF1 affects RdDM on a genome scale or how DTF1 functions in RdDM. In this study, we used second-generation sequencing to characterize the DNA methylome and 24-nt siRNAs in the dtf1 mutant. We found that DTF1 is required to promote CHH methylation at the majority of loci whose methylation level is dependent on Pol IV and DTF1 has an even more dramatic effect on Pol IV-dependent siRNAs. These results support that DTF1 is a core component of the RdDM pathway. Interestingly, we found that DTF1 is required for the accumulation of 24-nt siRNAs but not the DNA methylation level at a subset of Pol IV-dependent loci, suggesting that DTF1 plays an important role in a negative feedback mechanism regulating DNA methylation at some RdDM target loci. In addition, we show that DTF1 and its paralog DTF2 contain a conserved domain called SAWADEE that could specifically recognize histone H3 lysine 9 methylation. Using immunoaffinity purification, we show that DTF1 specifically associates with Pol IV but not Pol V and DTF1 directly interacts with CLSY1. These data suggest a model that DTF1 assists in recruiting Pol IV to RdDM target loci associated with H3K9 methylation, possibly through interacting with CLSY1.
Results
DTF1 Promotes DNA Methylation at the Majority of RdDM Target Loci.
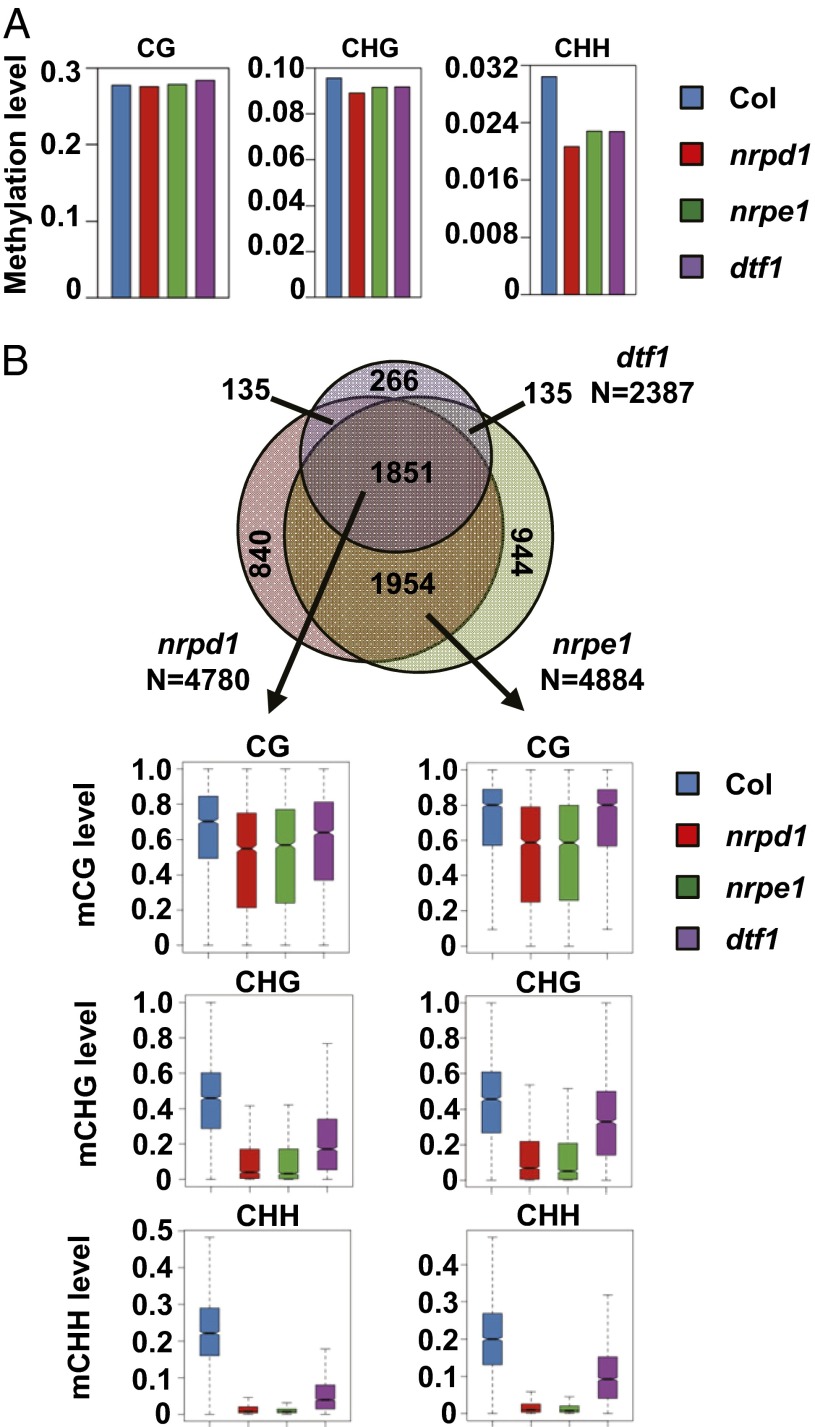
To determine the effect of DTF1 on targets whose methylation levels are dependent on the RdDM pathway, we performed whole genome bisulfite sequencing in 14-d-old wild-type and dtf1-2 seedlings. To identify loci affected by RdDM, we also sequenced the methylomes of nrpd1-3 (NUCLEAR RNA POLYMERASE D 1-3) and npre1-11 (NUCLEAR RNA POLYMERASE E 1-11), T-DNA knockout mutants in the largest subunit of Pol IV and Pol V, respectively. The average depth of sequenced Arabidopsis methylomes is 28–32x with an error rate of 0.3–0.8% (Dataset S1), indicating a high quality of the data. Calculations of the average CG, CHG, and CHH methylation across the whole genome show that the dtf1 mutation causes a decrease of CHH methylation by 25%, whereas it has no significant effect on the overall CG methylation level, consistent with it being a component of the RdDM pathway (Fig. 1A). As previously reported, knockout of NRPD1 or NRPE1 does not completely eliminate CHH methylation (16, 17). Instead CHH methylation in nrpd1 and nrpe1 decreases to similar levels as in dtf1.
Fig. 1.
The effect of dtf1 on genome cytosine methylation. (A) Average CG, CHG, and CHH methylation in Col, nrpd1, nrpe1, and dtf1 plants. Average methylation was calculated for all of the cytosines in the genome with four or more times coverage. (B) Overlap between NRPD1-, NRPE1-, and DTF1-dependent hypo DMRs and their DNA methylation levels. Venn diagram shows the number of hypo DMRs that overlap between nrpd1, nrpe1, and dtf1. Box plots show the distribution of average CG, CHG, or CHH methylation level calculated for hypo DMRs in the indicated subgroup in the Venn diagram.
To understand the effect of dtf1 on the methylation of genes and transposable elements (TEs), average DNA methylation level was calculated for each class (Fig. S1). The dtf1 mutation reduces CHH and CHG methylation levels in TE bodies as well as regions around them, but to a lesser extent compared with nrpd1 and nrpe1. CG methylation in TEs is also slightly reduced in nrpd1 and nrpe1 mutants but not in dtf1, suggesting that the RdDM pathway function is partially retained in the dtf1 mutant or DTF1 is involved in a negative feedback regulation of CG methylation. In contrast to TEs, genes are transcribed by Pol II and on average exhibit lower levels of DNA methylation compared with surrounding regions. Gene body CHH and CHG methylation decreases in dtf1, nrpd1, and nrpe1 mutants to roughly the same level. CG methylation adopts a bell-shaped distribution pattern with a peak toward the 3′ end of the gene. CG methylation is not affected by any of the three mutants examined, indicating that gene body CG methylation is regulated by other mechanisms.
We next identified methylated regions affected by DTF1. A method based on Fisher’s exact test was used to identify differentially methylated regions (DMRs) (18), which are defined as regions that show significant differences in the statistical test (P < 0.05) and more than 1.5-fold changes in DNA methylation levels compared with wild-type plants. We identified 2,387 hypomethylated regions in the dtf1 mutant, compared with 4,780 and 4,884 in nrpd1 and nrpe1, respectively (Dataset S2). Analyses indicated that 56% of the hypo DMRs in dtf1 overlaps with TEs (Fig. S1B). Hypo DMRs in nrpd1 and nrpe1 show rather similar distributions among TEs, TEs overlapping with genes, genes, and intergenic regions, with more than 65% of the hypo DMRs overlap with TEs. These results indicate that similar to other RdDM components, DTF1 has a strong tendency toward affecting TEs.
In contrast, the numbers of hyper DMRs identified are rather similar: 2,043 in dtf1, 1,972 in nrpd1, and 2,412 in nrpe1 (Fig. S2). It is well known that RdDM mutants exhibit a reduced expression of ROS1, a mC-specific DNA glycosylase that initiates active DNA demethylation (19). We found that the ROS1 expression level decreases to ∼6% of the wild-type level in the dtf1 mutant (Fig. S3). The identification of similar numbers of hyper DMRs in the three mutants may be attributed to their decreased ROS1 expression.
The hypo DMRs identified in dtf1 show a large overlap with those in nrpd1 and nrpe1 (Fig. 1B). Over 77% (1,851/2,387) of the hypo DMRs in dtf1 are also identified in nrpd1 and nrpe1. That ratio is similar to that observed between nrpd1 and nrpe1, 80%. Interestingly, a careful examination of the hypo DMRs that are considered to be specifically affected by NRPD1 and NRPE1 indicate that CHH methylation in these regions also decreases significantly in dtf1, although CG methylation remains unaffected (Fig. 1B). These results strongly suggest that DTF1 is a core component of the RdDM pathway and affects the DNA methylation of the majority of RdDM target loci.
The effects of the RdDM pathway on TEs of different sizes were investigated (Fig. S4). As TE size increases, the DNA methylation level increases, indicating that higher DNA methylation levels are required to silence longer transposons. Similar to nrpd1 and nrpe1, the dtf1 mutant shows the most dramatic defect in CHH methylation in small transposons and only causes a slight decrease of CHH methylation in TEs >4 kb, indicating that additional mechanisms may be involved in promoting CHH methylation in long transposons. Interestingly, at the two ends of longer transposons we observed a small peak of CHH methylation that clearly depends on the RdDM pathway function because it is absent in dtf1, nrpd1, and nrpe1 mutants. This pattern of DNA methylation resembles the Pol V targeting pattern in a recently reported Pol V ChIP-seq study (17). Further analyses of the DNA methylation level of different transposon families indicate that this pattern of CHH methylation only applies to the longer ones of the DNA transposon MuDR (>500 bp) family and the LTR transposon Copia (>4000 bp) family. It is possible that the RdDM machinery is targeted to the repeats at the ends of these two specific types of transposons and RdDM-dependent methylation at these regions may have important roles in repressing transposition of the TEs. Despite the strong effects of RdDM on smaller TEs, an enrichment of small TEs was not observed in the hypo DMRs in any of the mutant (Fig. S2C). The size distribution of TEs affected in dtf1, nrpd1, and nrpe1 is similar to that of total TEs in the genome. Thus, the RdDM pathway equally targets TEs of different sizes although it has stronger effects on the DNA methylation levels of small TEs.
DTF1 Is Required for Accumulation of Pol IV-Dependent 24-nt siRNAs.
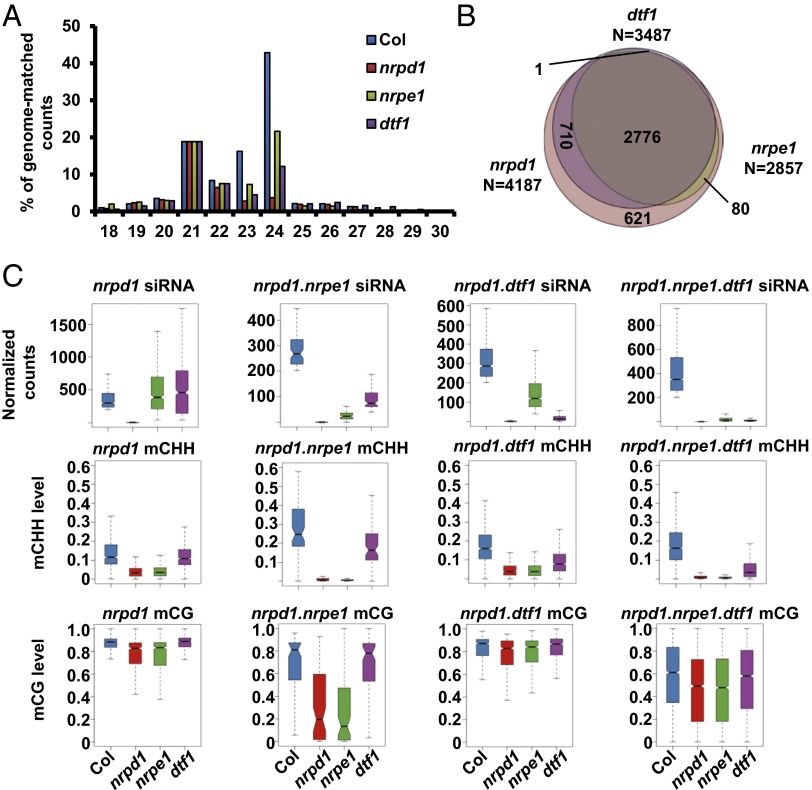
In parallel with whole genome bisulfite sequencing, we also characterized the 24-nt siRNAs in dtf1, nrpd1, and nrpe1 mutants. The total amount of 24-nt siRNA decreases by 91% and 50% in the nrpd1 and nrpe1 mutants, respectively (Fig. 2A and Dataset S3). The dtf1 mutant shows a 72% decrease of 24-nt siRNAs compared with the wild type. We identified 4,187 siRNA regions where 24-nt siRNAs are strongly decreased (more than fivefold decrease) in nrpd1 (Fig. 2B and Dataset S4). All of the siRNA clusters with reduced expression identified in dtf1 and nrpe1 are subsets of those identified in nrpd1 except one region, which shows only a fourfold decrease of 24-nt siRNA in nrpd1 and thus was missed by our method (Fig. 2B). DTF1- and NRPE1-dependent 24-nt siRNA clusters each correspond to 83% and 68% of NRPD1-dependent clusters and the clusters that are dependent on all three correspond to 66% of the NRPD1-dependent ones. We divided Pol IV-dependent siRNAs into four subgroups and investigated the siRNA and DNA methylation levels in each subgroup. The subgroup that only depends on NRPD1 for siRNA accumulation requires both NRPD1 and NRPE1 to maintain normal levels of DNA methylation (Fig. 2C), presumably because Pol V-dependent scaffold RNAs are required for RdDM. Thus, this subgroup fits well into the classic RdDM model in which Pol V functions downstream of siRNA biogenesis. The subgroup that depends on all three components for siRNA accumulation constitutes more than half of Pol IV-dependent siRNA clusters (Fig. 2C), supporting a rather big role of Pol V in siRNA accumulation. Interestingly, although dtf1 has a more dramatic effect on siRNA decrease than nrpe1 does, its effect on DNA methylation, especially CG methylation, is smaller.
Fig. 2.
The effect of dtf1 on 24-nt siRNAs. (A) Length distribution of the small RNAs. The level of small RNAs of each length is shown as the percentage of total small RNA counts and then normalized to 21-nt small RNAs in each library. (B) Venn diagram showing the number of differentially expressed 24-nt siRNA clusters that overlap between dtf1, nrpd1, and nrpe1. (C) Box plots showing comparisons of siRNA levels and average CHH and CG methylation levels of siRNA clusters in each subgroup shown in B.
For the subgroup that was categorized as dependent on Pol IV and Pol V (nrpd1.nrpe1) but not DTF1, we also observed a moderate decrease in 24-nt siRNAs in dtf1 (Fig. 2C). This is likely due to the high threshold (fivefold) used to identify differentially expressed siRNA clusters. A subset of clusters in this subgroup shows less than fivefold decrease in dtf1 and was thus not classified as DTF1 dependent. A similar situation is also observed for nrpe1 in the nrpd1.dtf1 subgroup (Fig. 2C). The patterns of DNA methylation in these two subgroups are similar, but can be distinguished by the levels of CHH methylation in nrpe1. The subgroups that depend on Pol V for siRNA accumulation tend to lose most of their CHH methylation in nrpe1, suggesting that de novo DNA methylation at these loci is mediated mostly by RdDM.
Loci Where siRNAs but Not DNA Methylation Is Reduced in the dtf1 Mutant.
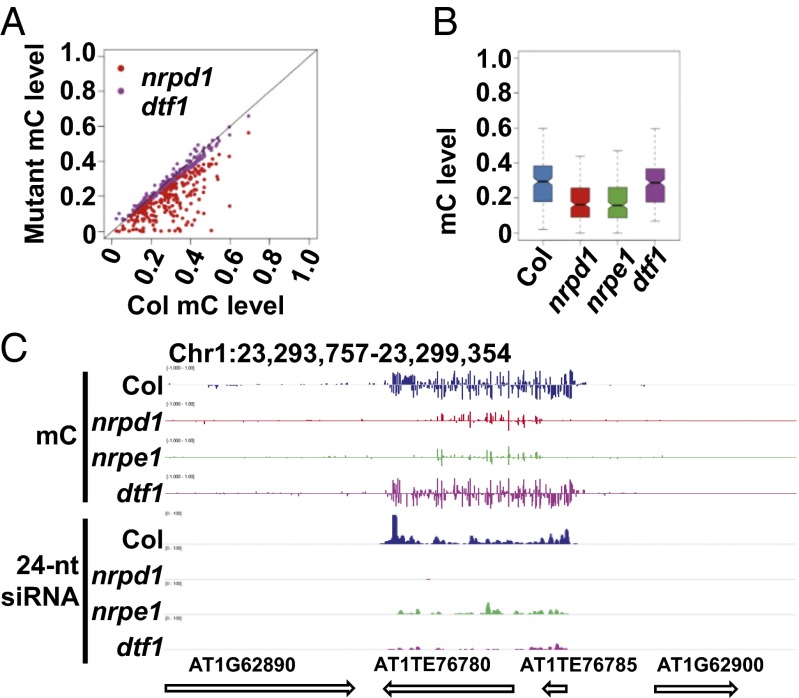
CHH methylation level at DTF1-dependent siRNA clusters is significantly lower than that in the wild type but is consistently higher than those in nrpd1 and nrpe1, especially for the siRNA subgroup that depends on NRPD1, NRPE1, and DTF1, where siRNAs decrease to minimum levels in dtf1 (Fig. 2C). We previously reported that the 24-nt siRNA level decreases and silencing is released at the solo-LTR locus but no significant change in DNA methylation was observed in dtf1 (14). We wondered whether the relatively small change in overall CHH methylation at DTF1-dependent siRNA regions is due to the presence of atypical RdDM loci like solo-LTR. Thus, we searched for DTF1-dependent siRNA regions where DNA methylation significantly decreases in nrpd1 but not in dtf1 and identified 211 regions that retain over 90% of wild-type methylation in the dtf1 mutant (Fig. 3A). Whereas the overall DNA methylation level at these regions decreases significantly in both nrpd1 and nrpe1, it remained essentially unchanged in the dtf1 mutant (Fig. 3B). An example of this type of loci is shown in Fig. 3C. This region covers AT1TE76780 and AT1TE76785 and generates 24-nt siRNAs that are dependent on NRPD1, NRPE1, and DTF1. The DNA methylation level of this region is decreased to below 10% in nrpd1 and nrpe1 but is virtually unchanged in dtf1. It is intriguing why siRNA depletion in nrpd1 but not dtf1 is sufficient to cause a decrease in de novo DNA methylation levels at these loci. The observation implicates an important role of DTF1 in a powerful feedback mechanism of DNA methylation regulation at some RdDM target loci.
Fig. 3.
Subset of DTF1-dependent siRNA clusters where DNA methylation decreases in nrpd1 but not dtf1. (A) Point plot showing the DNA methylation level of loci that behave like solo-LTR in wild-type Col vs. nrpd1 or dtf1. (B) Box plot showing the distribution of average DNA methylation levels of DTF1-dependent regions that behave like solo-LTR: DNA methylation level significantly decreases in nrpd1 and nrpe1, but not dtf1. (C) Snapshot in Integrative Genomics Viewer browser showing the methylation and 24-nt siRNA levels in 1 of the 211 regions identified in A and B.
DTF1/SHH1 Specifically Associates with Pol IV.
DTF1/SHH1, as well as RDR2, RDM4/DMS4, and CLSY1, was found to copurify with NRPD1 (15). In the NRPD1 copurified proteins, RDM4/DMS4 has been shown previously to also be a Pol II- and Pol V-interacting protein (20, 21). Thus, RDM4/DMS4 appears to be shared by Pol II, Pol IV, and Pol V. It is unclear whether DTF1, CLSY1, and RDR2 are associated specifically with Pol IV or are shared with the other RNA polymerases. We carried out affinity purification of DTF1 using transgenic dtf1 Arabidopsis plants expressing Flag-tagged DTF1 driven by the native DTF1 promoter. Mass spectrometric analysis identified NRPD1 and several subunits shared by Pol IV and Pol V in the proteins copurified with DTF1, but no NRPE1 or other Pol V-specific subunits (Table 1). Coimmunoprecipitation assays using DTF1-Myc and NRPE1-Flag also failed to detect any association between DTF1 and Pol V (Fig. S5A). These results strongly suggest that DTF1 is specifically associated with Pol IV in planta.
Table 1.
Mass-spectrometric analysis of DTF1 and CLSY1 copurifying proteins
| UniProt accession no. | AGI code | Protein | Mascot score | MW, Da | Matched queries | Matched peptides |
| DTF1 copurified proteins | ||||||
| IPI00520704 | AT1G15215 | DTF1/SHH1 | 4,700 | 29,487 | 247 | 29 |
| IPI00523111 | AT1G63020 | NRPD1 | 392 | 162,418 | 17 | 17 |
| IPI00546801 | AT3G23780 | NRPD2/E2 | 222 | 132,569 | 13 | 12 |
| IPI00544466 | AT2G15430 | NRPB3/D3/E3A | 115 | 35,439 | 6 | 6 |
| IPI00547850 | AT2G15400 | NRPD3B/E3B | 115 | 35,553 | 2 | 2 |
| IPI00535843 | AT4G11130 | RDR2 | 268 | 129,242 | 12 | 9 |
| IPI00544222 | AT3G42670 | CLSY1/CHR38 | 194 | 144,482 | 13 | 12 |
| IPI00518366 | AT5G20420 | CHR42 | 179 | 145,221 | 13 | 11 |
| CLSY1 copurified proteins | ||||||
| IPI00544222 | AT3G42670 | CLSY1 | 369 | 144,482 | 10 | 7 |
| IPI00523111 | AT1G63020 | NRPD1 | 79 | 162,418 | 2 | 2 |
| IPI00544466 | AT2G15430 | NRPB3/D3/E3A | 61 | 35,439 | 1 | 1 |
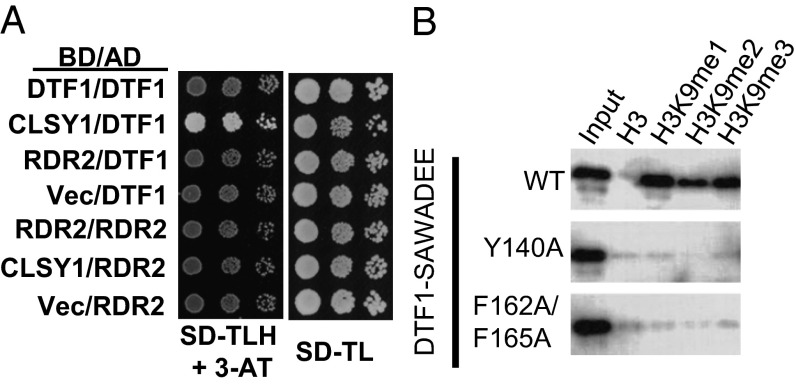
Furthermore, CLSY1 and its homolog CHROMATIN REMODELING 42 (CHR42) were also identified as DTF1-associated proteins, suggesting that DTF1 may cooperate with the chromatin remodeling proteins to facilitate Pol IV transcription on chromatin. We identified CLSY1-associated proteins through affinity purification of CLSY1-Myc and mass spectrometric analysis. The analyses revealed that CLSY1 is also associated specifically with Pol IV (Table 1). These results suggest that both DTF1 and CLSY1 are specifically associated with Pol IV but not Pol V. We then performed yeast two-hybrid assays to determine whether DTF1, CLSY1, and RDR2 may directly interact with each other. The results show that DTF1 directly interacts with CLSY1 but neither protein interacts directly with RDR2 (Fig. 4A). Coimmunoprecipitation results support that DTF1-Flag and CLSY1-Myc interact in vivo (Fig. S5A).
Fig. 4.
Characterization of the DTF1 protein. (A) DTF1 directly interacts with CLSY1. Yeast two-hybrid assay for interactions among DTF1, CLSY1, and RDR2. Positive interactions are indicated by above-background growth of cells on the synthetic dropout medium without His, Leu, Trp, and supplemented with 3-AT. (B) Anti-GST Western blot for the GST–pull-down assay. Interaction between the GST–SAWADEE fusion protein and histone peptides is indicated by a positive signal on the Western blot and “input” was used as a control.
Previous studies showed that the AGO4 protein level was reduced in the nrpd1, rdr2, and dcl3 mutants, which are defective in 24-nt siRNA biogenesis (22, 23). We performed Western blot analysis and found that the AGO4 protein level was also reduced in dtf1 and clsy1 mutant plants compared with that in the wild type or nrpe1 (Fig. S5B). The result is consistent with our model that DTF1 and CLSY1 associate specifically with Pol IV and are required for the accumulation of 24-nt siRNAs, which may be important in stabilizing the AGO4 protein.
SAWADEE Domain Specifically Recognizes H3K9 Methylation.
The Arabidopsis genome encodes a DTF1-like protein, which we named DTF2. Both DTF1 and DTF2 contain a homeodomain in their N terminus and a plant-specific SAWADEE domain in their C terminus (24) (Fig. S6A). The homeodomain is found in many transcription factors and has been shown to interact with specific DNA sequences (25), although we have not been able to identify sequences that may be recognized by the homeodomain of DTF1 or DTF2. SAWADEE is a plant-specific domain that has not been characterized functionally. We used the Phyre2 (protein homology/analogy recognition engine) server to predict the structure of the SAWADEE domain based on sequence homology, secondary structure prediction, and alignment to known protein structures (26). The amino acid sequences of 116 and 138 residues corresponding to the SAWADEE domains of DTF1 and DTF2, respectively, were submitted to the Phyre server in the intensive modeling mode. The results indicated that the SAWADEE domains could be divided into two conserved motifs, each of which may adopt a structure similar to that of the chromo barrel domain (Fig. S6C), a domain family that is similar to the classic chromo domain in sequence but adopts a β-barrel instead of the α + β fold of the chromo domain (27). A subset of the proteins containing chromo barrel or chromo domains has been shown to bind methylated histones and the interaction involves 3–4 conserved aromatic residues that form an “aromatic-cage” to pocket the methylated lysine from histones (28–30). Interestingly, the three aromatic residues are also conserved in the N-terminal half of the SAWADEE domains of DTF1 and DTF2 (Fig. S6B). Considering the role of DTF1 in transposon silencing, we tested whether the SAWADEE domain may interact with repressive histone marks in vitro. The SAWADEE domain of both DTF1 and DTF2 was fused with a GST tag and purified. We used the purified protein to probe a histone peptide array that contains various histone fragments and modified histones. The result revealed that the SAWADEE binds to histone H3 peptide with mono-, di-, or trimethylated lysine 9 (Fig. S6D). Interestingly, histone peptides containing both methylated lysine 4 and 9 failed to interact with the SAWADEE domain, indicating H3K4 methylation blocks the interaction between the SAWADEE and methylated H3K9 (Fig. S6D).
In vitro pull down assays confirmed that the SAWADEE domain from both DTF1 and DTF2 interacts with methylated H3K9, with a slight preference for H3K9me1 (Fig. 4B and Fig. S6E). We also tested whether the conserved aromatic residues that are predicted to directly interact with the methylated lysine are important for the observed interaction. Indeed, mutation of one or two of the three aromatic residues disrupted the interaction between SAWADEE and H3K9 methylation (Fig. 4B and Fig. S6D).
Discussion
In this study we characterized the DTF1 protein and found that it has an extensive and important role in 24-nt siRNA accumulation and DNA methylation at RdDM target loci. DTF1 interacts directly with CLSY1 and both proteins are specifically associated with Pol IV in planta. We found that the SAWADEE domain, which is present in both DTF1 and DTF2, could function as a unique histone-binding module that specifically recognizes H3K9 methylation, which in Arabidopsis mainly exists in the form of H3K9me1/2 and is associated with heterochromatic regions. Although genome-wide analysis of the relationship between H3K9me1/2 and RdDM targets has not been reported, multiple RdDM target loci were shown to associate with H3K9me2 (31, 32). Based on these observations we propose a model that DTF1 assists in recruiting Pol IV to genomic regions associated with H3K9me1/2, possibly through the interaction with CLSY1. In the absence of DTF1, Pol IV may not be efficiently recruited to RdDM loci and thus a large decrease of 24-nt siRNAs is observed. This model is consistent with the fact that 76% of Pol IV-dependent siRNA clusters are also dependent on DTF1. Unlike nrpd1, the dtf1 mutation does not abolish 24-nt siRNA accumulation at the affected loci, suggesting that Pol IV retains partial function in the dtf1 mutant. Probably other factors are also involved in recruiting Pol IV to RdDM target loci, because H3K9me1 and H3K9me2 are unlikely to be the sole determinant for RdDM target specificity. It has been shown that Pol IV is homologous to Pol II and the two share subunits (33). It is possible that like Pol II, Pol IV transcription also requires transcription factors that recognize cis-regulatory elements in the DNA sequence, whereas DTF1 assists in recruiting Pol IV by binding to histone H3 marked with H3K9me1/2 (Fig. S7). It is also possible that DTF2 functions redundantly and/or cooperatively with DTF1. Due to the absence of effective dtf2 mutants, it is presently difficult to test this hypothesis.
There is a subset of RdDM loci that behave like solo-LTR, where the siRNA level decreases dramatically but DNA methylation level remains little changed (Fig. 3) (14). Indeed, 211 DTF1-dependent siRNA clusters retain at least 90% of wild-type methylation in dtf1 while showing significant decreases (P < 0.01) in DNA methylation in nrpd1 (Fig. 3). Several possibilities could account for this observation. It is possible that DNA de novo methyltransferases other than DRM2 are targeted to these regions in the absence of 24-nt siRNAs and DTF1 may negatively regulate these other DNA methyltransferases. A potential role of DTF1 in the negative regulation of some DNA methyltransferases is also suggested by the overall lack of effect on CG methylation at the DTF1-dependent regions in dtf1 (Figs. 1B and 2C). It has been proposed that CMT3 could have some de novo methyltransferase activity in vivo (8). In addition, the drm1 drm2 cmt3 mutant still retains a substantial amount of CHH methylation in pericentromeric regions (34), indicating a yet unidentified DNA methyltransferase for CHH methylation. Alternatively, DTF1 could be involved in a negative feedback loop that counteracts RdDM at these regions. This feedback loop may be in addition to the regulation of ROS1 expression by RdDM components because there is a similar decrease in ROS1 expression in nrpd1 and nrpe1 as is in dtf1 but only dtf1 has unchanged DNA methylation levels. However, it is also possible that RdDM regulation of ROS1 expression alone can account for the negative feedback regulation, and the paradox may be explained because these regions in dtf1 still produce a residual level of siRNAs which may be sufficient to cause wild-type levels of DNA methylation in the absence of ROS1-mediated active DNA demethylation; whereas in nrpd1 there is a complete elimination of siRNAs such that RdDM is completely lacking, and in nrpe1 RdDM is also completely blocked due to a defective effector complex even though there is also a residual level of siRNAs.
Materials and Methods
Plant Materials and Growth Conditions.
All of the plant materials used in this study are in the Columbia-0 (Col) ecotype. And the mutants used include: dtf1-2 (SALK_074540), nrpd1-3 (SALK_128428), and nrpe1-11 (SALK_029919). All of the plants were grown under long day conditions at 22 °C. For 14-d-old seedlings, seeds were stratified for 2–3 d at 4 °C before being sown on 1/2 MS plates containing 2% (wt/vol) sucrose and 0.7% (wt/vol) agar.
Next Generation Sequencing and Data Analyses.
Whole genome bisulfite sequencing and small RNA sequencing were performed at Beijing Genomics Institute (Shenzhen, China). For details of data analyses, please see SI Materials and Methods.
Cloning and Purification of the SAWADEE Domain.
Primers used for cloning in this study are listed in Dataset S5. The C-terminal part of DTF1 and DTF2 were cloned into the pGEX-4T1 vector using the BamHI and NotI sites and overexpressed in BL21 (DE3) pLysS Escherichia coli strain. The cells were disrupted by sonication and GST fusion protein was purified from soluble fractions using glutathione sepharose Fast Flow beads (GE Healthcare Life Sciences). In brief total soluble protein was incubated with glutathione agarose beads at 4 °C for 3 h with rotation, washed with protein binding buffer (50 mM Tris⋅Cl, pH 7.5 at 25 °C, 300 mM NaCl and 0.05% Nonidet P-40) for three times and eluted twice with two bed volumes of elution buffer (50 mM Tris⋅Cl, pH8.0 at 25 °C, 10 mM reduced glutathione, 25% glycerol). The eluates were combined and used directly for downstream experiments.
Histone Peptide Array and in Vitro Pull Down Assay.
Histone peptide array was performed following a published protocol (35). For details please see SI Materials and Methods.
Western Blot Analysis.
Western blot analysis was performed as previously described (36). Briefly proteins were separated on a 12% SDS/PAGE gel with Tris-glycine running buffer, transferred to PVDF membrane and probed with anti-GST (Sigma-Aldrich; G7781) or anti-AGO4 (Agrisera; AS09 617) primary antibodies. Membranes were then washed with TBS-T, incubated with HRP-conjugated secondary antibodies, and exposed to X-ray films after reaction with ECL substrates.
Yeast Two-Hybrid Assay.
Yeast two-hybrid assays were performed using the Matchmaker Gold Yeast Two-Hybrid system from Clontech according to manufacturer’s instructions. In brief the cDNA sequences was cloned into pGADT7-AD or pGBKT7-BD vectors and the pair of genes that are to be tested for interaction were cotransformed into the Y2HGold yeast strain. Cotransformed yeast cells were first grown on synthetic dropout medium without Leu and Trp (DO −Leu/Trp) and then transferred to DO −Leu/Trp/His +3′AT to test for positive interactions.
Affinity Purification and Mass Spectrometry.
Affinity purification and mass spectrometry of purified proteins were performed as described previously (37). For detailed procedures, please see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Zhizhong Gong (China Agricultural University) for sharing biotin-labeled histone peptides and Dr. She Chen and other scientists (proteomics center of National Institute of Biological Sciences) for their technical support in mass spectrometry. This work was supported by the National Basic Research Program of China (973 Program) (2012CB910900) and the 973 Program (2011CB812600) from the Chinese Ministry of Science and Technology (to X.-J.H.), the Chinese Academy of Sciences, and the National Institutes of Health Grants R01GM070795 and R01GM059138 (to J.-K.Z.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE44209).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300585110/-/DCSupplemental.
References
- 1.He X-J, Chen T, Zhu J-K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21(3):442–465. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJM. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21(3):367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Zhu J-K. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14(2):142–147. doi: 10.1016/j.pbi.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12(8):483–492. doi: 10.1038/nrm3152. [DOI] [PubMed] [Google Scholar]
- 6.Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135(4):635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tariq M, Paszkowski J. DNA and histone methylation in plants. Trends Genet. 2004;20(6):244–251. doi: 10.1016/j.tig.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Du J, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151(1):167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson LM, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol. 2007;17(4):379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJM. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21(24):6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Searle IR, Pontes O, Melnyk CW, Smith LM, Baulcombe DC. JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev. 2010;24(10):986–991. doi: 10.1101/gad.579910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deleris A, et al. Involvement of a Jumonji-C domain-containing histone demethylase in DRM2-mediated maintenance of DNA methylation. EMBO Rep. 2010;11(12):950–955. doi: 10.1038/embor.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sridhar VV, et al. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447(7145):735–738. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, et al. An atypical component of RNA-directed DNA methylation machinery has both DNA methylation-dependent and -independent roles in locus-specific transcriptional gene silencing. Cell Res. 2011;21(12):1691–1700. doi: 10.1038/cr.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Law JA, Vashisht AA, Wohlschlegel JA, Jacobsen SE. SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet. 2011;7(7):e1002195. doi: 10.1371/journal.pgen.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wierzbicki AT, et al. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev. 2012;26(16):1825–1836. doi: 10.1101/gad.197772.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong X, et al. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol. 2012;19(9):870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausin I, et al. INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc Nat Acad Sci USA. 2012;109(22):8374–8381. doi: 10.1073/pnas.1206638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu J-K. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He X-J, et al. A conserved transcriptional regulator is required for RNA-directed DNA methylation and plant development. Genes Dev. 2009;23(23):2717–2722. doi: 10.1101/gad.1851809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanno T, et al. RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO Rep. 2010;11(1):65–71. doi: 10.1038/embor.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41(5):630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pontes O, et al. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126(1):79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee K, Brocchieri L, Bürglin TR. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol. 2009;26(12):2775–2794. doi: 10.1093/molbev/msp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolberger C. Homeodomain interactions. Curr Opin Struct Biol. 1996;6(1):62–68. doi: 10.1016/s0959-440x(96)80096-0. [DOI] [PubMed] [Google Scholar]
- 26.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen PR, et al. Structure of the chromo barrel domain from the MOF acetyltransferase. J Biol Chem. 2005;280(37):32326–32331. doi: 10.1074/jbc.M501347200. [DOI] [PubMed] [Google Scholar]
- 28.Schalch T, et al. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 2009;34(1):36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajakumara E, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43(2):275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Numa H, et al. Transduction of RNA-directed DNA methylation signals to repressive histone marks in Arabidopsis thaliana. EMBO J. 2010;29(2):352–362. doi: 10.1038/emboj.2009.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, et al. Antisilencing role of the RNA-directed DNA methylation pathway and a histone acetyltransferase in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(28):11425–11430. doi: 10.1073/pnas.1208557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tucker SL, Reece J, Ream TS, Pikaard CS. Evolutionary history of plant multisubunit RNA polymerases IV and V: Subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harb Symp Quant Biol. 2010;75:285–297. doi: 10.1101/sqb.2010.75.037. [DOI] [PubMed] [Google Scholar]
- 34.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian W, et al. A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science. 2012;336(6087):1445–1448. doi: 10.1126/science.1219416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He X-J, et al. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23(3):318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C-J, et al. IDN2 and its paralogs form a complex required for RNA-directed DNA methylation. PLoS Genet. 2012;8(5):e1002693. doi: 10.1371/journal.pgen.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.