Abstract
Reproductive genes and traits evolve rapidly in many organisms, including mollusks, algae, and primates. Previously we demonstrated that a family of glycine-rich pollen surface proteins (GRPs) from Arabidopsis thaliana and Brassica oleracea had diverged substantially, making identification of homologous genes impossible despite a separation of only 20 million years. Here we address the molecular genetic mechanisms behind these changes, sequencing the eight members of the GRP cluster, along with 11 neighboring genes in four related species, Arabidopsis arenosa, Olimarabidopsis pumila, Capsella rubella, and Sisymbrium irio. We found that GRP genes change more rapidly than their neighbors; they are more repetitive and have undergone substantially more insertion/deletion events while preserving repeat amino acid composition. Genes flanking the GRP cluster had an average Ka/Ks ≈ 0.2, indicating strong purifying selection. This ratio rose to ≈0.5 in the first GRP exon, indicating relaxed selective constraints. The repetitive nature of the second GRP exon makes alignment difficult; even so, Ka/Ks within the Arabidopsis genus demonstrated an increase that correlated with exon length. We conclude that rapid GRP evolution is primarily due to duplication, deletion, and divergence of repetitive sequences. GRPs may mediate pollen recognition and hydration by female cells, and divergence of these genes could correlate with or even promote speciation. We tested cross-species interactions, showing that the ability of A. arenosa stigmas to hydrate pollen correlated with GRP divergence and identifying A. arenosa as a model for future studies of pollen recognition.
Traits mediating reproduction often undergo rapid evolution, effectively restricting successful mating to a subset of available partners. Rapidly changing genes regulate sperm storage, sperm-egg binding, cell fusion, and spermatogenesis (1). In some cases, a selective advantage promotes divergence (positive selection); in other cases, relaxed selective constraints allow rapid change (neutral selection). On occasion, evolutionary changes are so great, or the species studied are sufficiently divergent, that homologs cannot be identified, and the nature of the selective pressures cannot be assessed (2, 3). Both mathematical models and experimental data demonstrate that rapid changes in sexual traits have the capacity to drive speciation and that the coevolution of genes encoding male and female traits can lead to reproductive isolation (4-9).
In some plant families, highly divergent genes limit inbreeding through self-incompatibility; many molecules required for self-pollen recognition have been identified (10, 11). In contrast, few components that allow plants to discriminate interspecific pollen are understood. Identifying molecules that regulate this selective process has agricultural applications; the ability to control gene flow between species would facilitate the creation of new hybrids and the containment of genetically modified varieties. Here, we examined a rapidly evolving gene family required for pollen recognition in the Brassicaceae (12), a family that includes several important oil and vegetable crops.
Reproductive interactions in flowering plants begin when pollen lands on the stigma, a specialized surface of the female reproductive organ. To become metabolically active, desiccated pollen grains must absorb water from the stigma. The Brassicaceae stigma surface is dry, and the extracellular pollen coat interacts with stigma cells to selectively trigger pollen hydration (13, 14). Brassicaceae pollen coats primarily contain proteins and long-chain lipids; in Arabidopsis thaliana the coat includes a glycine-rich protein (GRP) family specified by a tandem array of eight genes, each with two exons (3). The first exon encodes a lipid-binding oleosin domain, and the second exon encodes a glycine-rich repetitive domain (15). Before the coat is deposited on the pollen surface, the oleosin domain is cleaved, leaving the repetitive domain available for interaction with the stigma (16-18). Five of the eight A. thaliana GRPs have been detected in the pollen coat (GRP14 and GRP16-GRP19; ref. 3); two others (GRP20 and GRP22) have messages that are expressed during pollen development (19); an additional putative protein (GRP21) is identified in this study. Mutations that eliminate the most abundant protein, GRP17, result in delayed pollen hydration (12).
Although the major Arabidopsis pollen coat proteins have been identified, their roles in pollen hydration and speciation are not clear. Because they are similar, coexpressed and colocalized, GRPs likely have overlapping functions. Consequently, genetic dissection of their contributions requires simultaneously altering multiple family members, an approach limited by the absence of gene replacement technology in Arabidopsis and by the challenge of recombining tightly linked lesions. As an alternative, Mayfield et al. (3) examined the divergence of the GRP cluster between A. thaliana and Brassica oleracea, species separated by 20 million years (MY). Unfortunately, although the oleosin domains were maintained, the repetitive domains diverged so substantially that homologous relationships were obscured, making it difficult to infer the molecular genetic mechanisms contributing to sequence diversity or the selective pressure driving variation. Moreover, it was not clear whether these rapid changes were a consequence of a particularly dynamic genomic region or instead were specific to GRP genes. We address these questions, assessing changes in the GRPs relative to their genomic neighbors in near (5-8 MY diverged) and more distant relatives (15-20 MY diverged). This work provides insight into the molecular genetic mechanisms that drive the evolution of genes required for compatible mating and shows that rapid evolution of the GRPs is driven by their repetitive nature.
Methods
BAC Identification, Sequencing, and Assembly. BAC library filters (Amplicon Express, Pullman, WA) from Arabidopsis arenosa, Olimarabidopis pumila, Capsella rubella, and Sisymbrium irio (20) were probed with At5g07500 and At5g07580, two Arabidopsis genes flanking the GRP cluster. Of the BACs hybridizing to both probes, A. arenosa clones 1B10 and 6D24 (hereafter Aa1 and Aa2, respectively), O. pumila 37I22, C. rubella 33L6, S. irio 11E11, and B. oleracea 37N21 (TAMU BAC Center, College Station, TX) (3) were sequenced (GenBank accession nos. AY350710-AY350715); the BACs from A. arenosa, a self-incompatible tetraploid, were highly divergent, providing added insight into GRP diversity, either between the parental genomes of the tetraploid or within the population used to construct the library. BAC DNA was sheared and subcloned into pBluescript KS+. Automated DNA sequencing (≥8 times average coverage) was performed at the University of Chicago CRC DNA Sequencing Center or at Integrated Genomics (Chicago, IL). Gaps were filled by sequencing BAC DNA with flanking primers. Trimmed and assembled sequences (seqmanii, DNASTAR, Madison, WI) were manually edited to resolve ambiguities.
Annotation. Each sequence was manually scanned for coding regions homologous to A. thaliana genes between At5g07460 and At5g07630, a region that includes the GRPs and ≈5 kb on either side. Genes were identified by comparison with A. thaliana coding sequences confirmed with blast and tblastx matches to expressed sequences from A. thaliana and other plants (www.ncbi.nlm.nih.gov/blast and http://tigrblast.tigr.org/tgi/). Gene numbering was based on A. thaliana; newly identified genes were assigned intermediate numbers. In some cases, related species had intergenic regions that were larger than their A. thaliana counterparts; genes were identified in these regions by using blast. Intron and exon boundaries were predicted by using splice site rules for A. thaliana (21). B. oleracea GRP exon structure is supported by expressed sequence tag data (3) and was used to predict exon 1 in S. irio GRPs; A. thaliana exons were used for the other species. GRP exon 2 is highly variable, and A. thaliana similarities alone were inadequate for predictions in species other than A. arenosa. Consequently, analysis was supplemented by scanning the regions between oleosin-encoding exons for long ORFs containing (i) repetitive sequences, (ii) an enrichment of glycine, alanine, and/or proline codons, and (iii) a predicted isoelectric point between 9 and 12. GRP19 and GRP20 have second exons that are very short (73-91 and 139-271 bp, respectively) and have few repeats; these exons are more conserved across the species we analyzed, and their structures are supported by cDNA data in A. thaliana (GRP19 and GRP20) and B. oleracea (GRP19).
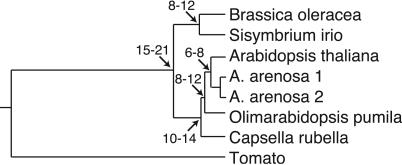
Analysis. DNA and protein alignments were generated with seqmanii and clustalw. Alignment of homologs of At5g07580, a gene flanking the GRPs, was used to generate a tree by using the Fitch-Margoliash least-squares method (Fig. 1). Estimated divergence times are based on synonymous mutation rates and an assumed 10-14 MY divergence between Arabidopsis and C. rubella (22). Phylogenetic relationships between GRPs are based on a Fitch-Margoliash least-squares analysis of a protein distance matrix of aligned GRPs (refs. 23-25; Fig. 6, which is published as supporting information on the PNAS web site). Substitutions/site was estimated by using the Jukes-Cantor method in dambe (26). tandem repeat finder (27) identified nucleotide repeats with a minimum alignment score of 50 and match, mismatch, and insertion/deletion (InDel) parameters of 2, 5, and 5. For overlapping repeats, the repeat with the highest alignment score was reported. Rates of nonsynonymous (Ka) and synonymous (Ks) substitutions were estimated for all genes with the method of Comeron (28) in k-estimator 6.0 (29). For the second GRP exon, confidence intervals for Ka and Ks were numerically derived by Monte Carlo simulations (1,000 replicates) with this software package.
Fig. 1.
Phylogenetic relationship between species. Estimated divergence (in MY) is indicated at nodes. Tomato (Lycopersicon esculentum, accession no. AY192370) was used as an outgroup.
Pollen Hydration Assay. Pollen from dehisced anthers was applied to mature stigmas from either male sterile (ms1) A. thaliana or emasculated A. arenosa flowers. The fraction of hydrated (swollen) pollen grains was counted with a Zeiss Axioskop light microscope 15 min after pollination, sufficient time for near-complete hydration in A. thaliana (12). Seeds were deposited in the Arabidopsis Biological Resource Center (Columbus, OH).
Results
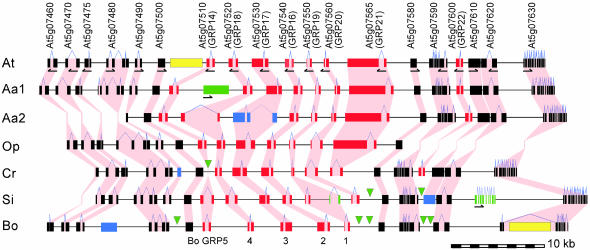
Structure of the GRP Cluster. To assess changes in the GRP pollen coat genes in their genomic context, we sequenced this region from four Brassicaceae species (A. arenosa, O. pumila, C. rubella, and S. irio; GenBank accession nos. AY350711-AY350715) and compared them with A. thaliana (NC003076) and B. oleracea (AY350710) sequences. Comparisons among homologs of At5g07580, which encodes a putative transcription factor (NC003076), verified evolutionary relationships among these species (Fig. 1; refs. 22 and 30). The GRP cluster, defined as the region between At5g07510 and At5g07565, ranged from 13.7 kb in C. rubella to 24.4 kb in A. arenosa (Aa2; Fig. 2). Differences in length were due to changes in intergenic regions and gene content rather than transposon insertions (Fig. 2). Phylogenetic analysis of all GRPs defined homologous relationships and demonstrated that GRP genes are in the same linear order (Figs. 2 and 6). Nevertheless, gene conversion may have contributed to changes in the GRPs, in particular, between 7510 and 7520 (GRP14 and GRP18), which group together in the dendrogram (Fig. 6) and between the oleosin domains, which are similar among all family members.
Fig. 2.
Genomic structure of the GRP region in six Brassicaceae species. Coding regions numbered as in A. thaliana (top) are indicated by filled boxes for each species (rows); At, A. thaliana; Aa1 and Aa2, two different sequences from A. arenosa; Op, O. pumila; Cr, C. rubella; Si, S. irio; Bo, B. oleracea. B. oleracea GRP names are indicated (bottom) (3). Pink shading connects homologous genes. Blue lines indicate introns. Exon shading for A. thaliana: GRPs, red; flanking genes, black; transposons, yellow. Green symbols represent deletions (triangles) or insertions (boxes) as compared with A. thaliana; blue boxes are pseudogenes. The direction of transcription is indicated for A. thaliana genes and for gene insertions in other species (arrows).
We identified GRP21 (gene 7565, GenBank accession no. BK001543), a family member not included in previous annotations. This gene contains an oleosin domain and a repetitive domain rich in proline, serine, and glycine. Thus A. thaliana has eight GRP genes, most of which were found in the other species. Exceptions include the following: (i) deletion of 7510 (GRP14) in C. rubella; (ii) duplication of 7550 (GRP19) to form an additional gene, 7555, in S. irio; (iii) deletion of 7560 (GRP20) in B. oleracea; (iv) absence of 7565 and 7600 (GRP21 and GRP22) in both S. irio and B. oleracea; and (v) degeneration of 7530 (GRP17) in one A. arenosa clone (Aa2) because of a 2-kb unique sequence insertion that introduces stop codons in exon 1 (Fig. 2). GRP17 plays a critical role in A. thaliana pollination (12); thus, A. arenosa, which is a tetraploid, may retain a functional copy at its second GRP locus, potentially represented by BAC clone Aa1. The similarities between GRPs suggest functional redundancy; however, maintenance of five to eight GRP genes across species indicates they have likely evolved unique functions.
Properties of GRP Genes and Predicted Proteins. Five types of conserved elements (I-V) that are candidate binding sites for transcriptional regulators have been identified upstream of A. thaliana and B. oleracea GRPs (3, 15). At least one of these elements was found 5′ of each GRP gene (Table 3, which is published as supporting information on the PNAS web site), consistent with coexpression during pollen development. The occurrence of individual elements varied across species. For example, the 5′ region of GRP14 (7510) contained elements I-V in A. arenosa, but only element IV in O. pumila and element III in S. irio. Similar patterns were observed throughout the cluster, except for GRP20 (7560) and GRP21 (7565), where a single 5′ element was predominant. This diversity across species could alter expression levels and, consequently, the relative abundance of GRPs.
The length of GRP exon 1 was highly conserved across all six species, consistent with prior comparisons of A. thaliana and B. oleracea (3). Exon 2 was more variable, in both length and composition of the predicted proteins (Table 3). These C-terminal domains are enriched in a few amino acids; >60% of this domain can be derived from as few as three amino acids, and glycine levels can approach 40%. These biases, coupled with a paucity of aspartate and glutamate, result in exceptionally basic proteins with predicted isoelectric points between 9 and 12 (Table 3).
The repetitiveness, abundance of InDel events, and the relatively low sequence complexity of the second GRP exon complicate analysis of the evolution of these genes. To better understand GRP composition and evolution, we examined the properties of individual repeat arrays identified with tandem repeat finder (27). The abundance and distribution of GRP repeats and predicted peptides were compared both within and between species [Table 1 (GRP14) and Table 4, which is published as supporting information on the PNAS web site (other GRPs)]. The repetitive content of exon 2 varied over a wide range: GRP14, 14-67%; GRP16, 51-89%; GRP17, 47-85%; GRP18, 20-64%; GRP19, 0%; GRP20, 22-51%; GRP21, 72-91%; and GRP22, 21-29%. The absence of GRP19 repeats is likely due to the small size of the second exon (25-31 aa, Table 3); other GRPs had one to five major repeat classes, and nucleotide repeats were not detected in the flanking genes. GRP repeats were 3-360 nucleotides in length (average = 30) and 2-87 in copy number (average = 8) and shared 60-100% identity within a species (average = 78%). Many repeat arrays overlapped, adding complexity to these patterns (e.g., O. pumila repeat II, GRP14), and many repeat borders were unclear. Although identical peptide sequences were not maintained across all six species, GRP18 and GRP20 had highly conserved motifs. Other proteins contained repeat types that were present in the closest relatives, but absent (repeat I, GRP14) or divergent (repeat III, GRP14) in more distant family members (Table 1). All InDels were multiples of 3 bp, indicating selection to maintain reading frame. These results indicate that the divergence of GRP exon 2 is dominated by changes in length, composition, and copy number of repetitive arrays, with preservation of particular sequences across species in only a few cases.
Table 1. Repeat type and abundance in GRP14.
| Repeat* | Species | Sequence† | Copy no. | % identity | Internal repeats‡ |
|---|---|---|---|---|---|
| I | At | GGACGTAGGAGATTTGGG | 3.2 | 90 | |
| G R R R F G | |||||
| Aa1 | GGAGGTAGGAGATTTGGG | 3.2 | 94 | ||
| G G R R F G | |||||
| Aa2 | GGAGGTAGGAGATTTGGG | 3.6 | 94 | ||
| G G R R F G | |||||
| II | At | GGTGGAGGTTTACCTGGAGGACTTGGAGGATTAGGA | 2.4 | 94 | 15, 5.4, 70 |
| G G G L P G G L G G L G | 9, 9.7, 67 | ||||
| Aa1 | GGGGGAGGTTTACCTGGAGGACTTGGAGGCCTTGGA | 3.2 | 94 | 9, 10.7, 69 | |
| G G G L P G G L G G L G | |||||
| Aa2 | GGGGGAGGTTTACCTGGAGGACTTGGAGGCCTTGGA | 3.1 | 94 | 9, 10.7, 69 | |
| G G G L P G G L G G L G | |||||
| Op | GGAGGTCTACCTGGAGCCGCAGGT | 6.0 | 91 | 12, 5.3, 64§ | |
| G G L P G A A G | |||||
| III | At | GAAACTGCACCTGCCGCT | 2.8 | 87 | |
| E T A P A A | |||||
| Aa1 | GGAGCTGCACCAGCCGCT | 6.2 | 90 | 6, 17.2, 58 | |
| G A A P A A | |||||
| Aa2 | GGAGCTGCACCTGCCGCT | 4.2 | 84 | ||
| G A A P A A | |||||
| Op | CCACCTGCTGCTGGAGCTGCTACACCACCTGCGCCTGGAGCTAGT | 2.9 | 82 | 21, 6.0, 71 | |
| P P A A G A A T P P A P G A S | |||||
| Si | GGAGGAGCTTCACCGGCAGGAGGAGCTGCACCAGCG | 3.2 | 92 | 18, 6.3, 80 | |
| G G A S P A G G A A P A | |||||
| Bo | GCACCTGCACCCGCG | 2.3 | 100 | ||
| A P A P A |
Repeated sequences in GRP14 exon 2, numbered in order of occurrence in the gene; species are abbreviated as in Fig. 2.
Consensus repeat sequences, adjusted to begin at the first position of a codon and to maintain periodicity between species; nucleotide sequence (upper row), protein sequence (lower row), and polymorphic sites (underlined).
For overlapping, internal repeats, sequence length (bp), copy number, and percent identity are shown, respectively.
In one case, the last three copies are interrupted with a 45-bp sequence, generating 2.4 copies of a 69-bp repeat sharing 98% identity.
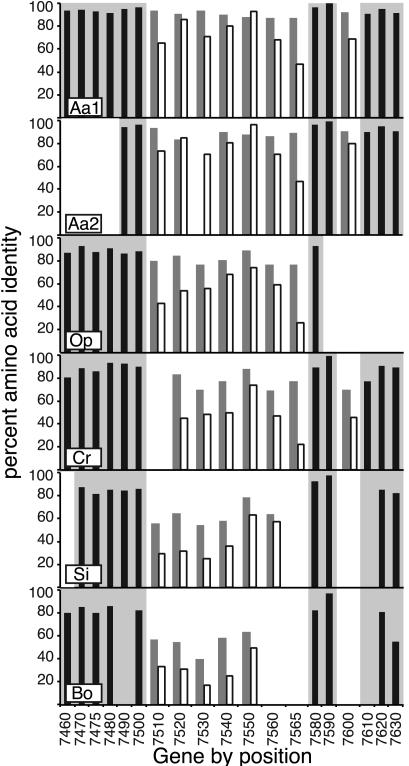
GRP Genes Evolve More Rapidly than Neighboring Loci. To understand the evolution of the genomic region containing the GRPs and to determine divergence levels for typical genes, we examined amino acid conservation across the region. Genes flanking the GRPs are highly conserved (Fig. 3) with amino acid identities to A. thaliana averaging 95 ± 3% (A. arenosa), 89 ± 5% (O. pumila and C. rubella), and 87 ± 6% (S. irio and B. oleracea). These values are consistent with comparisons with a 30-kb C. rubella region (87 ± 11%; ref. 31) or expressed sequence tags from B. oleracea (87 ± 7%; ref. 32). In contrast, the oleosin domains were less conserved, sharing an average of 89 ± 3% (A. arenosa), 78 ± 6% (C. rubella and O. pumila), and 59 ± 9% (S. irio and B. oleracea) identity with A. thaliana, whereas the repetitive domain was only 74 ± 14%, 51 ± 15%, and 36 ± 14% identical, respectively (Fig. 3). This variation differed significantly from that found between flanking genes (oleosin domain, P ≤ 0.05; repetitive domain, P ≤ 0.001, Kruskal-Wallis test).
Fig. 3.
Protein conservation in the GRP region. Percent amino acid identity (bars) for GRP exon 1 (gray), GRP exon 2 (white), and adjacent genes (black) as compared with A. thaliana. Species and gene names are as in Fig. 2; gray shading delineates regions adjacent to the GRP cluster.
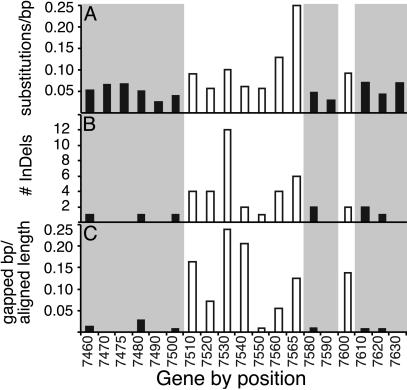
Changes in GRP sequences derive from both enhanced nucleotide divergence and InDels (Fig. 4). GRP nucleotide sequences from A. thaliana and A. arenosa were similar enough to be aligned reliably, revealing more substitutions than in neighboring genes (P = 0.007, Mann-Whitney test; Fig. 4). GRPs also had more InDels; flanking gene exons varied by an average of 5 bp, whereas GRP exons 1 and 2 varied by 17 and 290 bp, respectively. InDels are more frequent (P = 0.002), are larger (P = 0.01), and consume a greater portion of the aligned length (P = 0.0002) of the GRP genes (Fig. 4), distinguishing GRPs from typical genomic sequences.
Fig. 4.
Nucleotide divergence in GRP regions of A. thaliana and A. arenosa. Substitutions per site by using the Jukes-Cantor method (A), InDel number (B), and gaps introduced for alignment (C) were determined for each gene. Filled bars on gray background, flanking genes; open bars, GRPs.
The GRP Region Is Under Variable Selective Pressure. Coding sequences are typically conserved by purifying selection. Rapid changes indicate selective constraints are minimal or nonexistent (neutral evolution) or a selective advantage to divergence (positive selection). Evaluating selective pressures requires comparing the relative rates of nonsynonymous (Ka) and synonymous (Ks) nucleotide changes (33). Ka/Ks = 1 is expected under neutrality, Ka/Ks > 1 indicates positive selection, and Ka/Ks < 1 indicates purifying selection.
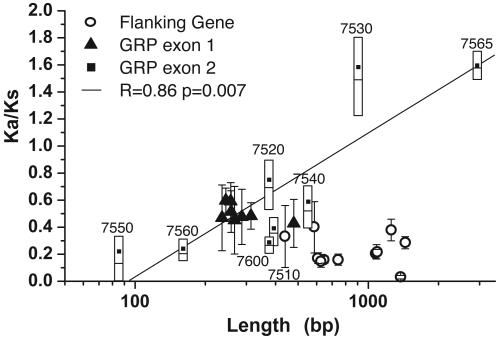
We estimated the pairwise Ka/Ks across all six Brassicaceae species for GRP exon 1 and the flanking genes (Fig. 5 and Table 5, which is published as supporting information on the PNAS web site). The repetitive nature of the second GRP exon limited robust nucleotide alignments; consequently, although protein similarities were assessed across all six species (Fig. 3), Ka/Ks estimations were limited to A. thaliana and A. arenosa (Fig. 5). Flanking genes have a mean Ka/Ks = 0.03-0.39, indicating strong purifying selection similar to averages from mammalian (34) and plant (35) genomes (0.20 and 0.14, respectively). GRP exon 1 is also under purifying selection, although to a lesser degree (mean Ka/Ks = 0.43-0.60; Fig. 5). Estimated Ka/Ks for GRP exon 2 was more variable (0.16-1.58), with the longer exons exhibiting more relaxed selective constraints (Fig. 5), and the longest exons (from GRP17 and GRP21) exhibiting Ka/Ks ratios > 1. For these genes, the 5% and 95% Ka/Ks confidence intervals are 0.97-2.58 and 1.32-1.88, respectively, suggesting neutral to weak-positive selection for GRP17 and weak-positive selection for the putative GRP21. Although our findings could be complicated by the challenge of aligning highly variable sequences containing numerous InDels, they more likely reflect the capacity of multiple repeats within a protein to buffer individual repeat unit divergence.
Fig. 5.
Selective pressures in the GRP region. The ratio of nonsynonymous and synonymous substitution rates (Ka/Ks) plotted versus sequence length. For GRP exon 1 and the flanking genes, all pairwise combinations from six Brassicaceae species were averaged; error bars indicate standard deviation. For GRP exon 2, Ka/Ks is based only on A. thaliana and A. arenosa comparisons, and the data presented represent 1,000 simulations. Black square, average; open boxes, 25th, 50th, and 75th percentiles (bottom, mid-line, and top, respectively). Values are listed in Table 5. Gene numbers for the second exons are noted. The diagonal line is a linear fit of GRP exon 2 data.
A Model for Testing GRP Function in Pollen Recognition. A direct test of GRP roles will require importing the genes into heterologous species and monitoring the species-specific recognition events that lead to pollen hydration. Unfortunately, because A. thaliana hydrates pollen from distantly related Brassicaceae species (36, 37), it cannot be used to test mating specificity within family boundaries. We demonstrated that self-incompatible A. arenosa could serve as an alternative system for GRP exchange, showing that it hydrated A. thaliana pollen efficiently, incompletely hydrated O. pumila pollen, and failed to hydrate pollen from C. rubella or S. irio (Table 2). Although these data do not point to GRPs as the sole cause of hydration differences, they do demonstrate a correlation between pollen coat divergence and the loss of pollen-stigma recognition. Future studies in A. arenosa, including deletion of the endogenous GRP genes, will likely clarify the molecular requirements for species specificity.
Table 2. Pollen hydration efficiency within and between species.
| Pollen
|
|||||
|---|---|---|---|---|---|
| Stigmas | At | Aa | Op | Cr | Si |
| At | +++ (4) | +++ (5) | +++ (5) | +++ (5) | ++ (5) |
| Aa | +++ (4) | +++ (4)* | ++ (7) | - (6) | - (6) |
Symbols indicate the extent of pollen hydration as measured by percent of grains that swell: +++, complete (>90%); ++, intermediate (1-90%); -, incomplete (<1%), averaged over (n) stigmas. Species are as indicated in Fig. 2. *Self-incompatible pollen was not hydrated (five trials).
Discussion
Here we performed a comparative analysis of a genomic region encoding rapidly evolving reproductive genes, examining their divergence among closely related species. We showed that the GRP genes evolve faster than neighboring loci, changing primarily through duplication, deletion, and divergence of repetitive domains. Despite these alterations, gene order was preserved and the GRP cluster retained five to eight genes, each beginning with a hydrophobic domain and terminating with a second domain of low sequence complexity and basic isoelectric point. GRPs contribute most of the pollen surface protein material, which is mobilized to form an interface on contact with the stigma. Their complete elimination in cer6 mutants causes sterility, and removal of the most abundant A. thaliana GRP reduces pollen fitness (3, 12, 13). Thus, the high rates of change we observed likely have an impact on the efficiency of pollen recognition and, consequently, the diversification of Brassicaceae species.
Mechanisms for Evolutionary Change in the GRP Region. The genes flanking the GRPs were highly conserved, contained few InDels, and showed strong, purifying selection (Figs. 3, 4, 5). Thus, the high levels of change observed in the GRPs are unusual and do not reflect a genomic region with an elevated divergence rate. The most variable feature of these genes is exon 2, which contains abundant overlapping repetitive motifs (Tables 1 and 4); the domain encoded by this exon resides in the pollen coat (16-18) and likely contacts the stigma.
Repetitive sequences can undergo unequal crossover and slipped-strand mispairing, altering the number of repeats in an array (38, 39). These mechanisms, combined with gene conversion, can act to homogenize repeats (38-41). Analysis of the GRP repetitive domain provided evidence for these types of changes. In-frame InDels were prevalent (Fig. 4), resulting in a highly variable number of repeats between homologs (Table 4); in the most extreme case (GRP14), repeat numbers ranged from 2.3 (B. oleracea) to 40.5 (A. thaliana). These repeats undergo substantial divergence, with nucleotide substitution rates 2-fold higher than the genes flanking the GRPs. Aligning the repetitive domains of A. thaliana and A. arenosa revealed that Ka/Ks increases with length, suggesting reduced selective pressures as repeat units increase and even positive selection in the longest and most repetitive genes (Fig. 5). The divergence and repetitiveness of the second exon was so substantial that alignments with A. thaliana nucleotides could not be extended to other species, preventing similar Ka/Ks calculations. Despite such high divergence rates, repeat nucleotide sequences within a species averaged 78% identity, suggesting repeat homogenization. Indeed, homogenization is apparent in the second GRP14 repeat where the last lysine codon is identical within both Arabidopsis species, but different between them (Table 1).
Differences in selective pressure throughout a gene are common and were observed in the GRPs. Exon 1 encodes a ≈72-aa oleosin domain that presumably anchors the proteins to lipid droplets in the developing pollen coat. Whereas comparisons across six species revealed that this domain changes faster than flanking genes (Ka/Ks ≈ 0.5 vs. Ka/Ks ≈ 0.2), it has accumulated neither repeats nor InDels, suggesting that maintenance of its unique hydrophobic structure is essential for function. Gene conversion between neighboring oleosin-encoding domains could contribute to the elevated changes observed in this exon.
Positive and Neutral Selection Drive Rapid Genetic Change. Homologous genes that function in reproduction have been sequenced in abalone, sea urchins, fruit flies, and mammals (1). Positive selection has been detected in at least 17 cases (33), including abalone sperm lysin (42) and mammalian egg proteins (43), and rapid evolution has been found in many others (1, 2, 44-48). Numerous genes involved in pathogenesis and pathogen defense are also under positive selection because of the advantages of change for both the host and the pathogen (33).
Repeated motifs are found in many reproductive proteins, potentially providing redundant binding sites that strengthen species-specific contacts. In such cases, selective pressure on an individual repeat could be reduced (49), allowing repeat units to evolve at rates approaching those expected under neutrality; positive selection could result when compensating mutations in the mating partner are favored. Consistent with this model, we found a positive correlation between GRP exon length and, therefore, number of repetitive motifs and Ka/Ks. The repetitive nature of the GRPs may allow critical interactions with female components to be maintained while allowing additional repeat copies to diversify. Mutations in interacting female proteins could afford a selective advantage to the altered repeats, and homogenization of the remaining repeats by concerted evolution could spread advantageous base changes.
Genetic Diversity and Plant Mating Interactions. Many plant reproductive traits undergo rapid change, potentially driving speciation. Pollen grains vary in size and outer cell wall (exine) structure, and stigma cells differ in form, cellular structure, and cuticle pattern. In addition, self-incompatible plants have capitalized on molecular differences to restrict interactions within a population (10, 11). Downstream interactions are also subject to rapid evolution; for example, pollen tubes from different Brassicaceae species display species-specific interactions with A. thaliana ovules (50). With the complete A. thaliana genome sequence and the BAC library resources described here, other rapidly evolving genes involved in plant reproduction may be identified.
Rapid GRP protein evolution is consistent with a role in species-specific signaling (1). These proteins modulate pollen hydration, potentially (i) providing identity tags recognized by the stigma, (ii) mediating pollen coat migration on the stigma, (iii) transporting water from the stigma to the desiccated pollen grain, or (iv) maintaining pollen coat integrity. GRP divergence correlates well with pollen hydration in A. arenosa (Table 2). A direct test of GRP roles in species-specific interactions will require deletion of the endogenous GRP region from a species such as A. arenosa and replacement with GRPs from another species. Such manipulations may ultimately make it possible to engineer transgenic plants that are limited in their breeding range, preventing the spread of transgenes between or within a species.
Supplementary Material
Acknowledgments
We thank G. Kettler for technical assistance; A. Hall for collaborating to make BAC libraries; I. Al-Shehbaz for aid in species identification; the University of Chicago DNA sequencing center; A. Hall, M. Johnson, J. Mayfield, and R. Palanivelu for careful reading of this manuscript; and S. Crosson for countless discussions and critical feedback. This work was supported in part by the National Science Foundation and the Howard Hughes Medical Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GRP, glycine-rich protein; InDel, insertion/deletion; MY, million years.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY350710-AY350715 and BK001543).
References
- 1.Swanson, W. J. & Vacquier, V. D. (2002) Nat. Rev. Genet. 3, 137-144. [DOI] [PubMed] [Google Scholar]
- 2.Ferris, P. J., Pavlovic, C., Fabry, S. & Goodenough, U. W. (1997) Proc. Natl. Acad. Sci. USA 94, 8634-8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayfield, J. A., Fiebig, A., Johnstone, S. E. & Preuss, D. (2001) Science 292, 2482-2485. [DOI] [PubMed] [Google Scholar]
- 4.Arnqvist, G., Edvardsson, M., Friberg, U. & Nilsson, T. (2000) Proc. Natl. Acad. Sci. USA 97, 10460-10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Doorn, G. S., Luttikhuizen, P. C. & Weissing, F. J. (2001) Proc. R. Soc. London Ser. B 268, 2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker, G. A. & Partridge, L. (1998) Phil. Trans. R. Soc. London B 353, 261-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu, C. I. (1985) Evolution (Lawrence, Kans.) 39, 66-82. [Google Scholar]
- 8.Gavrilets, S. (2000) Nature 403, 886-889. [DOI] [PubMed] [Google Scholar]
- 9.Palumbi, S. R. (1999) Proc. Natl. Acad. Sci. USA 96, 12632-12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiscock, S. J. & McInnis, S. M. (2003) Plant Biol. 5, 23-32. [Google Scholar]
- 11.Nasrallah, J. B. (2000) Curr. Opin. Plant Biol. 3, 368-373. [DOI] [PubMed] [Google Scholar]
- 12.Mayfield, J. A. & Preuss, D. (2000) Nat. Cell Biol. 2, 128-130. [DOI] [PubMed] [Google Scholar]
- 13.Preuss, D., Lemieux, B., Yen, G. & Davis, R. W. (1993) Genes Dev. 7, 974-985. [DOI] [PubMed] [Google Scholar]
- 14.Heslop-Harrison, Y. & Shivanna, K. R. (1977) Ann. Bot. 41, 1233-1254. [Google Scholar]
- 15.de Oliveira, D. E., Franco, L. O., Simoens, C., Seurinck, J., Coppieters, J., Botterman, J. & Van Montagu, M. (1993) Plant J. 3, 495-507. [DOI] [PubMed] [Google Scholar]
- 16.Ruiter, R. K., van Eldik, G. J., van Herpen, R. M. A., Schrauwen, J. A. M. & Wullems, G. J. (1997) Plant Cell 9, 1621-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross, J. H. E. & Murphy, D. J. (1996) Plant J. 9, 625-637. [DOI] [PubMed] [Google Scholar]
- 18.Ting, J. T. L., Wu, S. S. H., Ratnayake, C. & Huang, A. H. C. (1998) Plant J. 16, 541-551. [DOI] [PubMed] [Google Scholar]
- 19.Kim, H. U., Hsieh, K., Ratnayake, C. & Huang, A. H. C. (2002) J. Biol. Chem. 277, 22677-22684. [DOI] [PubMed] [Google Scholar]
- 20.Hall, A. E., Fiebig, A. & Preuss, D. (2002) Plant Physiol. 129, 1439-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown, J. W. S., Smith, P. & Simpson, C. G. (1996) Plant Mol. Biol. 32, 531-535. [DOI] [PubMed] [Google Scholar]
- 22.Koch, M., Haubold, B. & Mitchell-Olds, T. (2001) Am. J. Bot. 88, 534-544. [PubMed] [Google Scholar]
- 23.Jones, D. T., Taylor, W. R. & Thornton, J. M. (1992) Comput. Appl. Biosci. 8, 275-282. [DOI] [PubMed] [Google Scholar]
- 24.Fitch, W. M. & Margoliash, E. (1967) Science 155, 279-284. [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein, J. (1988) Annu. Rev. Genet. 22, 521-565. [DOI] [PubMed] [Google Scholar]
- 26.Xia, X. & Xie, Z. (2001) J. Hered. 92, 371-373. [DOI] [PubMed] [Google Scholar]
- 27.Benson, G. (1999) Nucleic Acids Res. 27, 573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Comeron, J. M. (1995) J. Mol. Evol. 41, 1152-1159. [DOI] [PubMed] [Google Scholar]
- 29.Comeron, J. M. (1999) Bioinformatics 15, 763-764. [DOI] [PubMed] [Google Scholar]
- 30.Yang, Y. W., Lai, K. N., Tai, P. Y. & Li, W. H. (1999) J. Mol. Evol. 48, 597-604. [DOI] [PubMed] [Google Scholar]
- 31.Rossberg, M., Theres, K., Acarkan, A., Herrero, R., Schmitt, T., Schumacher, K., Schmitz, G. & Schmidt, R. (2001) Plant Cell 13, 979-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cavell, A. C., Lydiate, D. J., Parkin, I. A. P., Dean, C. & Trick, M. (1998) Genome 41, 62-69. [PubMed] [Google Scholar]
- 33.Ford, M. J. (2002) Mol. Ecol. 11, 1245-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makalowski, W. & Boguski, M. S. (1998) Proc. Natl. Acad. Sci. USA 95, 9407-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tiffin, P. & Hahn, M. W. (2002) J. Mol. Evol. 54, 746-753. [DOI] [PubMed] [Google Scholar]
- 36.Hulskamp, M., Kopczak, S. D., Horejsi, T. F., Kihl, B. K. & Pruitt, R. E. (1995) Plant J. 8, 703-714. [DOI] [PubMed] [Google Scholar]
- 37.Hiscock, S. J. & Dickinson, H. G. (1993) Theor. Appl. Genet. 86, 744-753. [DOI] [PubMed] [Google Scholar]
- 38.Smith, G. P. (1976) Science 191, 528-535. [DOI] [PubMed] [Google Scholar]
- 39.Levinson, G. & Gutman, G. A. (1987) Mol. Biol. Evol. 4, 203-221. [DOI] [PubMed] [Google Scholar]
- 40.Elder, J. F. & Turner, B. J. (1995) Q. Rev. Biol. 70, 297-320. [DOI] [PubMed] [Google Scholar]
- 41.Tautz, D., Trick, M. & Dover, G. A. (1986) Nature 322, 652-656. [DOI] [PubMed] [Google Scholar]
- 42.Lee, Y. H. & Vacquier, V. D. (1992) Biol. Bull. 182, 97-104. [DOI] [PubMed] [Google Scholar]
- 43.Swanson, W. J., Zhang, Z. H., Wolfner, M. F. & Aquadro, C. F. (2001) Proc. Natl. Acad. Sci. USA 98, 2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biermann, C. H. (1998) Mol. Biol. Evol. 15, 1761-1771. [DOI] [PubMed] [Google Scholar]
- 45.Gao, Z. & Garbers, D. L. (1998) J. Biol. Chem. 273, 3415-3421. [DOI] [PubMed] [Google Scholar]
- 46.Hellberg, M. E., Moy, G. W. & Vacquier, V. D. (2000) Mol. Biol. Evol. 17, 458-466. [DOI] [PubMed] [Google Scholar]
- 47.Metz, E. C. & Palumbi, S. R. (1996) Mol. Biol. Evol. 13, 397-406. [DOI] [PubMed] [Google Scholar]
- 48.Yang, Z. H., Swanson, W. J. & Vacquier, V. D. (2000) Mol. Biol. Evol. 17, 1446-1455. [DOI] [PubMed] [Google Scholar]
- 49.Kajava, A. V. (2001) J. Struct. Biol. 134, 132-144. [DOI] [PubMed] [Google Scholar]
- 50.Shimizu, K. K. & Okada, K. (2000) Development (Cambridge, U.K.) 127, 4511-4518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.