Abstract
ZEBReplication Activator (ZEBRA), a viral basic zipper protein that initiates the Epstein–Barr viral lytic cycle, binds to DNA and activates transcription through heptamer ZEBRA response elements (ZREs) related to AP-1 sites. A component of the biologic action of ZEBRA is attributable to binding methylated CpGs in ZREs present in the promoters of viral lytic cycle genes. Residue S186 of ZEBRA, Z(S186), which is absolutely required for disruption of latency, participates in the recognition of methylated DNA. We find that mutant cellular AP-1 proteins, Jun(A266S) and Fos(A151S), with alanine-to-serine substitutions homologous to Z(S186), exhibit altered DNA-binding affinity and preferentially bind methylated ZREs. These mutant AP-1 proteins acquire functions of ZEBRA; they activate expression of many viral early lytic cycle gene transcripts in cells harboring latent EBV but are selectively defective in activating expression of some viral proteins and are unable to promote viral DNA replication. Transcriptional activation by mutant c-Jun and c-Fos that have acquired the capacity to bind methylated CpG challenges the paradigm that DNA methylation represses gene expression.
ZEBReplication Activator (ZEBRA) is related to cellular DNA-binding transcription factors, c-Fos and c-Jun of the AP-1 family, with a basic zipper (bZIP) structural motif (1–7). The DNA-binding specificity of ZEBRA and cellular AP-1 proteins overlap (8, 9). An early hypothesis was that ZEBRA represented a mutated cellular AP-1 protein that was “captured” by a virus, in the manner of the oncogenes of RNA tumor viruses (10). While exploring this hypothesis, we showed that a chimeric protein in which the DNA recognition domain of c-Fos was substituted for that of ZEBRA did not disrupt latency (11).
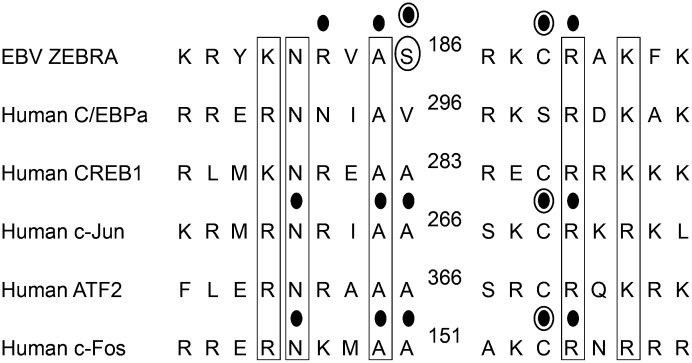
The crystal structure of the bZIP region of c-Fos/c-Jun heterodimer bound to an AP-1 site (12, 13) showed that five amino acids made hydrogen bonds or van der Waals interactions with bases in the AP-1 site in DNA (Fig.1). Four of these five amino acids were positionally conserved in the basic domain of ZEBRA. The exceptions were c-Fos (A151) and the homologous c-Jun (A266); the residue at this position in ZEBRA is S186 (Fig. 1). We studied the phenotype of a missense mutant Z(S186A) in which S186 in ZEBRA was changed to an alanine to resemble c-Fos/c-Jun (14). The Z(S186A) mutant was a potent transcriptional activator of plasmid-based reporters bearing the promoters of EBV lytic cycle genes such as BRLF1 and BMRF1; however, it was unable to drive expression of these genes from the latent viral genome. The primary defect of the Z(S186A) mutant was traced to its inability to activate expression of the R transactivator (Rta) protein from the latent virus. The capacity of Z(S186A) to activate early viral lytic cycle genes, such as BMRF1, a synergistic target, could be rescued by overexpression of Rta (15, 16).
Fig. 1.
Comparison of amino acid sequences in the basic domains of EBV ZEBRA and five cellular bZIP proteins. Boxed amino acids are identical or similar in all of the bZIP proteins. Dots indicate amino acids that contact bases in the crystal structure of ZEBRA (5) and the c-Fos/c-Jun heterodimer (12). Circled dots indicate amino acids that were mutated in the crystal structures. ZEBRA (S186) was mutated to alanine. The cysteines at positions ZEBRA (C189), c-Jun (C269), and c-Fos (C154) were all changed to serine. The unique serine 186 of ZEBRA is circled.
The finding that the Z(S186A) mutant was a potent activator of transcription of plasmid reporters containing promoters of viral lytic cycle genes, but was unable to activate expression of lytic genes from the latent viral genome, pointed to an important function of Z(S186) in targeting a viral genome with epigenetic modifications of DNA or chromatin. Because the latent EBV genome is extensively methylated at CpGs (17, 18), a plausible explanation for the defect in Z(S186A) came with the seminal discovery that ZEBRA binds preferentially to methylated viral DNA (19). ZEBRA binds preferentially to two methylated ZREs in Rp, the promoter of the BRLF1 gene, designated ZRE-2 and ZRE-3; binding of Z(S186A) to methylated ZRE-2 and methylated ZRE-3 was reduced or abolished (20).
Important biologic questions about the relationship between ZEBRA and cellular AP-1 proteins, and, in particular, the unique importance of residue S186 of ZEBRA, were left unanswered in a crystal structure of the bZIP portion of ZEBRA bound to DNA (5). The structure was solved with a ZEBRA mutant in which S186 was changed to alanine. The structure shows ZEBRA binding to an AP-1 site, whereas the biologic activity of ZEBRA is dependent on binding to ZREs not to AP-1 sites. Moreover, the DNA sequence in the solved structure was not methylated.
Here, we extend the comparison between ZEBRA and cellular AP-1 proteins: we studied the phenotype of cellular AP-1 proteins containing reciprocal alanine-to-serine substitutions in the basic domain by analyzing the capacity of these mutants to activate the EBV lytic cascade. We investigated whether the mutations were accompanied by changes in DNA-binding affinity and by the acquisition of the capacity to bind methylated DNA. Our experiments demonstrate that single alanine-to-serine changes in the two AP-1 proteins produce a profound gain-of-function associated with the capacity to drive viral lytic gene expression and to bind preferentially to methylated DNA.
Results
Mutants Jun (A266S) and Fos(A151S) Substitute for ZEBRA to Initiate the EBV Lytic Cycle.
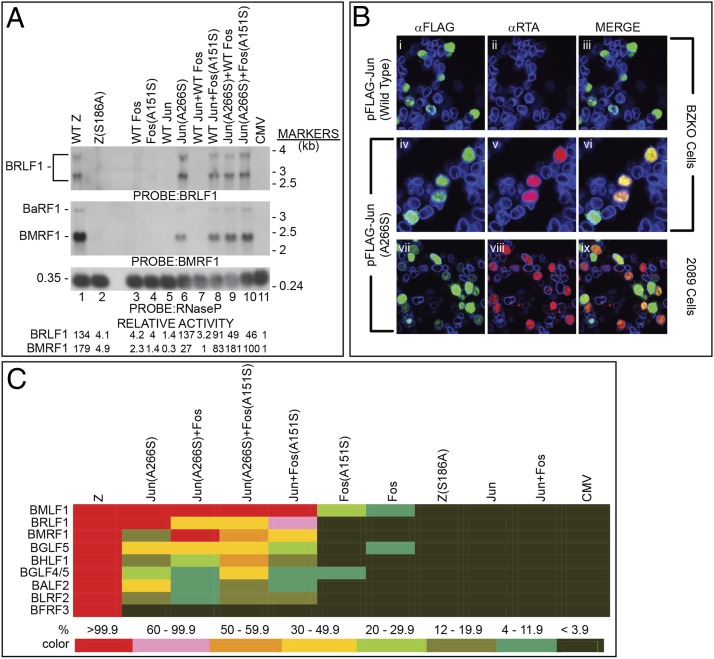
When viewed against the extensive conservation of basic and nonbasic residues in the regions of bZIP proteins that contact DNA, the serine at position 186 of ZEBRA is unusual (Fig. 1). The corresponding position is alanine in four of five cellular bZIP proteins and valine in CCAAT/enhancer-binding protein alpha (C/EBPα). We investigated the capacity of missense point mutants in the basic domain of the AP-1 proteins, in which the amino acids of c-Jun and c-Fos corresponding in position to S186 of ZEBRA were converted from alanine to serine, to promote synthesis of BRLF1 mRNA from a latent EBV genome (Fig. 2A). Plasmids encoding wild-type (wt) ZEBRA, c-Jun, and c-Fos and their corresponding mutants were transfected into 293 cells carrying an EBV bacmid in which the EBV gene BZLF1, encoding ZEBRA, was insertionally inactivated (BZKO cells) (21) (Fig. S1). Providing wt ZEBRA to BZKO cells led to synthesis of BRLF1 mRNA detected on a Northern blot (Fig. 2A, lane 1). BRLF1 mRNA was not detected when wt c-Fos or wt c-Jun were introduced individually or together (Fig. 2A, lanes 3, 5, and 7). However, Jun(A266S) promoted synthesis of BRLF1 mRNA to levels comparable to those induced by wt ZEBRA (Fig. 2A, lane 6). The mutant Fos(A151S) did not by itself promote expression of BRLF1 mRNA but was able to do so when cotransfected with wt Jun (Fig. 2A, lane 8). It is known that maximal DNA-binding activity of AP-1 requires heterodimerization of c-Fos with c-Jun (22).
Fig. 2.
Point mutants in the basic domain of c-Jun and c-Fos substitute for ZEBRA in initiation of the EBV lytic cascade. (A) Plasmids encoding the indicated proteins were transfected in BZKO cells. The transfected cells were assessed for expression of EBV BRLF1, BMRF1, and BaRF1 mRNAs by Northern blotting. Relative activity was determined by densitometry of autoradiographs. (B) Jun(A266S) but not wt c-Jun activates expression of EBV Rta in individual cells. FLAG-tagged wt c-Jun (i–iii) or FLAG-tagged Jun(A266S) (iv–ix) were transfected into BZKO or 2089 cells that were fixed 43 h after transfection and incubated with primary antibodies to FLAG, Rta, and lamin B and appropriate secondary antibodies conjugated with FITC, DyLight 549, or DyLight 645. Images were obtained by confocal microscopy. (C) Heat map illustrating the relative capacity of wt and mutant forms of ZEBRA and cellular AP-1 proteins to activate expression of eight representative EBV lytic genes The primary data were generated from Northern blots presented in A and Fig. S2; the kinetic class and functions of these genes are described in Table S1.
Jun(A266S) alone, or in combination with Fos(A151S), yielded two- to fourfold higher levels of Rta protein than that obtained with ZEBRA (Fig. S2A). To determine whether Jun(A266S) promoted a high level of Rta expression in a few cells or expression of Rta in many cells, the Jun(A266S) mutant was introduced into 293 cells containing EBV bacmids; the cells were examined simultaneously for expression of the Jun(A266S) and Rta proteins by immunofluorescence (Fig. 2B). Approximately 40% (88/219) of BZKO cells that expressed Jun(A266S) also expressed Rta. Rta was not seen in cells that expressed wt c-Jun. In a representative profile of 293 cells containing an intact EBV bacmid (2089 cells), more than 70% (88/120) of cells that expressed Jun(A266S) protein also expressed Rta protein.
The EBV gene BMRF1, encoding DNA polymerase-processivity factor, is activated in synergy by ZEBRA and Rta (23). Because Jun(A266S), and Fos(A151S) in combination with wt Jun, could substitute for ZEBRA to stimulate synthesis of Rta protein, we asked whether the AP-1 mutants could act synergistically with Rta synthesized from the EBV genome in producing expression of BMRF1 mRNA. Jun(A266S) by itself, Fos(A151S) together with wt c-Jun, and the combination of both AP-1 protein mutants all activated expression of BMRF1 mRNA (Fig. 2A, lanes 6, 8, 10). The combination of the two AP-1 mutants was 56% as active as wt ZEBRA.
Jun(A266S) by itself, the Fos(A151S) plus wt c-Jun mixture, and the combination of the two AP-1 mutants activated expression of early antigen-diffuse (EA-D) protein, the product of the BMRF1 gene, considerably less efficiently than did ZEBRA. The level of EA-D protein produced by Jun(A266S) by itself was only 1% the level stimulated by ZEBRA (Fig. 3, lane 7); the combination of Jun(A266S) and Fos(A151S) activated expression of the EA-D protein to 9% the level produced by ZEBRA (Fig. S2B). Thus, the AP-1 mutants activated expression of EBV lytic cycle proteins in a gene-specific manner.
Fig. 3.
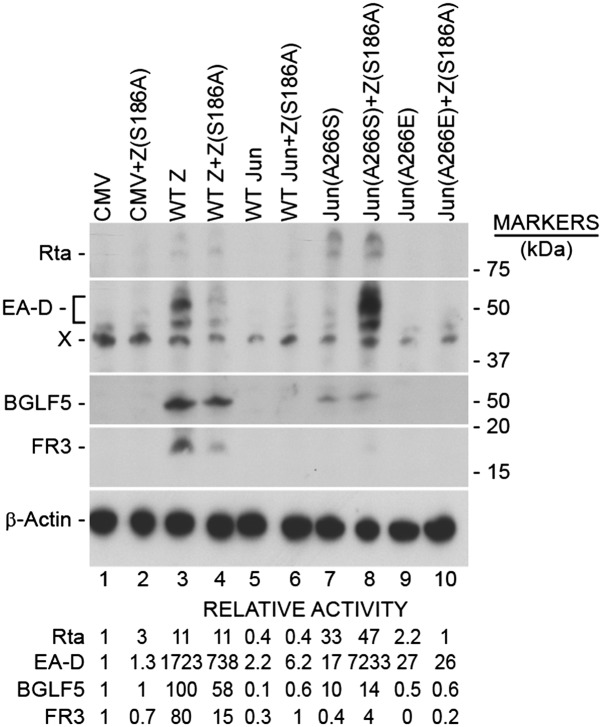
Defect in the capacity of c-Jun(A266S) to activate expression of EBV early protein EA-D can be complemented by ZEBRA mutant Z(S186A). BZKO cells were transfected with plasmids encoding wt or mutant c-Jun(A266S) in the presence or absence of Z(S186A), a ZEBRA mutant that by itself is unable to activate the lytic cycle. The transfected cells were examined by Western blot for Rta, EA-D, and BGLF5 early proteins and BFRF3 late protein.
Jun and Fos Mutants Activate Early but Not Late EBV Gene Expression in the Absence of ZEBRA.
The experiments illustrated in Fig. 2A were expanded to examine the capacity of Jun(A266S) and Fos(A151S) to activate selected early and late EBV genes in BZKO cells. (These data are represented as a heat map in Fig. 2C; the Northern blots from which the heat map was derived are presented in Fig. 2A and Fig. S3; the functions of the genes analyzed are summarized in Table S1.) The AP-1 mutants substituted for ZEBRA in activating expression of mRNAs of at least six early viral lytic-cycle genes encoding proteins. These included the BMLF1 gene, the product of which plays a role in the processing and transport of viral mRNAs, BRLF1, the Rta gene, BGLF5, encoding a protein with alkaline exonuclease and host shutoff activity (24), BMRF1, BGLF4 encoding a kinase (25), and BALF2, an early gene encoding the single-stranded DNA-binding protein. The AP-1 mutants also promoted expression of BHLF1, a noncoding RNA that is important in lytic viral replication (26). BMRF1, BALF2, BGLF5, and BHLF1 are synergistic targets of ZEBRA and Rta. The AP-1 mutants, however, were significantly impaired in activation of late gene expression. The Jun mutant was only 14% as active as ZEBRA in its capacity to promote synthesis of the transcript of BLRF2, a gene encoding a viral tegument protein. This gene can be weakly activated by Rta alone during early stages of the viral lytic cycle but is only strongly activated late in the viral life cycle following lytic viral DNA replication (27, 28). The AP-1(A/S) mutants did not activate either the transcript (Fig. S3B) or the protein (Fig. 3) of BFRF3, a true late gene (29). Because BFRF3 was not expressed, a likely hypothesis is that in the absence of ZEBRA protein the Jun and Fos mutants were unable to activate lytic DNA replication that is a requirement for late gene expression. The experiments suggest that mutant AP-1 proteins can assume the functions of ZEBRA as a transcription factor but not its functions in viral DNA replication.
ZEBRA Mutant Z(S186A) Complements Defective Expression of an Early Viral Protein by the Jun(A266S) Mutant.
Although the mixture of Jun and Fos (A-to-S) mutants induced expression of the BMRF1 mRNA at levels averaging 54% of those induced by wt ZEBRA (Fig. 2A, lane 10), they were markedly deficient at promoting expression of EA-D protein, the product of the BMRF1 mRNA (Fig. 3 and Fig. S3B). These results suggested that the AP-1 mutants lacked a function that is possessed by ZEBRA in promoting expression of the early protein encoded by BMRF1. To address this hypothesis, BZKO cells were cotransfected with wt or mutant Jun genes with or without the ZEBRA mutant Z(S186A) (Fig. 3). The Z(S186A) mutant by itself is unable to activate BMRF1 expression but when Rta is provided in trans, Z(S186A) synergizes with Rta to activate BMRF1 mRNA and EA-D protein (15, 16). Because Jun(A266S) is competent to activate expression of endogenous Rta protein, we reasoned that Jun(A266S) might synergize with Z(S186A) to activate EA-D. Coexpression of Z(S186A) with Jun(A266S) did not enhance the amount of BRLF1 mRNA (Fig. S2C, lanes 7 and 8) but caused a 3.5-fold increase in BMRF1 mRNA compared with Jun(A266S) alone (Fig. S2C, lanes 7 and 8). In noted contrast, the stimulatory effect of the combination of ZEBRA mutant Z(S186A) and Jun mutant (A266S) on the EA-D protein was dramatic (>1,000-fold) (Fig. 3, lane 8). When mutant ZEBRA and c-Jun proteins were coexpressed, the amounts of EA-D protein were stimulated to levels greater than seen with wt ZEBRA protein (Fig. 3, compare lanes 3 and 8). In five replicate experiments, the combination of Z(S186A) and Jun(A266S) induced expression of EA-D protein to levels 0.35- to 4.2-fold the level activated by wt ZEBRA. The level of EA-D protein stimulated by Jun(A266S) alone was only 1% the level produced by ZEBRA (Fig. S2B); the level of EA-D after introduction of Z(S186A) was at background levels.
A surprising result was that whereas coexpression of Z(S186A) with the Jun mutant markedly enhanced expression of EA-D protein, this combination did not enhance the level of BGLF5 protein (Fig. 3, lanes 7 and 8). Because the defect of Jun(A266S) in expression of some, but not all, early lytic proteins was enhanced by Z(S186A), ZEBRA selectively exerts a posttranscriptional role in promoting the translation of certain viral mRNAs or stabilizing certain viral proteins.
The amount of viral DNA measured after introduction of Jun(A266S) and Z(S186A) was not above the background (Fig. S2D). The failure of the Z(S186A) protein to complement the defect of the Jun mutant in activating viral DNA amplification (Fig. S2D) or BFRF3 late gene expression may be linked to the observation that addition of Z(S186A) suppressed DNA amplification (Fig. S2D, lanes 3 and 4) and late gene expression (Fig. 3) by wt ZEBRA.
Mutant AP-1 Proteins Activate the EBV Lytic Cycle in Cultured Cells from Burkitt Lymphoma.
The previously described experiments were conducted in the genetically tractable but biologically artificial system of 293 human embryonic kidney cells containing EBV bacmids. We also studied whether the mutants Jun(A266S) and Fos(A151S) could activate the EBV lytic cycle in natural host cells for EBV, a B-cell line from Burkitt lymphoma (BL). In HH514-16 BL cells, ZEBRA activates synthesis of BRLF1 mRNA and Rta activates expression of BZLF1 mRNA. Thus, each of the two viral activators is competent to initiate and to complete the EBV lytic cycle in this cell background (30). If introduction of the AP-1 mutants were to activate either BZLF1 or BRLF1 in HH514-16 cells, there should be evidence of late gene expression.
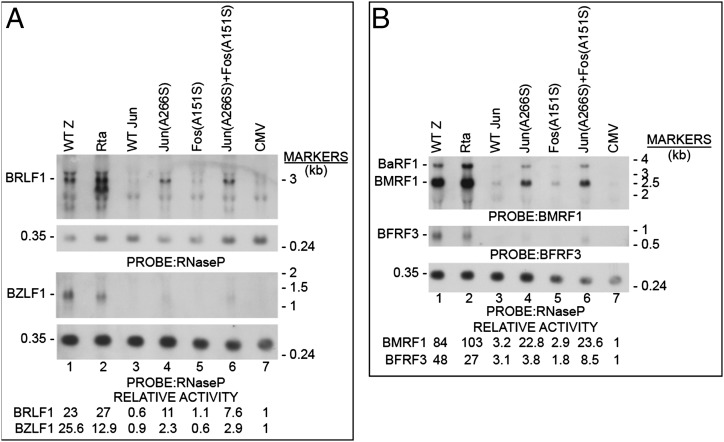
Fig. 4A shows that transfection of HH514-16 cells with Jun(A266S) activated BRLF1 mRNA expression approximately one-third to one-half as efficiently as did transfection of ZEBRA. Although Jun(A266S) by itself did not detectably activate expression of Rta protein, the combination of Jun(A266S) and Fos(A151S) activated expression of Rta protein (Fig. S4A, lane 6). Jun(A266S) and the combination of AP-1(A/S) mutant proteins also activated low levels of BZLF1 mRNA (Fig. 4A, lanes 4 and 6) and ZEBRA protein (Fig. S4A, lane 6). The mutant Jun(A266S) alone or in combination with Fos(A151S) activated expression of the mRNA of the early gene BMRF1 but only weakly activated EA-D protein (Fig. S4A, lane 6), as had been observed in BZKO cells (Fig. 3). Transfection of the combination of Jun(A266S) and Fos(A151S) led to expression of BFRF3 mRNA (Fig. 4B, lane 6) and BFRF3 protein (Fig. S4A, lane 6). A Southern blot showed that Jun(A266S) by itself and in combination with Fos(A151S) caused a low but reproducible increase viral DNA above background levels (Fig. S4B).
Fig. 4.
c-Jun(A266S) activates the EBV lytic cycle in a cell line derived from BL. HH514-16 cells were nucleofected with plasmids encoding wt ZEBRA, Rta, wt Jun, or the basic domain mutants c-Jun(A266S) or c-Fos(A151S). The cells were assessed for expression of BRLF1 and BZLF1 mRNAs (A) and BMRF1, BaRF1, and BFRF3 mRNAs (B).
In a subsequent experiment, cells transfected with Jun(A266S) were cotransfected with mGFP and sorted; the GFP-positive cells expressed BFRF3 mRNA to a 124-fold higher level than GFP-negative cells (Fig. S4C). These experiments allowed two conclusions that were not possible in the 293 cell/EBV bacmid system: first, the AP-1 mutants were active at inducing lytic gene expression from the EBV genome in the setting of a natural host cell; and second, under conditions in which introduction of the AP-1 mutants stimulated expression of both Rta and ZEBRA protein, they could promote low levels of lytic viral DNA replication and late gene expression.
Alanine-to-Serine Mutations in AP-1 Proteins Alter Their DNA-Binding Affinity to ZREs in the Promoter of BRLF1.
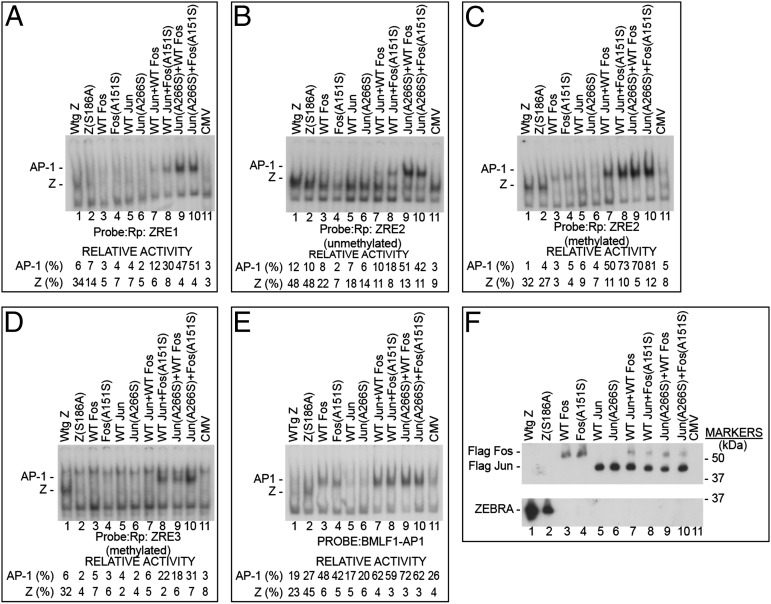
ZEBRA binds to well-defined ZREs, designated ZRE-1, ZRE-2, and ZRE-3, in the promoter of BRLF1 (Rp). ZRE-2 and ZRE-3 both contain CpG motifs that can be methylated. ZEBRA binds unmethylated and methylated ZREs in Rp (19). Binding by ZEBRA, Z(S186A), and wt and mutant AP-1 proteins to the three ZREs in Rp was compared using electrophoretic mobility-shift assays (EMSAs) with duplex oligonucleotides 20 bp in length (Fig. 5 and Table S2). ZRE-2 and ZRE-3 were studied in an unmethylated and methylated state. wt ZEBRA bound more avidly to ZRE-1 (Fig. 5A) and methylated ZRE-3 (Fig. 5D and Fig. S5A) than did the mutant Z(S186A), which is incapable of activating BRLF1 expression. The Jun(A266S) mutant that by itself strongly activates BRLF1 did not detectably bind to any of the ZREs. Therefore, the DNA-binding activity revealed by EMSA does not account for the activating phenotype of the Jun(A266S) mutant. However, the mixture of Jun(A266S) mutant with wt Fos or with mutant Fos(A151A) bound strongly to ZRE-1, unmethylated and methylated ZRE-2, and methylated ZRE-3 (Fig. 5 A–D, lanes 9 and 10, and Fig. S5A). The Fos mutant (A151S) did not bind ZREs by EMSA; however, when accompanied by wt Jun, Fos(A151S) bound to ZRE-1, unmethylated and methylated ZRE-2, and methylated ZRE-3 (Fig. 5 A–D, lane 8). A surprising result was that the mixture of wt Jun and wt Fos, which does not activate BRLF1 expression, bound strongly to methylated ZRE-2 DNA (Fig. 5C, lane 7). Therefore, binding to methylated ZRE-2 does not explain the phenotypic difference between wt and mutant AP-1. Neither ZEBRA nor AP-1 proteins bound to unmethylated ZRE-3 (Fig. S5B).
Fig. 5.
Comparative binding of wt ZEBRA, c-Fos, and c-Jun and basic domain mutants Z(S186A), Fos(A151S), and Jun(A266S) to unmethylated and methylated ZEBRA response elements. Cell extracts of BZKO cells transfected with the indicated expression plasmids were prepared for EMSA. The probes were derived from sequences in the promoter of the BRFL1 gene (Rp) or the promoter of the BMLF1 gene (Table S2). These were Rp ZRE-1 (A), unmethylated Rp ZRE-2 (B), methylated Rp ZRE-2 (C), methylated ZRE-3 (D), and BMLF1p AP-1 (E). Relative activity was determined by densitometry of autoradiographs. (F) Immunoblot with antibody to FLAG in extracts prepared for the EMSA reactions.
The EMSA data demonstrated that DNA-binding affinity was altered as the result of mutations in c-Fos and c-Jun. Because the mixture of Jun(A266S) and Fos(A151S) bound to all of the promoter sequences tested in a methylated state, the results provide evidence that enhanced binding to methylated ZREs can be acquired as the result of specific alanine-to-serine mutations in cellular AP-1 proteins.
Both wt and Mutant AP-1 Proteins Bind with Similar Affinity to a Classical AP-1 Site.
Additional experiments addressed the question of whether the same mutations that affected binding to ZREs in Rp also altered binding to a classical AP-1 site, TGACTCA, such as that found in the promoter of the EBV BMLF1 gene (2). On a classical AP-1 site, the Z(S186A) mutant was enhanced in binding relative to wt ZEBRA (Fig. 5E). The binding of wt and mutant AP-1 proteins was similar on the classical AP-1 site. The results of Fig. 5, therefore, show that the alterations in DNA-binding affinity attributable to alanine-to-serine mutations of the AP-1 proteins are specific to some binding sites and not to others.
Fos/Jun Mutants Bind Preferentially to Methylated DNA.
Because ZRE-1 is not methylated, and neither ZEBRA nor the Fos/Jun mutants bind to unmethylated ZRE-3, binding by ZEBRA and the AP-1 mutants to the same site in an unmethylated or methylated state can be compared only on ZRE-2. We used EMSA with cold competitors to compare binding of ZEBRA and the Fos/Jun mutants to unmethylated and methylated ZRE-2 (Fig. S5 C–E). In these experiments, the probe was unmethylated ZRE-2; the cold competitors were titrated from 5× to 80× molar excess relative to the probe. Competition with 20× methylated ZRE-2 inhibited binding by the AP-1(A/S) mutants by 83%, but competition with 20× unmethylated ZRE-2 did not inhibit binding (Fig. S5C). In a similar titration using cell extracts that expressed ZEBRA (Fig. S5D), 20× cold competitor strongly inhibited binding whether or not the competitor was methylated. These findings suggested that ZEBRA bound equally to both to methylated and unmethylated DNA, whereas the AP-1(A/S) mutants bound preferentially to methylated ZRE-2. By mixing cell extracts expressing either ZEBRA or the Fos/Jun mutants, the effects of cold unmethylated or methylated competitor could be compared on the same gel (Fig. S5E). A 20× excess of unmethylated ZRE-2 and a 10× excess of methylated ZRE-2 inhibited binding by ZEBRA by more than 50%. A 40× excess of unmethylated ZRE-2 and a 20× excess of methylated ZRE-2 inhibited binding by the AP-1 mutants to a comparable extent. From these experiments, we conclude that the AP-1 mutants and ZEBRA each preferentially bind to methylated ZRE-2 but they also bind, with lower avidity, to unmethylated ZRE-2. The binding affinity of ZEBRA appears to be somewhat higher than the AP-1 mutants.
Discussion
Mutants of two cellular AP-1 proteins, c-Jun and c-Fos, containing single alanine-to-serine missense mutations in the DNA-recognition domain assume critical functions of ZEBRA that are required for activation of the EB viral lytic cascade. The AP-1 mutants strongly activate the EBV BRLF1 gene, the product of which, Rta, stimulates transcription of many viral genes and participates in viral DNA replication during the lytic cycle (28, 31). A plausible mechanism for the ability of the AP-1 proteins to activate expression of Rta is the acquisition the capacity to bind methylated ZEBRA response elements embedded in the promoter of the BRLF1 gene encoding Rta. The mutant AP-1 proteins also activate expression of several viral early genes that are involved in lytic-cycle processes such as viral DNA replication (BMRF1, BHLF1), mRNA processing (BMLF1), host cell shutoff (BGLF5), and phosphorylation of viral and cellular proteins (BGLF4) (Table S1). Many of these genes are activated in synergy between the mutant AP-1 proteins and Rta.
The mutant AP-1 proteins, however, do not substitute for all functions of ZEBRA. The AP-1 mutants do not efficiently stimulate lytic viral DNA replication and late gene expression. They are profoundly defective in expression of an early viral lytic protein, EA-D. Because this defect can be complemented by a ZEBRA mutant that by itself does not stimulate EA-D, the experiments with the AP-1 mutants have revealed previously unrecognized roles of ZEBRA in facilitating protein expression from certain viral lytic transcripts.
Mutant AP-1 Proteins Are Defective in Promoting DNA Amplification and Late Gene Expression.
A number of hypotheses might account for the defect of the mutant AP-1 proteins in stimulating lytic DNA replication and late gene expression. The defect of the AP-1 mutants in lytic DNA synthesis may be linked to the defect in promoting expression of early proteins, such as EA-D, which is involved in DNA replication. If this were the case, it should be possible to rescue lytic DNA replication by providing one or more of the group of viral proteins that are essential for lytic replication in trans. A second possibility is that although the AP-1 mutants can substitute for the transcriptional activity of ZEBRA, they may not function as origin binding proteins and promote assembly of the complex of replication proteins on oriLyt (origin of lytic replication). Future experiments must examine whether Jun(A266S) can directly interact with oriLyt and replication proteins. A third possibility is that the AP-1 mutants are unable to stimulate expression of transcripts, such as BHLF1 and BHRF1, that are adjacent to oriLyt and have been found to facilitate the function of oriLyt (26). However, the AP-1 mutants did induce the BHLF1 mRNA (Fig. S3C). A fourth possibility is that the AP-1 mutants, although having gained function as activators of transcription of viral lytic genes, behave as repressors of lytic DNA replication.
Experimental Observations Linking the Capacity of the AP-1 Mutants to Bind to Methylated ZREs with Their Capacity to Initiate the EBV Lytic Cycle.
Alterations in DNA-binding affinity occurred as the result of introduction of alanine-to-serine mutations into c-Fos and c-Jun at positions corresponding to ZEBRA S186. When expressed under conditions that favor the formation of Fos/Jun heterodimers, the AP-1 mutants bound more strongly to unmethylated ZRE-1 and to unmethylated ZRE-2 than did wt AP-1 proteins (Fig. 5 A and B). When the mutants Jun(A266S) and Fos(A151S) were coexpressed, they also bound to oligonucleotides consisting of methylated ZRE-2 and methylated ZRE-3 from Rp, the promoter of the BRLF1 gene, more strongly than wt AP-1 proteins (Fig. 5 C and D and Fig. S5A). Particularly striking were the results showing that the mutant, but not wt AP-1 proteins, bound to methylated ZRE-3 (Fig. S5A). These observations show that a single missense mutation in a cellular protein can promote the capacity to recognize methylated DNA. They clearly demonstrate that the serine in the DNA-binding domain of the mutated bZIP proteins is crucial in recognizing methylated DNA.
In cell lines from BL, the EBV genome is extensively methylated (32). Interaction of both viral and cellular transcription factors with methylated ZRE-3 in Rp may be one of the crucial events for BRLF1 activation. Notably, the Jun/Fos mutants are not different from wt AP-1 proteins in their capacity to bind a classical AP-1 site (Fig. 5E). Earlier experiments with ZEBRA/Fos chimeras demonstrated that binding to and transcriptional activation through an AP-1 site were not linked to disruption of EBV latency (11). Therefore, there is a strong correlation between binding to methylated ZREs, but not to AP-1 sites, and capacity to disrupt latency.
Although the mutant Jun/Fos proteins bind five- to sevenfold more strongly to methylated ZRE-3 than do wt AP-1 proteins (Fig. 5D and Fig. S5A), the AP-1 mutants also bind more strongly to the unmethylated ZRE-1 probe than do wt Jun/Fos (Fig. 5A). Therefore, a significant change in DNA-binding affinity of the AP-1 mutants is observed on both unmethylated and methylated ZREs. The activity required to disrupt EBV latency is likely to require interaction with both methylated and unmethylated DNA. For example, the promoters of the BHLF1 and BHRF1 genes that overlap oriLyt and are required for replication (26) contain ZREs without CpGs.
Our EMSA experiments did not evaluate interactions with more than one ZRE-binding site on the same probe or DNA containing ZREs and binding sites for other transcription factors. For example, EGR1 binds near ZRE-1, YY1 near ZRE-2, and Sp1/Sp3 near ZRE-3 (33-36). Detailed studies of other “enhancesomes” emphasize the importance of many transcription factors bound together on adjacent regions of DNA (37).
Broader Implications and Further Questions.
The experiments presented here suggest the hypothesis that mutations in cellular genes in some individuals may account for high levels of viral replication that are known to precede most cancers associated with EBV. Although EBV infection is nearly universal in the human population, EBV-associated cancers are relatively rare. When followed prospectively, the sera of patients who develop EBV-associated cancers, such as nasopharyngeal cancer, BL, and Hodgkin disease, contain higher antibody titers to lytic EBV gene products than those of control subjects (38–41). These observations from seroepidemiology imply that more active lytic viral replication and a higher viral load precede the development of the cancer in susceptible individuals. The conventional explanation for this observation is that a defect in immunosurveillance or an exposure to an environmental agent promotes lytic viral replication in patients who develop cancer. An alternative hypothesis is that patients who develop virally induced cancer might have activating mutations in genes encoding cellular proteins that promote lytic viral reactivation or inactivating mutations of cellular proteins that repress viral reactivation. Such mutations could also occur in genes that modify the levels of the proteins or their posttranslational modifications.
A second hypothesis of general interest is that because the mutations introduced into the AP-1 proteins changed their capacity to interact with binding sites containing methylated CpGs, some cellular genes might be activated rather than repressed by promoter DNA methylation. The mutant AP-1 proteins might aid in the discovery of a class of cellular genes that could be positively regulated by AP-1 mutants competent to bind methylated DNA.
Materials and Methods
Human embryonic kidney 293 cells containing EBV bacmids and the HH514-16 BL cell line were transfected with plasmids expressing wt or mutant ZEBRA, c-Fos, or c-Jun under control of the CMV immediate-early promoter. Activation of the EBV lytic cycle was assessed by Northern, Western, and Southern blotting and by indirect immunofluorescence. DNA-binding affinity was evaluated by EMSAs with radiolabeled duplex oligonucleotides that were synthesized in an unmethylated or methylated state (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank Sidney Altman, Daniel DiMaio, Ayman El-Guindy, Kelly Gorres, Anthony Koleske, John Kolman, Tom Lawrence, Mark Ptashne, and Joan Steitz for critical readings and helpful discussions and Susan Prisley for preparation of the manuscript. This work was supported by National Institutes of Health (NIH) Grants CA12055 and CA16038. R.W. was supported by NIH Medical Scientist Training Program Grant 2T43GM07205 and the Horton Hallowell Graduate Fellowship (Wellesley College).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301577110/-/DCSupplemental.
References
- 1.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 2.Farrell PJ, Rowe DT, Rooney CM, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8(1):127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flemington E, Speck SH. Evidence for coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc Natl Acad Sci USA. 1990;87(23):9459–9463. doi: 10.1073/pnas.87.23.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouzarides T, Packham G, Cook A, Farrell PJ. The BZLF1 protein of EBV has a coiled coil dimerisation domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene. 1991;6(2):195–204. [PubMed] [Google Scholar]
- 5.Petosa C, et al. Structural basis of lytic cycle activation by the Epstein-Barr virus ZEBRA protein. Mol Cell. 2006;21(4):565–572. doi: 10.1016/j.molcel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Bohmann D, et al. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science. 1987;238(4832):1386–1392. doi: 10.1126/science.2825349. [DOI] [PubMed] [Google Scholar]
- 7.Vogt PK, Bos TJ, Doolittle RF. Homology between the DNA-binding domain of the GCN4 regulatory protein of yeast and the carboxyl-terminal region of a protein coded for by the oncogene jun. Proc Natl Acad Sci USA. 1987;84(10):3316–3319. doi: 10.1073/pnas.84.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieberman PM, Hardwick JM, Sample J, Hayward GS, Hayward SD. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64(3):1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehman AM, Ellwood KB, Middleton BE, Carey M. Compensatory energetic relationships between upstream activators and the RNA polymerase II general transcription machinery. J Biol Chem. 1998;273(2):932–939. doi: 10.1074/jbc.273.2.932. [DOI] [PubMed] [Google Scholar]
- 10.Bishop JM. Enemies within: The genesis of retrovirus oncogenes. Cell. 1981;23(1):5–6. doi: 10.1016/0092-8674(81)90263-4. [DOI] [PubMed] [Google Scholar]
- 11.Kolman JL, Taylor N, Gradoville L, Countryman J, Miller G. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J Virol. 1996;70(3):1493–1504. doi: 10.1128/jvi.70.3.1493-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995;373(6511):257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 13.Fujii Y, Shimizu T, Toda T, Yanagida M, Hakoshima T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat Struct Biol. 2000;7(10):889–893. doi: 10.1038/82822. [DOI] [PubMed] [Google Scholar]
- 14.Francis AL, Gradoville L, Miller G. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J Virol. 1997;71(4):3054–3061. doi: 10.1128/jvi.71.4.3054-3061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adamson AL, Kenney SC. Rescue of the Epstein-Barr virus BZLF1 mutant, Z(S186A), early gene activation defect by the BRLF1 gene product. Virology. 1998;251(1):187–197. doi: 10.1006/viro.1998.9396. [DOI] [PubMed] [Google Scholar]
- 16.Francis A, Ragoczy T, Gradoville L, El-Guindy A, Miller G. Amino Acid substitutions reveal distinct functions of serine 186 of the ZEBRA protein in activation of lytic cycle genes and synergy with the EBV Rta transactivator. J Virol. 1999;73(6):4543–4551. doi: 10.1128/jvi.73.6.4543-4551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ernberg I, et al. The role of methylation in the phenotype-dependent modulation of Epstein-Barr nuclear antigen 2 and latent membrane protein genes in cells latently infected with Epstein-Barr virus. J Gen Virol. 1989;70(Pt 11):2989–3002. doi: 10.1099/0022-1317-70-11-2989. [DOI] [PubMed] [Google Scholar]
- 18.Schaefer BC, Strominger JL, Speck SH. Host-cell-determined methylation of specific Epstein-Barr virus promoters regulates the choice between distinct viral latency programs. Mol Cell Biol. 1997;17(1):364–377. doi: 10.1128/mcb.17.1.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat Genet. 2004;36(10):1099–1104. doi: 10.1038/ng1424. [DOI] [PubMed] [Google Scholar]
- 20.Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. BZLF1 activation of the methylated form of the BRLF1 immediate-early promoter is regulated by BZLF1 residue 186. J Virol. 2005;79(12):7338–7348. doi: 10.1128/JVI.79.12.7338-7348.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feederle R, et al. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 2000;19(12):3080–3089. doi: 10.1093/emboj/19.12.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakabeppu Y, Ryder K, Nathans D. DNA binding activities of three murine Jun proteins: Stimulation by Fos. Cell. 1988;55(5):907–915. doi: 10.1016/0092-8674(88)90146-8. [DOI] [PubMed] [Google Scholar]
- 23.Quinlivan EB, et al. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 1993;21(14):1999–2007. doi: 10.1093/nar/21.8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rowe M, et al. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc Natl Acad Sci USA. 2007;104(9):3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen MR, Chang SJ, Huang H, Chen JY. A protein kinase activity associated with Epstein-Barr virus BGLF4 phosphorylates the viral early antigen EA-D in vitro. J Virol. 2000;74(7):3093–3104. doi: 10.1128/jvi.74.7.3093-3104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rennekamp AJ, Lieberman PM. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. J Virol. 2011;85(6):2837–2850. doi: 10.1128/JVI.02175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Guindy AS, Miller G. Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta. J Virol. 2004;78(14):7634–7644. doi: 10.1128/JVI.78.14.7634-7644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ragoczy T, Miller G. Role of the epstein-barr virus RTA protein in activation of distinct classes of viral lytic cycle genes. J Virol. 1999;73(12):9858–9866. doi: 10.1128/jvi.73.12.9858-9866.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serio TR, Kolman JL, Miller G. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J Virol. 1997;71(11):8726–8734. doi: 10.1128/jvi.71.11.8726-8734.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72(10):7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Guindy A, Ghiassi-Nejad M, Golden S, Delecluse HJ, Miller G. Essential role of Rta in lytic DNA replication of Epstein-Barr virus. J Virol. 2013;87(1):208–223. doi: 10.1128/JVI.01995-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalla M, Göbel C, Hammerschmidt W. The lytic phase of epstein-barr virus requires a viral genome with 5-methylcytosine residues in CpG sites. J Virol. 2012;86(1):447–458. doi: 10.1128/JVI.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragoczy T, Miller G. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J Virol. 2001;75(11):5240–5251. doi: 10.1128/JVI.75.11.5240-5251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zalani S, Coppage A, Holley-Guthrie E, Kenney S. The cellular YY1 transcription factor binds a cis-acting, negatively regulating element in the Epstein-Barr virus BRLF1 promoter. J Virol. 1997;71(4):3268–3274. doi: 10.1128/jvi.71.4.3268-3274.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zalani S, Holley-Guthrie E, Kenney S. The Zif268 cellular transcription factor activates expression of the Epstein-Barr virus immediate-early BRLF1 promoter. J Virol. 1995;69(6):3816–3823. doi: 10.1128/jvi.69.6.3816-3823.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zalani S, Holley-Guthrie EA, Gutsch DE, Kenney SC. The Epstein-Barr virus immediate-early promoter BRLF1 can be activated by the cellular Sp1 transcription factor. J Virol. 1992;66(12):7282–7292. doi: 10.1128/jvi.66.12.7282-7292.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panne D, Maniatis T, Harrison SC. Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer. EMBO J. 2004;23(22):4384–4393. doi: 10.1038/sj.emboj.7600453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller N, et al. 1991. Epstein-Barr virus antibody patterns preceding the diagnosis of non-Hodgkin’s lymphoma. Int J Cancer 49(3):387–393.
- 39.de-Thé G, et al. Epidemiological evidence for causal relationship between Epstein-Barr virus and Burkitt’s lymphoma from Ugandan prospective study. Nature. 1978;274(5673):756–761. doi: 10.1038/274756a0. [DOI] [PubMed] [Google Scholar]
- 40.Zeng Y, et al. 1985. Prospective studies on nasopharyngeal carcinoma in Epstein-Barr virus IgA/VCA antibody-positive persons in Wuzhou City, China. Int J Cancer 36(5):545–547.
- 41.Old LJ, et al. Precipitating antibody in human serum to an antigen present in cultured burkitt’s lymphoma cells. Proc Natl Acad Sci USA. 1966;56(6):1699–1704. doi: 10.1073/pnas.56.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.