Abstract
Viral microRNAs (miRNAs) play an important role during infection by posttranscriptionally regulating both host and viral gene expression. However, the function of many viral miRNAs remains poorly understood. In this study, we investigated the role of the BK polyomavirus (BKPyV) miRNA in regulating virus replication. The function of the polyomavirus miRNA was investigated in archetype BKPyV, which is the transmissible form of the virus and thought to establish a persistent infection in the host urinary tract. In agreement with previous studies, we show that the BKPyV miRNA targets early mRNAs. Importantly, we show that the miRNA plays a significant role in limiting archetype BKPyV replication in a natural host cell model of infection. This regulation is accomplished through the balance of regulatory elements located within the noncoding control region that control early gene expression and miRNA expression before genome replication. We therefore provide evidence for a unique function of the polyomavirus miRNA that may have important implications for the mechanism of viral persistence.
MicroRNAs (miRNAs) are small noncoding RNAs that posttranscriptionally regulate gene expression by repressing translation or directing cleavage of target mRNAs (1). miRNAs regulate diverse cellular processes and are generally considered fine tuners of gene expression (2). In animal cells, miRNAs usually recognize targets with imperfect complementarity, therefore, an individual miRNA may have hundreds of targets. However, in plants and some viruses, miRNAs are able to recognize a target with perfect complementarity and direct cleavage of the target in a manner similar to small interfering RNAs (siRNAs). Virally encoded miRNAs have been described that target host gene expression, viral gene expression, or both. However, viral miRNA functions are generally not well understood. Viruses such as herpesviruses autoregulate their gene expression through the use of miRNAs (3), and autoregulation of viral genes has been implicated in regulating viral latency.
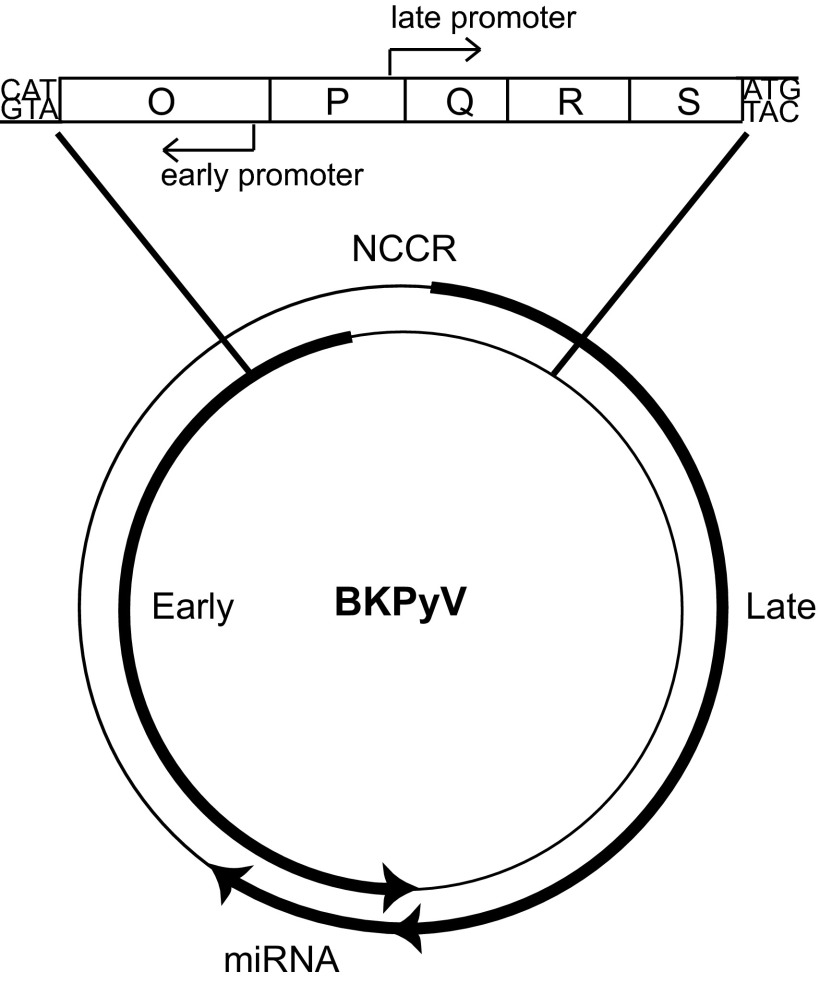
Polyomaviruses are species-specific viruses with small double-stranded DNA genomes. BK polyomavirus (BKPyV) is the causative agent of polyomavirus-associated nephropathy and hemorrhagic cystitis in kidney transplant and bone marrow transplant recipients, respectively (4). There are currently no FDA-approved drugs to treat BKPyV-associated diseases. There are two forms of the BKPyV genome, designated archetype virus and rearranged variants. These forms are distinguished by the DNA sequence of their noncoding control region (NCCR). The archetype virus is thought to be the transmissible form of the virus, because it is found in both healthy people, in which the virus establishes a persistent subclinical infection, and in diseased patients. The NCCR structure of archetype virus is divided into five sequence blocks termed O, P, Q, R, and S (Fig. 1) (5). The O block is named for the origin of replication, whereas the P, Q, R, and S blocks are the enhancer region and contain transcription factor binding sites for the divergent early and late promoters. The early promoter drives the early coding region products large T antigen (TAg), small t antigen, and truncated T antigen, whereas the late promoter drives the late coding region products agnoprotein and structural viral proteins 1 (VP1), VP2, and VP3. Rearranged variants are characterized by duplications and deletions of the blocks of sequence that make up the NCCR and are associated with BKPyV disease (6). Rearranged variants, but not archetype virus, produce progeny in primary renal proximal tubule epithelial (RPTE) cells, a natural cell culture model of lytic replication (7). We have previously shown that archetype virus produces undetectable levels of TAg and very low, if any, viral DNA replication in RPTE cells (8). Therefore, there is currently no natural cell culture model of archetype virus replication.
Fig. 1.
Schematic of the BKPyV genome. The viral genome is a double-stranded circular DNA molecule. The early primary transcript (counter clockwise bold arrow) and the late primary transcript (clockwise bold arrow) are divergently transcribed from the NCCR by the early and late promoters, respectively. The miRNA is complementary to the early coding region and transcribed from the late strand.
Many polyomaviruses including SV40, murine polyomavirus (MPyV), JC polyomavirus (JCPyV), BKPyV, and Merkel cell polyomavirus encode a miRNA that targets early coding region products, such as large TAg mRNA (9–12). One of the major functions of TAg is driving DNA replication of the viral genome at the origin of replication (13). A conserved functional role of the polyomavirus miRNA regulation of TAg remains unclear, however. The SV40 miRNA was the first polyomavirus miRNA described and was shown to down-regulate early viral gene expression (12). Cells infected with an SV40 miRNA mutant are more sensitive to TAg-specific cytotoxic T-cell (CTL) lysis in vitro, suggesting that the miRNA is important in viral evasion of the host immune response. However, the miRNA mutant SV40 produces the same amount of infectious progeny as wild-type (WT) virus. A naturally occurring deletion mutant of the MPyV miRNA has no defects with respect to replication, transformation efficiency, or establishment or clearance of infection in vivo (10). Consequently, the miRNA was deemed to be nonessential for infection in vitro and in vivo. Unlike the SV40 miRNA, the MPyV miRNA does not limit the host CTL response during either the acute or persistent phases of in vivo infection. Therefore, no clearly defined role, conserved among all polyomavirus miRNAs, has yet been described.
JCPyV and BKPyV encode one precursor (pre)-miRNA on the late strand with perfect complementarity to the 3′ coding end of the TAg mRNA (Fig. 1) (9). The miRNA may be expressed from the late promoter, because it is located on the late strand, although it is possible that it has a unique promoter located within or outside the NCCR. In animal cells, normally one arm of the miRNA duplex is preferentially loaded into the RNA-induced silencing complex (RISC), whereas the other arm is degraded (1). However, modified 5′ RACE analysis showed that both arms of the BKPyV and JCPyV miRNA direct degradation of the viral TAg mRNA target, in an siRNA-like fashion (9). No consequence of TAg down-regulation has yet been identified for the JCPyV or BKPyV miRNAs. However, because TAg is responsible for initiating viral DNA replication (13), we reasoned that the miRNA may play a role in limiting viral replication. Whereas the previous study with SV40 used a rearranged variant, we were interested in evaluating the functional role of the miRNA in the archetype form of the virus, which establishes a persistent infection in the urinary tract of the host (14).
This study is unique in examining the role of a polyomavirus miRNA using an archetype virus. Although previous studies have indicated a role for the polyomavirus miRNA in limiting host immune system recognition, we show that a polyomavirus miRNA controls viral replication early in the course of infection. Previously, the miRNA was assumed to regulate early viral gene expression late in the infection because it is expressed from the late strand and is complementary to early mRNAs (9). However, we provide evidence that the miRNA can regulate early mRNA expression before genome replication. This regulation is accomplished through the control of early gene and miRNA expression influenced by the delicate balance of elements within the NCCR. Thus, we define a role for the polyomavirus miRNA in regulating viral replication that is unique to the form of the virus that establishes persistence in the human host. This work implicates the miRNA as an important viral factor in the mechanism of polyomavirus persistence.
Results
BKPyV miRNA Mutant Virus Is Unable to Regulate Its Target Early mRNA.
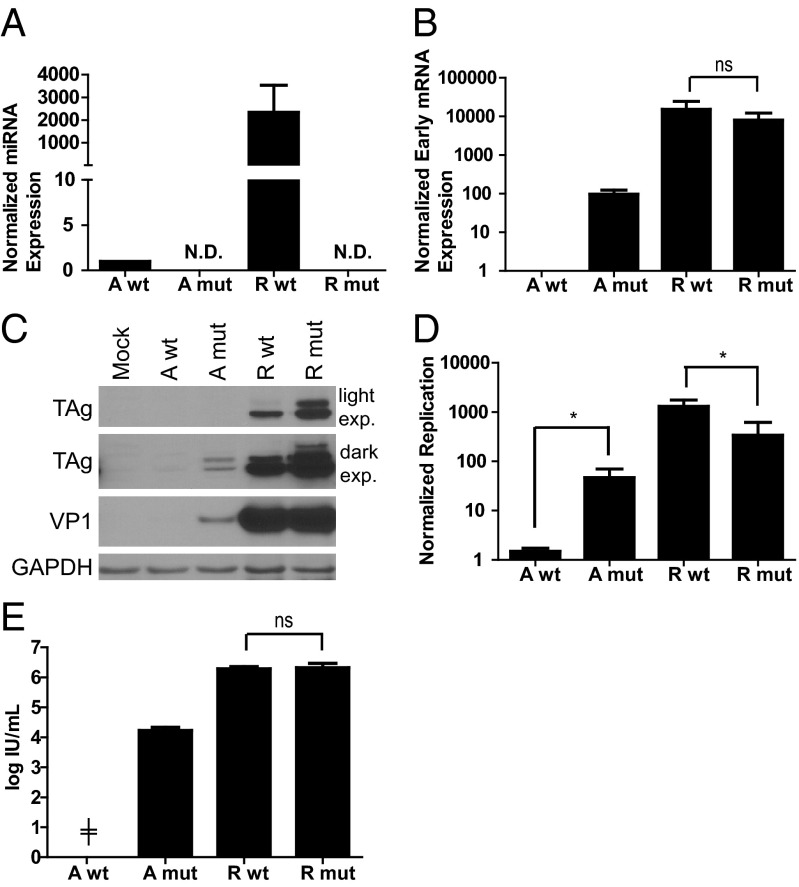
BKPyV expresses a miRNA that targets its early mRNAs in an siRNA-like manner (9). To examine the role of the miRNA in limiting viral replication, we created BKPyV mutant viruses in the archetype and rearranged backgrounds that are unable to express both the mature 5 prime (5P) and 3P miRNAs (Fig. 2B). Because the miRNA is complementary to the 3′ end of the TAg coding sequence, it was necessary to mutate the miRNA sequence at wobble positions in the TAg coding sequence to conserve TAg function. Previously, an SV40 miRNA mutant was created by mutating a majority of the wobble positions in the pre-miRNA sequence, a total of 18 point mutations (12). We chose to create a BKPyV archetype virus miRNA mutant with fewer point mutations to minimize any potential off-target effects due to the mutations. Three positions in the pre-miRNA sequence were mutated, based on the predicted creation of a bubble in the pre-miRNA folded structure that would interfere with miRNA processing (Fig. 2A). The WT and miRNA mutant viruses were purified from 293TT cells, which allow both archetype virus and rearranged variants to replicate (8). RNA was harvested from infected 293TT cells and Northern blotting confirmed that the mutant virus does not produce its mature 5P and 3P miRNAs (Fig. 2B). To ensure that the wild-type probe can recognize the mutant miRNA sequence despite the presence of the mutations, we probed a Southern blot containing plasmid DNAs corresponding to the wild-type and mutant genomes and saw no difference in hybridization (Fig. S1). Additionally the mature 5P miRNA, the more abundant arm (9), was not detected in the mutant virus infection of 293TT cells by stem-loop quantitative (q)PCR of RNA (Fig. 2C), a more sensitive detection method (15). Mature miRNA expression is likely higher in the rearranged variant compared with archetype virus due to increased replication ability of the rearranged variant in 293TT cells.
Fig. 2.
The BKPyV miRNA mutant is unable to target early mRNAs. (A) Predicted fold of the WT and the mutant (mut) pre-miRNAs (29). Point mutations are indicated by circles and bases recognized by the stem-loop RT primer are in bold. (B) The BKPyV miRNA mutant virus does not produce mature miRNA. 293TT cells were infected with purified WT or miRNA mutant virus at an MOI of 0.01. Total cell RNA was harvested 5 dpi. Mature 5P and 3P miRNAs were detected by Northern blotting. The blot was hybridized using the 5P probe (which recognizes the 5P arm), stripped, and then rehybridized using the 3P probe. Ethidium-stained rRNA bands are shown as a loading control. (C) BKPyV 5P miRNA expression was quantified by stem-loop RT qPCR and normalized to the cellular control hsa-let-7a. The A WT sample was arbitrarily set to 1. No miRNA expression was detected in mock, mock infected; A, archetype; R, rearranged; WT, WT miRNA; mut, mutant miRNA; ND, not detected. (D) Relative luciferase levels from 293 cells cotransfected with the luciferase reporter plasmid and a plasmid expressing the WT BKPyV miRNA, mutant BKPyV miRNA, or empty vector (EV). Renilla luciferase (Rluc) values were normalized to firefly luciferase (Ffluc) values as a transfection control. Each bar is the average from three (C) or four (D) independent experiments and the error bars are SD. ND, not detected; **P < 0.01. (The two-tailed, unpaired Student t test was performed for statistical analyses).
To confirm the role of the BKPyV miRNA in targeting early mRNAs, we used a luciferase assay to quantify the miRNA inhibitory activity (9). Plasmids expressing the WT or mutant BKPyV miRNAs were cotransfected into 293 cells with a plasmid encoding the miRNA target sequence cloned into the 3′-untranslated region (UTR) of the Renilla luciferase gene. The cotransfection of the target plasmid and the BKPyV WT miRNA plasmid resulted in a significant decrease in luciferase levels compared with the miRNA mutant (Fig. 2D). These results demonstrate that the BKPyV miRNA mutant is functionally unable to recognize the early mRNA target sequence.
Archetype BKPyV Replication Is Limited by the Viral miRNA.
No previous study of polyomavirus miRNAs showed a role in regulating replication; however, previous studies were performed using rearranged variants as opposed to archetype virus (9, 10, 12). To examine the role of the BKPyV miRNA in archetype virus replication in a natural host cell for BKPyV, RPTE cells were infected with WT or miRNA mutant viruses purified from 293TT cells. We confirmed that the WT viruses recapitulated results seen previously with transfection; i.e., that rearranged variants but not archetype virus are able to replicate in RPTE cells (16). We hypothesized that early mRNA levels would increase in the miRNA mutant archetype virus and this would result in an increase in viral replication. We measured mature 5P miRNA expression by stem-loop qPCR in the mutant virus infection of RPTE cells: miRNA was detected in the WT but not the mutant viruses (Fig. 3A). Mature miRNA expression is higher in the rearranged variant compared with archetype virus because only the rearranged variant can replicate in RPTE cells. We next examined early transcript and TAg protein production in WT vs. miRNA mutant virus infection at 3 d postinfection (dpi) by harvesting total RNA and proteins, respectively. Consistent with the luciferase results, early mRNA and TAg protein expression, measured by quantitative reverse transcriptase PCR (qRT-PCR) and Western blotting, respectively, were increased in the archetype miRNA mutant virus infection compared with WT (Fig. 3 B and C). However, mutating the miRNA in the rearranged variant did not increase early mRNA production but resulted in increased TAg protein expression compared with WT rearranged variant (Fig. 3 B and C), similar to what had been previously reported for mutant SV40 and MPyV. Because early mRNA expression was increased by 100-fold in the archetype virus miRNA mutant virus infection, we also measured viral DNA replication by qPCR. Low molecular weight DNA harvested at 3 dpi was normalized to low molecular weight DNA harvested at 1 dpi (input). Viral DNA replication was ∼50-fold higher in the archetype virus miRNA mutant virus infection compared with WT (Fig. 3D). We also noted increased VP1 protein expression (Fig. 3C) in the miRNA mutant virus. This is likely the result of increased replication capacity because it is well known that polyomavirus genome amplification results in increased late gene expression (7, 17). In contrast, the rearranged variant miRNA mutant did not display increased replication ability and was actually slightly reduced, compared with WT (Fig. 3D). Lastly, progeny production was assessed by a VP1 infectious unit (IU) assay at 3 dpi (8). We detected only 1–2 foci per field of view in the WT archetype virus assay in each experimental repeat, which is below the effective limit of detection of the assay (20 foci or 8.8 × 103 IU/mL). In contrast, the archetype miRNA mutant virus had a measurable titer of 1.7 × 104 IU/mL (Fig. 3E). Notably, the rearranged variant miRNA mutant once again behaved similarly to other rearranged polyomavirus miRNA mutants, producing the same amount of progeny compared with WT rearranged virus. Increased early mRNA and TAg protein expression shows that the archetype virus miRNA negatively regulates early mRNA expression in RPTE cells. Notably, early transcript and replication were ∼100-fold and 50-fold higher, respectively, in the miRNA mutant compared with the WT virus, indicating that the miRNA is largely responsible for limiting archetype virus replication in RPTE cells.
Fig. 3.
The BKPyV miRNA limits viral replication in RPTE cells. RPTE cells were infected with purified WT or miRNA mutant virus at an MOI of 0.01. Total cell RNA was harvested 3 dpi. (A) BKPyV 5P miRNA expression was quantified by stem-loop RT qPCR and normalized to the cellular control hsa-let-7a. (B) Early transcript was measured by qRT-PCR normalized to GAPDH transcript. The A WT sample was arbitrarily set to 1. (C) Total protein was harvested at 3 dpi and analyzed by Western blotting for the expression of TAg, VP1, and GAPDH. Light and dark exposures of TAg are shown. Blots shown are representative of three independent experiments. (D) Low molecular weight DNA was harvested at 1 dpi and 3 dpi and BKPyV DNA was quantified by qPCR. Replicated DNA (3 dpi) was normalized to input DNA (1 dpi). (E) Viral lysates were harvested from cells 3 dpi and progeny were quantified by an IU assay. Each bar is the average from three independent experiments and the error bars are SD. No mature miRNA, early transcript, viral genome, or progeny were detected in mock-infected samples. ND, not detected; A, archetype; R, rearranged; WT, WT miRNA; mut, mutant miRNA; ╪, below the limit of detection; *P < 0.05.
miRNA Expression Is Dictated by the Balance of Regulatory Elements Within the NCCR.
The steady-state level of the early transcript is largely the product of mRNA synthesis, driven by the early promoter, and degradation, driven by the miRNA. Therefore, we hypothesized that the relative levels of early mRNA and miRNA transcription dictate the difference between archetype virus and rearranged variant replication in RPTE cells. In the archetype virus, early mRNA production would be restricted by low early promoter activity combined with high miRNA expression, whereas the opposite would be true for rearranged variants. The results in Fig. 3B demonstrate that the archetype virus produces ∼100-fold less early mRNA compared with the rearranged variant. This suggests low levels of early promoter activity in the archetype virus compared with the rearranged variant. Additionally, we hypothesized that the archetype virus would have relatively high levels of miRNA expression, potentially driven by the late promoter even early in infection, compared with early promoter activity. This would limit viral replication because the early transcript product TAg is responsible for initiating viral DNA synthesis (13). In contrast, in the rearranged variant, which readily replicates in RPTE cells, we predicted there would be low miRNA expression and high early promoter activity driving target early mRNAs.
Previously, Gosert et al. developed a bidirectional reporter vector to investigate the functional outcome of BKPyV NCCR rearrangements with respect to early and late gene expression outside the context of infection (6). This study showed that rearranged NCCR structures have high early gene expression and low late gene expression, whereas archetype virus NCCR structure has the opposite pattern. In this study, rearranged variants were associated with increased cytopathology in patients and increased replication capacity as a result of high early gene expression. We tested our archetype and rearranged NCCRs in this bidirectional reporter assay and found that archetype virus has low early and high late promoter activity, whereas the rearranged variant has high early and low late promoter activity, consistent with previous results (Fig. S2). These data support our hypothesis that archetype virus and rearranged variants have differential promoter activity controlling early and late strand expression.
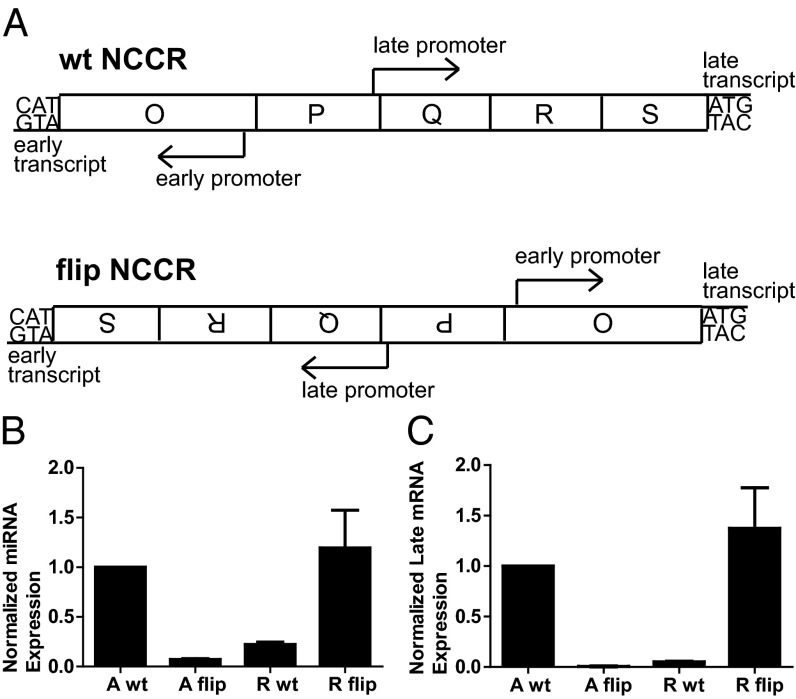
To test this hypothesis in a system more relevant to viral infection, we created a flipped NCCR structure mutant in both the archetype (A flip) and rearranged (R flip) genomes (Fig. 4A). In these mutants the late promoter drives early gene expression and the early promoter drives late gene expression. Therefore, we were able to measure both early and late promoter activity within the same assay by measuring the late transcript, which is driven by the late promoter in the WT virus and the early promoter in the flip mutant virus. To examine promoter activity during the early phase of infection, without the complication of genome replication, DNA replication was blocked with 40 µg/mL cytosine arabinoside (AraC) (18) to assess initial miRNA expression in these viruses. RPTE cells were infected with equal genomes of the flip and WT viruses. Total RNA was harvested at 48 h postinfection (hpi), and mature 5P miRNA, the dominant arm, was measured by stem-loop qRT-PCR. Consistent with our hypothesis, the miRNA expression of archetype (A WT) and rearranged (R flip) was high, whereas miRNA expression of R WT and A flip was low (Fig. 4B). Viral late transcript was also measured by qRT-PCR, and it paralleled the miRNA expression results (Fig. 4C). The data from the flip experiment suggest that miRNA expression is controlled by sequences in the NCCR, in a manner similar to late mRNA transcripts. Additionally, because DNA replication was blocked in this experiment, it is clear that both the miRNA and late transcript can be expressed before the onset of DNA replication. Together, these results demonstrate that early in the viral infection, the delicate balance of regulatory elements within the NCCR controls the expression of miRNA and early mRNAs, and this balance dictates the ability to control archetype virus replication in RPTE cells.
Fig. 4.
Balance of NCCR regulatory elements in archetype virus vs. rearranged variant. (A) Schematic of the WT and flip NCCR constructs. RPTE cells were infected with 1 × 109 genomes of crude stock virus in the presence of AraC. Total cell RNA was harvested 2 dpi. (B) BKPyV 5P miRNA expression was quantified by stem-loop qPCR and normalized to the cellular control hsa-let-7a. (C) Late transcript was measured by qRT-PCR and normalized to GAPDH transcript. The A WT sample was arbitrarily set to 1. Each bar represents the average from three independent experiments and the error bars are SEM. No mature miRNA or late transcript was detected in mock-infected samples. A, archetype; R, rearranged.
Discussion
Archetype BKPyV is the transmissible form of BKPyV and the form that establishes persistence in the human host. In this study we set out to investigate the mechanism controlling archetype virus replication in RPTE cells, to begin to provide insights into the mechanism of polyomavirus persistence. We show here that the BKPyV miRNA regulates early mRNA expression, and by extension, viral replication. Significantly, a mutant virus unable to produce mature miRNAs showed a striking increase in early transcript expression, DNA replication, and progeny production, indicating that the miRNA is largely responsible for controlling archetype viral replication. This high degree of regulation is atypical for miRNAs, which are generally considered “fine tuners” of gene expression (2). Furthermore, we show that the balance of regulatory elements driving expression of high levels of miRNA and low levels of early mRNA uniquely defines archetype virus. We suggest this balance dictates the unique ability to control archetype virus replication through the action of the miRNA. To our knowledge, this is a unique result demonstrating that the polyomavirus miRNA is capable of regulating viral replication.
Polyomavirus miRNAs have many unique attributes. Outside of plants, miRNAs typically form an imperfect duplex with their targets; one arm of the mature miRNA is favored and incorporated into RISC, whereas the other is degraded. However, the polyomavirus miRNAs pair perfectly with early mRNAs that are encoded on the opposite strand, and both the 5P and 3P mature miRNAs are incorporated into RISC for the same target (9). Many miRNAs recognize the 3′-UTR of a target transcript; however, many polyomaviruses target the transcripts within the 3′-coding region. TAg, one of the products of the early mRNA that is targeted by the miRNA, plays a major role in the initiation of viral DNA replication at the origin (13). Unexpectedly, mutation or deletion of the SV40 or MPyV miRNA results in increased TAg protein levels but no measurable enhancement of viral replication (10, 12). As a result, previous studies of the polyomavirus miRNAs focused on their roles in regulating the host immune response by limiting viral antigen (TAg) or directly targeting mediators of immune recognition (12, 19).
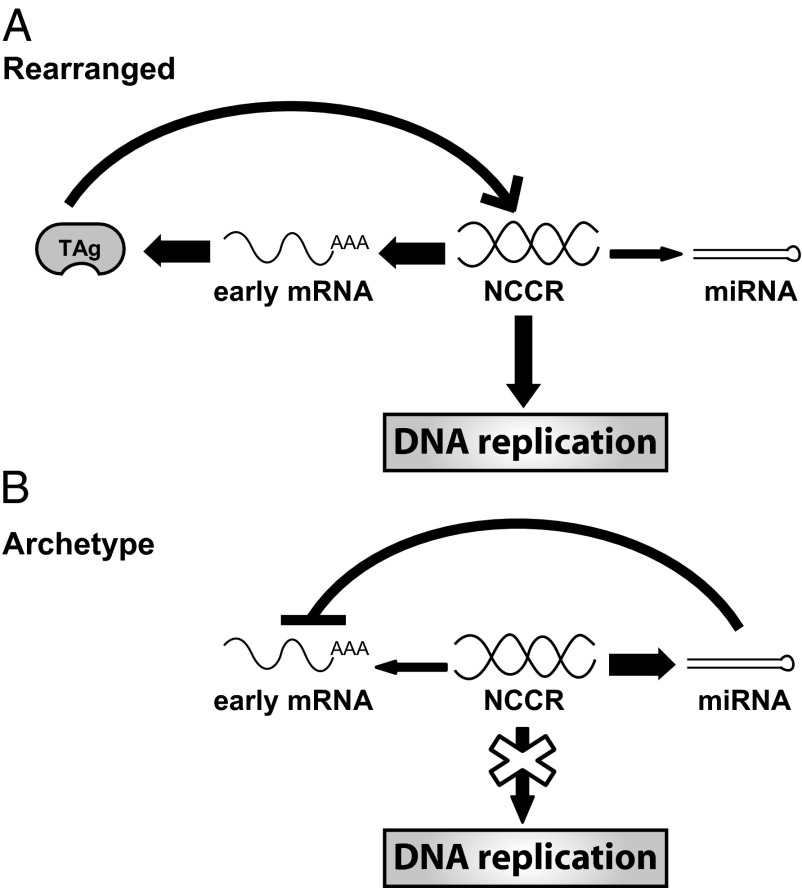
In this study we define a unique role of the polyomaviruses miRNA in controlling archetype viral replication. Previous work with an SV40 rearranged variant and MPyV laboratory stain with duplications in the enhancer region showed no measurable effect on production of infectious progeny of mutating the miRNA, which was recapitulated in this study (Fig. 3E). We also did not detect an increase in replication capacity of the rearranged BKPyV variant miRNA mutant, and in fact saw a small but reproducible decrease (Fig. 3D). However, we detected a dramatic phenotype in the archetype virus miRNA mutant, resulting in a 100-fold increase in early transcript production and a 50-fold increase in replicated viral DNA. This regulation by the miRNA is accomplished through the differential regulation of promoter activity controlling the miRNA and target early mRNA expression in archetype virus compared with rearranged variants. High miRNA expression is effective in limiting archetype virus replication, because this virus also has low early promoter activity driving the early mRNA target (Fig. 5). In contrast, the rearranged variants have high early promoter activity driving early mRNA expression and miRNA expression that is low relative to archetype virus. We surmise that the limited amount of miRNA present during rearranged variant infection is unable to impact the level of early mRNA, which is abundant. Indeed, these results are in agreement with our previous work, which shows that the archetype virus NCCR is sufficient to confer loss of replication in a rearranged variant background (16). Because prior work on polyomavirus miRNAs used rearranged variants, this explains why no previous studies have observed a functional role for polyomavirus miRNA in regulating viral replication. Our results with rearranged BKPyV also support this conclusion.
Fig. 5.
Model of miRNA control of archetype virus replication. (A) In rearranged variants, high levels of early mRNA are expressed from high early promoter activity, whereas the miRNA is only weakly expressed. TAg, translated from early mRNA, binds to the origin of replication and drives DNA replication. (B) Archetype virus early mRNA is weakly expressed from the early promoter, whereas miRNA is robustly expressed and targets early mRNA for degradation. Therefore, DNA replication is blocked in archetype virus in RPTE cells.
We hypothesize that miRNA expression is controlled by elements in the NCCR that overlap, or are the same as, the viral late promoter, the only known promoter on the late strand. The late promoter has no clearly defined elements, which is likely a mechanism to cope with the large number of natural NCCR rearrangements that occur within this region (20). As a result, late transcription has to be able to initiate at a variety of sites depending on which sequence blocks are present and in what order and take into account the distance to the start codon. The results of the NCCR flip experiment are consistent with the miRNA being expressed from the late promoter or a unique promoter within the NCCR. However, it is also possible that sequence elements in the NCCR are able to regulate miRNA expression or that there is a cryptic promoter on the late strand that can drive miRNA expression. Alternatively, the miRNA may be able to use more than one promoter for its expression. Fine mapping of the miRNA promoter is an interesting area for future study.
Many viral miRNAs were discovered in cells latently infected with herpesviruses, and as a result, viral miRNAs have been suggested to play an essential role in establishing or maintaining viral latency or persistence (21). Moreover, several Kaposi sarcoma herpesvirus miRNAs are vital for directly and indirectly controlling lytic transcript expression (22–24). Whereas previous studies have described the polyomavirus miRNAs as being expressed late during infection (9, 12), our data show that the miRNA is expressed before the onset of DNA replication. Thus, the archetype virus miRNA promoter is active early, before DNA replication. We also noted in the Northern blot that there was an accumulation of high molecular weight unprocessed miRNA precursors and a decrease in the pre-miRNA band in the miRNA mutant virus, indicating that the defect in miRNA biogenesis likely occurs before pre-miRNA processing (25).
We show here that the balance of regulatory activity controlling miRNA and its target early mRNA expression regulates archetype BKPyV replication. Because archetype virus is the form of BKPyV that is believed to be transmitted and establish persistence in the host, the regulation of replication by the miRNA is likely essential to the ability of this form of the virus to persist in the host. We suggest that the polyomavirus miRNA plays a major role in the establishment of viral persistence because we noted a dramatic reduction of regulation in archetype virus replication in the miRNA mutant virus. Polyomaviruses have miRNAs antisense to their early coding regions, and therefore miRNA regulation of viral replication may be a common mechanism used by polyomaviruses to establish a life-long persistent infection (21). This has implications for the increasing number of immunocompromised patients affected by polyomavirus-associated diseases.
Materials and Methods
Cell Culture.
Primary human RPTE cells (Lonza) were maintained in renal epithelial growth medium (REGM) for up to six passages (26). 293TT cells were obtained from Chris Buck at the National Cancer Institute and were maintained under 400 μg/mL hygromycin selection (27). 293 (28) and 293TT cells were maintained in Dulbecco’s modified Eagle medium with 10% FBS and 100 units/mL penicillin, 100 μg/mL streptomycin. All cells were grown in a 5% CO2 environment in a humidified 37 °C incubator.
miRNA Folding Prediction.
The mfold RNA folding form was used to predict the BKPyV miRNA WT and mutant RNA structures (http://mfold.rna.albany.edu/) (29).
DNA Constructs, Transfection, Infection, Drug Treatment, and Luciferase Assays.
Archetype virus and rearranged variant miRNA mutated viruses were created using site-directed mutagenesis as described in SI Materials and Methods and a list of primers used is presented in Table S1. The WT and mutant viral genomes were transfected into 293TT cells to produce crude lysates (8). Viral stocks were expanded in 293TT cells and purified by density centrifugation on Cesium chloride gradients (30). Viral titers were determined by VP1 IU assay (8). We set the limit of detection for the IU assay as a minimum of 20 foci per field of view.
The luciferase target construct was created as described (9) and detailed in SI Materials and Methods. The miRNA expression construct, pcDNA3.1BKV miRNA, (received from G. Seo and C. Sullivan, University of Texas at Austin, Austin, TX) was mutated as described in SI Materials and Methods. The 293 cells were transfected and luciferase assays were performed as described in SI Materials and Methods using the Promega dual-luciferase reporter assay system.
The archetype virus and rearranged variant NCCR were cloned into the phRG promoter construct (6), kindly provided by H. Hirsch (University of Basel, Basel, Switzerland), as described in SI Materials and Methods. 293 cells were transfected with TransIT LT-1 transfection reagent (Mirus Bio) according to the manufacturer's instructions and analyzed at 2 d posttransfection dpt (6).
NCCR flip constructs were created in the Dik-3 site (archetype virus) and Dunlop-3 site (rearranged variant), which replicates equivalently to WT Dunlop in RPTE cells (16); backbones are as described in SI Materials and Methods. Religated genomes were transfected into 293TT cells and crude lysates were harvested 7 dpt. Lysates were freeze/thawed three times and genomes were quantified by real-time PCR (8).
For infection, cells were prechilled for 15 min at 4 °C. Cells were exposed to virus diluted into media (DMEM for 293TT cells or REGM for RPTE cells) at a multiplicity of infection (MOI) of 0.01 or 1 × 109 genomes per cell (NCCR flip experiments) and incubated for 1 h at 4 °C. The viral inoculum was then removed and infection was initiated by adding warm media and transferring the cells to 37 °C.
Northern and Western Blotting.
Total RNA was harvested from cells with TRIzol (Life Technologies) and processed using the Direct-zol RNA MiniPrep (Zymo Research). RNA was run on a Tris-borate-EDTA-urea-15% polyacrylamide gel and transferred to Hybond H+ membrane as previously described (31). The blot was probed with Starfire probes according to the manufacturer’s instructions (IDT). The following probes were used: BKV 5P probe 5′-CAATCACAATGCTCTTCCCAAGTCTCAGATACTTCA-3′ and BKV 3P probe 5′-ACTGAAGACTCTGGACATGGATCAAGCACTGAATCC-3′.
E1A lysis buffer was used to harvest total cell proteins, which were quantified and immunoblotted as previously described (8, 30). The following antibodies and concentrations were used: pAb416 (32) for TAg at 1:3,000; P5G6 for VP1 at 1:10,000; and Ab9484 (Abcam) for GAPDH at 1:10,000.
Quantitative PCR.
The miRNA stem-loop qPCR protocol for the BKPyV 5P miRNA, kindly provided by C. Sullivan, was adapted from previously published protocols (15, 33) and is detailed in SI Materials and Methods. Results are presented as fold change in miRNA transcript levels relative to levels in WT archetype virus-infected samples, normalized using the 2−ΔΔC(t) method (34) to a human miRNA control (hsa-let-7a), which is abundantly expressed in human kidney (35). miRNA copy number was determined by comparison with a standard curve using RNA oligos (IDT) corresponding to the BKPyV 5P or hsa-let-7a 5P miRNAs (Table S1). Average Ct value and copy number/nanogram RNA for each miRNA are given in Table S2 for the experiments presented in Figs. 2C, 3A, and 4B.
To quantify virus DNA replication by qPCR, low molecular weight DNA was isolated using a Hirt isolation protocol. Real-time PCR was performed and the data were analyzed as previously described (18, 36). Viral DNA replicated at 3 dpi was normalized to input DNA harvested 1 dpi using a ΔCt method. To quantify early and late transcripts, total RNA was harvested as above at the indicated time points and qRT-PCR was performed as detailed in SI Materials and Methods. Results are presented as fold change in transcript levels relative to levels in WT archetype virus-infected samples, normalized to GAPDH using the 2−ΔΔC(t) method (34).
Supplementary Material
Acknowledgments
We thank members of the M.J.I. laboratory and Adam Lauring for comments and discussion; the S. Camper laboratory for use of their luminometer; C. Sullivan and G. Seo for the miRNA luciferase reporter assay constructs; H. Hirsch and Rainer Gosert for the phRG promoter construct; and C. Sullivan for advice in designing the miRNA mutant virus, for the 5P BKPyV miRNA stem-loop qPCR assay, and other helpful suggestions. This work was supported by National Institutes of Health (NIH) Grant AI060584 (to M.J.I.) and in part by NIH Grant CA046592 (to the University of Michigan Cancer Center). N.M.B. was supported in part by NIH National Research Service Award T32-GM07544 from the National Institute of General Medical Sciences and a Rackham predoctoral fellowship from the University of Michigan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301907110/-/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3(6):375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahsan N, Shah KV. Polyomaviruses and human diseases. Adv Exp Med Biol. 2006;577:1–18. doi: 10.1007/0-387-32957-9_1. [DOI] [PubMed] [Google Scholar]
- 5.Moens U, Van Ghelue M. Polymorphism in the genome of non-passaged human polyomavirus BK: Implications for cell tropism and the pathological role of the virus. Virology. 2005;331(2):209–231. doi: 10.1016/j.virol.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 6.Gosert R, et al. Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J Exp Med. 2008;205(4):841–852. doi: 10.1084/jem.20072097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Low J, Humes HD, Szczypka M, Imperiale M. BKV and SV40 infection of human kidney tubular epithelial cells in vitro. Virology. 2004;323(2):182–188. doi: 10.1016/j.virol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 8.Broekema NM, Imperiale MJ. Efficient propagation of archetype BK and JC polyomaviruses. Virology. 2012;422(2):235–241. doi: 10.1016/j.virol.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo GJ, Fink LH, O’Hara B, Atwood WJ, Sullivan CS. Evolutionarily conserved function of a viral microRNA. J Virol. 2008;82(20):9823–9828. doi: 10.1128/JVI.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sullivan CS, et al. Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology. 2009;387(1):157–167. doi: 10.1016/j.virol.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seo GJ, Chen CJ, Sullivan CS. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 2009;383(2):183–187. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435(7042):682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 13.Fanning E, Zhao K. SV40 DNA replication: From the A gene to a nanomachine. Virology. 2009;384(2):352–359. doi: 10.1016/j.virol.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heritage J, Chesters PM, McCance DJ. The persistence of papovavirus BK DNA sequences in normal human renal tissue. J Med Virol. 1981;8(2):143–150. doi: 10.1002/jmv.1890080208. [DOI] [PubMed] [Google Scholar]
- 15.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broekema NM, et al. A system for the analysis of BKV non-coding control regions: Application to clinical isolates from an HIV/AIDS patient. Virology. 2010;407(2):368–373. doi: 10.1016/j.virol.2010.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imperiale MJ, Major EO. Polyomaviruses. Philadelphia: Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 18.Jiang M, Entezami P, Gamez M, Stamminger T, Imperiale MJ. Functional reorganization of promyelocytic leukemia nuclear bodies during BK virus infection. MBio. 2011;2(1):e00281–e10. doi: 10.1128/mBio.00281-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauman Y, et al. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe. 2011;9(2):93–102. doi: 10.1016/j.chom.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Moens U, Johansen T, Johnsen JI, Seternes OM, Traavik T. Noncoding control region of naturally occurring BK virus variants: Sequence comparison and functional analysis. Virus Genes. 1995;10(3):261–275. doi: 10.1007/BF01701816. [DOI] [PubMed] [Google Scholar]
- 21.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411(2):325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei X, et al. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat Cell Biol. 2010;12(2):193–199. doi: 10.1038/ncb2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu F, Stedman W, Yousef M, Renne R, Lieberman PM. Epigenetic regulation of Kaposi’s sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J Virol. 2010;84(6):2697–2706. doi: 10.1128/JVI.01997-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu CC, et al. MicroRNAs encoded by Kaposi’s sarcoma-associated herpesvirus regulate viral life cycle. EMBO Rep. 2010;11(10):784–790. doi: 10.1038/embor.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kincaid RP, Burke JM, Sullivan CS. RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci USA. 2012;109(8):3077–3082. doi: 10.1073/pnas.1116107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abend JR, Low JA, Imperiale MJ. Inhibitory effect of gamma interferon on BK virus gene expression and replication. J Virol. 2007;81(1):272–279. doi: 10.1128/JVI.01571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buck CB, Pastrana DV, Lowy DR, Schiller JT. Efficient intracellular assembly of papillomaviral vectors. J Virol. 2004;78(2):751–757. doi: 10.1128/JVI.78.2.751-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 29.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang M, Abend JR, Tsai B, Imperiale MJ. Early events during BK virus entry and disassembly. J Virol. 2009;83(3):1350–1358. doi: 10.1128/JVI.02169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClure LV, Lin YT, Sullivan CS. Detection of viral microRNAs by Northern blot analysis. Methods Mol Biol. 2011;721:153–171. doi: 10.1007/978-1-61779-037-9_9. [DOI] [PubMed] [Google Scholar]
- 32.Harlow E, Crawford LV, Pim DC, Williamson NM. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39(3):861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.