Abstract
Cold acclimation is the major process that prepares plants for freezing tolerance. In addition to extensive transcription regulation by cold-inducible master transcription factors, oxidative stress signaling has been postulated to play a role in freezing tolerance. Activation of oxidative signaling through the expression of an active mitogen-activated protein kinase kinase kinase provided benefits in transgenic tobacco at freezing temperature bypassing cold acclimation. Because involvement of the mitogen-activated protein kinase cascade in oxidative stress signaling is evolutionarily conserved in eukaryotes from yeast to mammals, we tested the effect of expressing a heterologous tobacco mitogen-activated protein kinase kinase kinase (Nicotiana PK1), which can mimic H2O2 signaling, in a major cereal crop. We demonstrate that low-level but constitutive expression of the Nicotiana PK1 gene enhances freezing tolerance in transgenic maize plants that are normally frost sensitive. Our results suggest that a new molecular approach can be designed to genetically enhance freezing tolerance in important crops.
Keywords: Nicotiana protein kinase, stress tolerance, Zea mays
Plants acclimate to environmental stresses by activating cascades or network events initiating with stress perception and ending with the expression of many effector genes (1). The alterations of extensive gene expression by these acclimation processes commonly result in enhancing stress tolerance in plants (2-7). Overexpression of master transcription activators such as dehydration-responsive element bindings (DREBs)/cold-inducible master transcription factors (CBFs) that bind to the DNA regulation element (the C-repeat) in the promoter region of cold-responsive genes could improve freezing tolerance in transgenic Arabidopsis (6, 7). In addition to cold-induced transcription regulation, oxidative stress signaling also plays a role in freezing tolerance. Under different abiotic and biotic stresses, plants accumulate active oxygen species, including hydrogen peroxide (H2O2) (8). Whereas large amounts of H2O2 accumulation lead to programmed cell death, a relatively small amount of H2O2 modifies gene expression and results in enhanced plant stress responses (9). Maize and tomato plants pretreated with H2O2 showed improved chilling tolerance (10, 11). One explanation for this is that H2O2 can induce the synthesis of heat shock proteins (HSPs), which in turn protect plants from high- and low-temperature stresses (10). Increased expression of HSPs, either in response to high-temperature stress or by transgenic approaches were found to be associated with a significant increase in chilling tolerance in tomato (12, 13). Kovtun et al. (14) found that H2O2 induced the activity of an Arabidopsis mitogen-activated protein kinase kinase kinase (MAPKKK). Transgenic tobacco constitutively expressing Nicotiana PK1 (NPK1), an Arabidopsis MAPKKK orthologue, showed enhanced tolerance to freezing, salt, and heat stresses (14). Because the activation of a mitogen-activated protein kinase (MAPK) signal transduction pathway by H2O2 is conserved among different organisms (15), we hypothesized that the expression of NPK1 would increase the stress tolerance in other plant species, such as the major agronomic crop, maize.
Maize originated from tropical regions and is a frost-sensitive plant (16). Low-temperature stresses, including chilling and frost, greatly affect the germination and growth of the plant, and limit the geographical distribution of the crop. In this study, we show that expression of the tobacco NPK1 gene expressing an active form of protein kinase enhances the freezing tolerance in transgenic maize plants. To our knowledge, this is the first report in which the trait of freezing tolerance was achieved through constitutive expression of an MAPKKK gene in maize.
Materials and Methods
DNA Constructs and Maize Transformation. Agrobacterium strain EHA101 harboring a binary vector pSHX004 (Fig. 1) was used to transform Hi II maize immature embryo as described (18). R0-transgenic plants were crossed with hybrid Hi II or inbred line B73 to produce R1 seeds.
Fig. 1.
T-DNA fragment of pSHX004. LB, left border; RB, right border; bar, phospinothricin acetyl transferase gene; NPK1 gene; P35S*, 2× CaMV 35S promoter (45); P35S**, a modified 35S promoter (35SC4PPDK; ref. 17); TEV, tobacco etch virus 5′ UTR (46); Tnos, nopaline synthase terminator (47); Tvsp, soybean vegetative storage protein terminator (48).
Freezing Tolerance Tests. Maize seeds were germinated and grown in a growth chamber (PGV36, Conviron Instruments, Winnipeg, MB, Canada) at 25°C with 14 h of light. At the three-leaf stage, maize seedlings were exposed to a graduated freezing condition or to a constant freezing condition in a growth chamber (dark). In the graduated freezing treatment, the temperature of the growth chamber was set at -1°C and was programmed to decrease to minus 6°C at the rate of 1°C per hour. In the constant freezing treatment, the temperature of the growth chamber was set at -5°C. Before moving the plantlets into the freezing condition, the seedlings were sprayed with a layer of water on the leaves to initiate the formation of ice.
Electrolyte Conductivity. For each treatment, six leaves from three plants per event (two leaves per plant, six leaves per event) were measured for electrolyte leakage (EL). Sampled leaves were rinsed with ddH2O to remove possible ion contamination on the surface. Leaf discs (7 mm in diameter) were punched by using a standard three-hole punch and were placed into a 20 × 150-mm glass test tube (Fisher Scientific) containing 15 ml of distilled water. The test tubes were subjected to vacuum three times at 5-min intervals at 60 psi to remove air bubbles adhered to the surface of the leaves. Tubes were then shaken at 300 rpm for 2 h in a slanted position. After shaking, conductivity of the solution was measured by using a conductivity meter (Thermo Orion 150A+, Orion Research, Beverly, MA). The solutions were then autoclaved at 122°C for 20 min to completely lyse the plant cell walls. The electrolyte conductivities of autoclaved solutions were recorded as the absolute conductivity. The percentage of EL was calculated by dividing the initial conductivity by the absolute conductivity.
Soluble Carbohydrate Content. All plants were grown to three-leaf stage at 25°C before subjected to cold treatment (4°C) for 24 and 48 h. Total soluble extracts were prepared from the second leaf of transgenic plants and their negative siblings. Leaf samples were lyophilized and homogenized. Soluble carbohydrates were extracted with 20 ml of 80% ethanol for 24 h at 4°C. The insoluble residual was removed by centrifuging at 5,000 × g for 10 min. Total carbohydrate content was determined by the phenol-sulfuric acid method (21). Fructose, sucrose, and glucose contents were measured by using the corresponding assay kits from Sigma, following the manufacturer's instructions.
Sequences of Maize Genes for Microarray. Protein sequences of stress-related Arabidopsis or tobacco genes (Table 1) were used to run a blast search against the maize genome database (ZMDB; which can be accessed at www.maizegdb.org) for maize homologs.
Table 1. Genes used for microarray.
| Genes | EST or GenBank accession no. | Putative function | Ref. |
|---|---|---|---|
| 18S RNA | AF168884 | Housekeeping gene | |
| NPK1 | D26601 | Transgene, signal transduction in oxidative pathway | 15 |
| Alcohol dehydrogenase | X00580 | Stress defense | 36 |
| Auxin-regulated protein | TUC02-10-16-619.2 | Signal transduction | 33 |
| Blue copper protein | TUC01-09-30-1263.1 | Cellular organization and biogenesis | 34 |
| CCR4-associated factor | TUC02-07-26-9731.1 | Transcription factor | 34 |
| COR47 (cold-regulated gene) | BM337639 | Stress defense | 36 |
| DREB1 | BM953485 | Transcriptional factor | 34 |
| EREBP (ethylene-responsive element-binding protein) | BU092546 | Transcriptional factor | 34 |
| EREBP-1 (ethylene-responsive element-binding protein) | BU572129 | Transcriptional factors | 34 |
| GPX2 (glutathione peroxidase 2) | TUC02-07-26-1389.2 | Cellular protection and detoxification | 3 |
| GR (glutathione reductase) | TUC02-04-28-7376.1 | Cellular protection and detoxification | 3 |
| GST | TUC02-07-26-9410.1 | Cellular protection and detoxification | 38 |
| GST1 | TUC02-09-09-5639.1 | Cellular protection and detoxification | 35 |
| HSP17.4 | BU499390 | Cellular protection and detoxification | 34 |
| HSP17.5 | BM267274 | Cellular protection and detoxification | 34 |
| HSP17.8 | BU097877 | Cellular protection and detoxification | 34 |
| HSP83 | TUC02-10-16-3582.1 | Cellular protection and detoxification | 34 |
| HSP101 | AW257917 | Cellular protection and detoxification | 34 |
| Jasmonic acid-inducible protein | AW562671 | Cellular protection and detoxification | 34 |
| LOX1 (lipoxygenase) | TUC02-10-16-1078.1 | Oxylipin metabolism | 38 |
| P5CS (1-pyrroline-5-carboxylate synthetase) | TUC02-10-16-983.3 | Cellular protection and detoxification | 37 |
| Peroxidase | TUC02-09-09-2229.1 | Cellular protection and detoxification | 32 |
| PR1 | TUC02-07-26-1442.1 | Cellular protection and detoxification | 32 |
| PR2 | TUC02-04-28-4452.2 | Cellular protection and detoxification | 32 |
| PR5 | TUC02-07-25-12188.1 | Cellular protection and detoxification | 32 |
| Proline transporter | TUC02-02-07-8772.1 | Cellular protection and detoxification | 33 |
| Pyruvate decarboxylase | TUC02-02-07-15716.1 | Metabolism | 34 |
| RD22 (dehydration-responsive) | BM078186 | Stress defense | 36 |
| WRKY11 | TUC02-02-07-15126.1 | Transcription factor | 38 |
Further information can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Results
Performance of NPK1-Transgenic Maize. A cDNA fragment encoding the 268-aa kinase domain of NPK1 driven by a modified CaMV 35S promoter (ref. 17 and Fig. 1) was introduced into maize by using Agrobacterium-mediated transformation (18). Twenty-two transgenic maize events, designated as A4, were produced from Hi II immature zygotic maize embryos.
Plant height (PH) and leaf number (LN) of R1 plants from 22 NPK1-transgenic events were evaluated in the field during the summer of 2002. Five transgenic plants and five null segregants per event were evaluated (Table 2). Statistical analysis showed that significant differences existed among different transgenic events in these two agronomic traits (P < 0.0001 for both PH and LN). Because the hybrid Hi II was used in transformation experiments, and the R1 seeds were generated through either crossing with nontransgenic Hi II plants or inbred line B73 (one of the parent lines for the generation of Hi II) (25), the differences in PH and LN in R1 generation can be attributed to the segregation of these traits. However, no significant difference was detected between transgenic plants and their null segregants of the same event (P = 0.3841 for PH and P = 0.6943 for LN), suggesting that the NPK1 transgene has no impact on maize plant growth under normal growth conditions. Therefore, the null segregants served as a negative control for the evaluation of this transgene in maize plants.
Table 2. Transgene expression levels, PH, and LNs in R1 transgenic plants and nontransgenic segregants.
| Transgene expression levels
|
PH, cm
|
LN
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Transgenic
|
Nontransgenic
|
Transgenic
|
Nontransgenic
|
|||||||
| Events | Pedigree | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| A4-1 | 0.1197 | R0 × Hi II | 127.67 | 3.58 | 135.64 | 6.05 | 11.00 | 1.00 | 12.00 | 1.87 |
| A4-2 | 0.1955 | B73 × R0 | 143.00 | 12.49 | 159.51 | 5.17 | 11.00 | 1.00 | 11.40 | 1.34 |
| A4-3 | 0.1348 | R0 × Hi II | 106.17 | 9.05 | 88.27 | 1.06 | 10.80 | 2.17 | 10.00 | 0.00 |
| A4-4 | 1.8421 | B73 × R0 | 131.80 | 19.60 | NA | NA | 10.40 | 0.55 | NA | NA |
| A4-5 | 0.6551 | R0 × Hi II | 121.67 | 2.22 | 119.63 | 4.28 | 10.60 | 0.55 | 10.60 | 0.55 |
| A4-8 | 0.2171 | R0 × Hi II | 136.14 | 3.90 | 144.27 | 4.49 | 13.20 | 1.48 | 12.80 | 1.10 |
| A4-9* | 0.5401 | B73 × R0 | 164.34 | 5.54 | 152.15 | 3.32 | 12.60 | 1.67 | 11.40 | 0.89 |
| A4-10 | 0.1686 | Hi II × R0 | 110.49 | 2.55 | 122.34 | 3.69 | 9.40 | 1.14 | 11.00 | 1.00 |
| A4-13 | 0.6960 | R0 × Hi II | 126.37 | 2.33 | 123.95 | 4.92 | 10.00 | 0.82 | 10.40 | 0.55 |
| A4-14 | 0.7887 | R0 × Hi II | 110.24 | 6.65 | 111.76 | 5.28 | 11.00 | 0.00 | 11.40 | 0.55 |
| A4-15* | 2.0981 | R0 × Hi II | 112.01 | 1.39 | 116.59 | 3.42 | 11.60 | 0.89 | 11.20 | 0.84 |
| A4-16 | 0.1492 | R0 × Hi II | 109.22 | 4.29 | 105.66 | 5.03 | 10.00 | 0.71 | 10.20 | 0.45 |
| A4-17 | 0.6560 | R0 × Hi II | 102.87 | 5.68 | 120.14 | 9.78 | 10.00 | 1.22 | 10.60 | 1.14 |
| A4-18 | 0.2832 | R0 × Hi II | 99.82 | 8.19 | 98.55 | 4.96 | 10.00 | 0.71 | 9.80 | 1.64 |
| A4-19 | 1.1698 | R0 × Hi II | 106.93 | 6.39 | 102.87 | 11.78 | 10.40 | 1.14 | 10.20 | 0.45 |
| A4-20 | 0.0890 | R0 × Hi II | 124.46 | 4.58 | 107.95 | 4.77 | 11.33 | 0.58 | 11.33 | 1.53 |
| A4-22 | 0.9031 | R0 × Hi II | 122.68 | 2.97 | 118.11 | 7.34 | 11.40 | 1.14 | 10.75 | 1.71 |
| A4-23 | 0.2190 | R0 × Hi II | 121.67 | 7.02 | 135.26 | 4.65 | 10.80 | 1.30 | 10.50 | 0.58 |
| A4-24 | 0.1056 | R0 × Hi II | 144.53 | 4.51 | 146.30 | 1.98 | 10.80 | 1.10 | 10.60 | 1.34 |
| A4-28 | 0.1336 | B73 × R0 | 151.13 | 4.21 | 156.46 | 4.10 | 10.40 | 0.55 | 10.20 | 0.84 |
| A4-29 | 0.2126 | B73 × R0 | 151.64 | 4.87 | 158.75 | 5.07 | 11.40 | 1.67 | 11.00 | 1.22 |
| A4-32 | 0.1277 | B73 × R0 | 158.75 | 3.71 | 165.42 | 1.89 | 11.00 | 0.71 | 11.50 | 1.91 |
NA, not available.
Events were selected for freezing test.
A quantitative real-time RT-PCR analysis was carried out to determine expression levels of the NPK1 transgene in the transgenic events. Results showed that all transgenic events expressed the NPK1 gene although the expression levels of the transgene were different (Table 2). Transgenic events A4-9 and A4-15, representing medium and high levels of NPK1 gene expression, respectively, were selected for further freezing tests.
Effect of NPK1 on Freezing Tolerance. To investigate whether constitutive expression of the NPK1 gene in maize provides protection against freezing damage, we performed two freezing tests: a graduated freezing test and a constant freezing test, on transgenic events A4-9 and A4-15. Giving the intrinsic genetic variation among these transgenic events, we used the null segregants from each transgenic event as the negative control. Cellular damage of treated seedlings due to freeze-induced membrane lesions was estimated by measuring EL from the leaves of treated plants. The higher the EL, the more severe the damage to the plant membrane, and the less tolerant the plants were to freezing challenging.
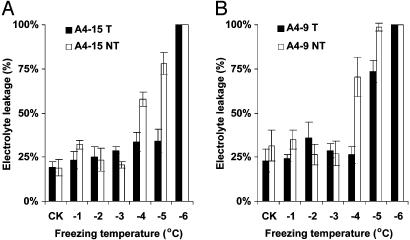
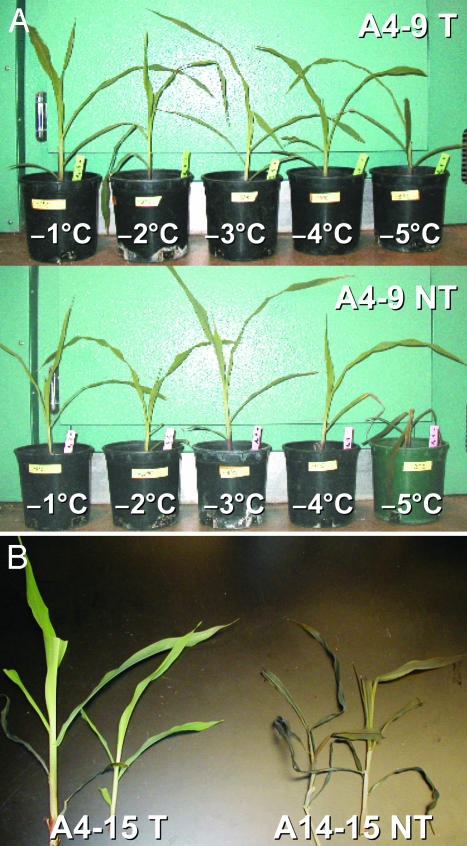
For each treatment, six leaf samples from three plants per event were assayed for EL. The leaf EL increased with a decrease in the environmental temperature (Fig. 2). When the temperature dropped to -4°C, the EL of negative segregants increased sharply, indicating that severe membrane damage had been caused by freezing stress. The EL of the A4-15-transgenic plants, on the other hand, did not increase until the temperature dropped to -6°C (Fig. 2A). This result indicates that transgenic maize event A4-15 was able to tolerate 2°C lower freezing temperature than its negative control siblings. Similarly, the EL of transgenic event A4-9 did not increase until the environmental temperature reached -5°C, which is 1°C lower than that of its negative siblings (Fig. 2B). Fig. 3A illustrates that nontransgenic seedlings of A4-9 started to show freezing damage when the temperature was reduced to -4°C and were dead at -5°C. Their transgenic counterparts showed no damage at -4°C and were only partially damaged at -5°C.
Fig. 2.
ELs of transgenic and nontransgenic plants under graduated freezing conditions. (A) Event A4-15. (B) Event A4-9. CK, control leaf samples before freezing treatment; T, transgenic plants; NT, nontransgenic plants.
Fig. 3.
Freezing tolerance of NPK1-transgenic maize. (A) A4-9-transgenic and -nontransgenic seedling under graduated freezing treatment. (B) A4-15 transgenic and -nontransgenic seedlings after 4-h constant freezing treatment (-5°C). T, transgenic plants; NT, nontransgenic plants.
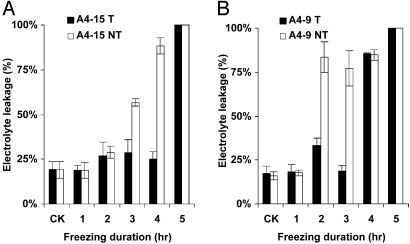
The increased freezing tolerance in transgenic plants of events A4-15 and A4-9 was confirmed in the constant freezing test (Fig. 4). The EL of A4-15-transgenic plants did not increase until 5 h at -5°C temperature. The EL of A4-15 negative control siblings, however, increased to 57% and 90% after 3 and 4 h of -5°C treatment, respectively (Fig. 4A). This result indicates that transgenic A4-15 plants survived 1-2 more hours than did their negative control siblings at a constant freezing temperature of -5°C. Fig. 3B shows that the null segregants of A4-15 were killed after 4 h at -5°C freezing treatment, whereas the transgenic seedlings survived. A similar difference in freezing tolerance between transgenic plants and their nontransgenic siblings was observed in event A4-9, in which transgenic plants survived for 3 h at -5°C, whereas the negative siblings survived for only 1 h (Fig. 4B).
Fig. 4.
ELs of transgenic and nontransgenic plants under-5°C constant freezing conditions. CK, control leaf samples before freezing treatment. (A) Event A4-15. (B) Event A4-9. T, transgenic plants; NT, nontransgenic plants.
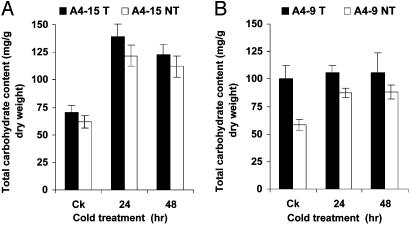
Effects of NPK1 on Sugar Metabolism in Maize. A positive correlation between freezing tolerance and soluble sugar content has been documented in a number of studies (26-29). Soluble sugars function as cryoprotectants and osmolytes that protect cells from freezing damage (30). We measured the sugar content in transgenic plants and their null segregants of events A4-9 and A4-15 before cold acclimation, and 24 and 48 h after 4°C cold acclimation. The second leaves of five three-leaf stage seedlings were pooled for measuring the contents of total soluble carbohydrate (TSC) content, as well as individual soluble carbohydrate content, such as sucrose, glucose, and fructose. As shown in Fig. 5, NPK1-expressing transgenic plants from both A4-9 and A4-15 have higher TSC content compared with their negative siblings in all treatments. Twenty-four- and 48-h cold acclimations significantly increased the TSC levels in all plants (P < 0.01). Under normal growth conditions, transgenic plants from event (NA) A4-9 contained significantly higher TSC compared with their null segregants (P < 0.02). The level of TSC in the nonacclimated (NA) A4-9-transgenic plants was higher than that in their cold-acclimated (CA), nontransgenic siblings. Further analysis indicated that the difference of TSC between transgenic and nontransgenic plants resulted from the difference in glucose and fructose content (data not shown). No difference in sucrose content between transgenic and nontransgenic seedlings was detected (data not shown).
Fig. 5.
Effect of NPK1 expression on the levels of TSCs. (A) Event A4-15. (B) Event A4-9. T, transgenic plants; NT, nontransgenic plants.
Effect of NPK1 on the Expression of Other Stress-Related Genes. It has been documented that constitutively active NPK1 mimics an oxidative stress signal and induces MAPK cascades in tobacco (14). To investigate whether the same mechanism was responsible for the enhanced freezing tolerance in NPK1-transgenic maize, we compared the expression levels of several stress-induced genes between transgenic and nontransgenic plants with or without cold acclimation by using a fiberoptic array technology (22, 23, 31).
We selected 28 maize EST (Table 1) sequences from the Maize Genome database, based on the protein sequences of putative stress-related Arabidopsis or tobacco orthologues (3, 32-38). The housekeeping gene (18S rRNA gene) and the transgene NPK1 also were included.
The relative expression levels of the selected genes were detected as signals on the array. The ratio of the gene expression in transgenic plants over their null segregants was used to determine whether the gene was up-regulated due to the presence of the NPK1 gene in transgenic plants. When the ratio is >1.5, the gene is defined as being up-regulated by the NPK1 gene expression in the sample.
According to the gene expression pattern in the two transgenic events under NA or CA conditions, genes were categorized into four groups. Genes in group I had at least 1.5 times higher expression in transgenic plants than in the null segregants in all four pairs of samples (events A4-9 and A4-15, both NA and CA conditions). The group included a GST gene, a small HSP (HSP17.8) gene, and a PR1 gene (Table 3).
Table 3. Difference in gene expression between NPK1 transgenic plants and their null segregants.
| Ratio*
|
||||
|---|---|---|---|---|
| Nonacclimated
|
48-h acclimated
|
|||
| Genes | A4-15 | A4-9 | A4-15 | A4-9 |
| Group I: Up-regulated in all samples | ||||
| GST | 1.8 | 1.9 | 1.7 | 2.1 |
| HSP17.8 | 3.4 | 12.6 | 2.4 | 3.8 |
| PR1 | 7.1 | 3.2 | 2.3 | 20.2 |
| Group II: Up-regulated in three of four samples | ||||
| Alcohol dehydrogenase | 1.2 | 1.8 | 1.8 | 5.0 |
| Blue copper protein | 1.0 | 2.6 | 2.1 | 2.1 |
| DREB1 | 1.9 | 1.4 | 2.8 | 2.1 |
| EREBP | 1.3 | 1.8 | 1.9 | 2.5 |
| GPX2 | 1.0 | 1.5 | 1.9 | 2.0 |
| HSP17.4 | 1.3 | 1.7 | 1.6 | 4.8 |
| HSP17.5 | 1.2 | 7.4 | 1.6 | 6.2 |
| HSP101 | 1.2 | 1.5 | 1.7 | 22.6 |
| Jasmonic acid-inducible protein | 1.2 | 1.7 | 4.0 | 8.4 |
| P5CS | 1.4 | 1.8 | 3.2 | 4.1 |
| Pyruvate decarboxylase | 1.0 | 1.6 | 1.6 | 2.4 |
| WRKY11 | 2.8 | 0.7 | 2.8 | 1.8 |
| Group III: Up-regulated in one or two of four samples | ||||
| CCR4-associated factor | 1.0 | 1.1 | 1.5 | 1.5 |
| COR47 | 2.6 | 1.4 | 1.1 | 1.2 |
| EREBP-1 | 1.1 | 1.9 | 2.2 | 1.1 |
| GST1 | 1.1 | 1.3 | 1.7 | 2.2 |
| HSP83 | 1.0 | 1.2 | 1.9 | 2.1 |
| Proline transporter | 1.1 | 1.2 | 1.8 | 2.3 |
| RD22 | 0.9 | 2.3 | 4.1 | 1.4 |
| Group IV: Genes not detected or no changes | ||||
| 18S RNA | 1.0 | 1.0 | 1.0 | 1.1 |
| Auxin-regulated protein | 1.0 | 1.0 | 1.1 | 1.1 |
| GR | 1.2 | 1.3 | 1.4 | 1.4 |
| LOX1 | 1.1 | 1.2 | 1.1 | 1.4 |
| PR2 | 1.0 | 1.0 | 1.3 | 0.7 |
| Peroxidase | NA | NA | NA | NA |
| PR5 | NA | NA | NA | NA |
Ratio was calculated by the hybridization signal detected in transgenic plants divided by that in the null segregants. NA, no hybridization signal was detected in these genes.
Group II included genes that had higher expression levels in three of four transgenic samples than in the negative counterparts. It contains an alcohol dehydrogenase, a blue copper protein, DREB1, an ethylene-responsive element-binding protein (EREBP gene), a glutathione peroxidase, three HSPs (HSP17.4, HSP17.5, and HSP101), genes encoding jasmonic acid-inducible protein, 1-pyrroline-5-carboxylate synthase (P5CS), pyruvate decarboxylase, and a WRKY11 gene.
Group III was comprised of seven genes: CCR4-associated factor, COR47, EREBP-1, GST1, HSP83, proline transporter, and RD22. Genes in this group had higher expression in one or two transgenic samples, compared with their null counterparts. The remaining seven genes (group IV) were either similarly expressed in transgenic and nontransgenic plants or not detected in all of the samples (Table 3). Expression of the housekeeping gene (18S RNA gene) was constant in all samples.
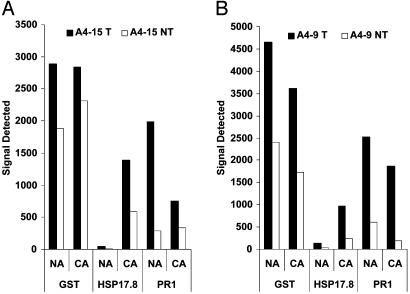
As shown in Table 3, under normal growth conditions, expression level of the GST gene (homolog to Arabidopsis clone gi:15221302) in transgenic plants of event A4-15 and A4-9 was 1.8 and 1.9 times higher than in their null segregants, respectively, indicating that the constitutively expressing NPK1 gene induced the GST gene expression in transgenic maize plants. Cold acclimation, on the other hand, did not affect GST gene expression (Table 3 and Fig. 6). Gene expression levels in the CA-transgenic plants of events A4-15 and A4-9 were 1.7 and 2.1 times higher than their negative counterparts, respectively (Table 3). Similarly, constitutive expression of the NPK1 gene induced the expression of a small HSP (HSP17.8) and the PR1 gene. Under normal growth conditions, the ratios of HSP17.8 gene expression in transgenic A4-15 and A4-9 plants versus their null segregants were 3.4 and 12.6, respectively. The ratios of PR1 gene expression for the same plants were 7.1 and 3.2, respectively.
Fig. 6.
Genes up-regulated by NPK1 overexpression. The array intensity (y axis) was shown for the three group I genes (x axis) in both NA and CA plants. Acclimation treatment was performed at 4°C for 48 h. (A) Event A4-15. (B) Event A4-9. T, transgenic plants; NT, nontransgenic plants.
In group II, nine genes were expressed at higher levels in three of four transgenic samples than were their negative counterparts. For example, the blue copper protein gene was up-regulated in A4-9 under both NA and CA conditions, and in A4-15 under CA conditions. However, this gene showed no enhancement in A4-15 under NA condition. We observed that both transgenic events had enhanced expression for these genes after CA treatment. In addition, the ratios of these up-regulated genes (groups I and II) in event A4-9 were higher than in event A4-15 under both NA and CA treatments. These differences could be attributed to the intrinsic genetic variability between these transgenic events.
To confirm this array result, a quantitative RT-PCR experiment was conducted on the transgene (NPK1), all three group I genes (GST, HSP17.8, and PR1), and two group IV genes (auxin-regulated protein and PR2). The RT-PCR result matched the array result in all of the tested samples with a correlation coefficient of 0.94-0.96 (P < 0.01).
Discussion
Originally a tropical crop, maize is naturally frost-sensitive. We show here, for the first time, to our knowledge, that freezing tolerance in maize can be improved by constitutively expressing the active version of a tobacco MAPKKK gene, NPK1, which is an activator of the oxidative signaling pathway. Two NPK1-transgenic maize events were able to withstand up to 2°C lower freezing temperature compared with their nontransgenic siblings. The 2°C improvement in the freezing tolerance would dramatically minimize yield loss due to frost damage that often occurs in spring and fall seasons, thereby stabilizing the productivity of maize (16).
By using genetic engineering strategies, genes involved in cold acclimation and their transcriptional activators have been introduced into plants to improve freezing tolerance (5-7, 26). Fowler and Thomashow (39) showed that 12% of the cold-responsive genes are members of the CBF regulon and at least 28% of the cold-responsive genes were not regulated by CBF transcription factors, indicating that regulatory pathways other than the CBF regulon are involved in the cold-acclimation process. Alteration of any of these multiple stress regulatory pathways could provide an adaptive response in plants, which could lead to stress protection. In this study, the transgene used for engineering maize, NPK1, is involved in an H2O2 signaling pathway (14). Transgenic maize plants expressing this gene displayed enhanced freezing tolerance. Our data demonstrates that the oxidative signaling pathway is one of the multiple pathways regulating plant response to stress. Prasad et al. (10) observed that the cold-acclimation process created a mild oxidative stress in maize seedlings, which consequently induced chilling tolerance. Pretreating maize seedlings with H2O2 can achieve similar chilling tolerance (10). Our results show that at least two genes (GST and HSP17.8), documented in the oxidative signaling pathway (14), were up-regulated in both of the NPK1-transgenic events studied. This result indicates that a dicotyledonous MAPKKK is able to activate the oxidative signaling pathway in a monocotyledonous plant. This activation, in turn, provided protection to plants from freezing damage. Arabidopsis CBF3 (26) and CBF1 (6) overexpressing lines were able to tolerate temperatures 3.5°C and 3.3°C, respectively, lower than those tolerated by nontransgenic control plants. Our best NPK1-transgenic maize event (A4-15) was able to tolerate 2°C lower temperature than did negative control plants. Considering the fact that maize is a thermophilic crop and Arabidopsis is a winter annual, the freezing tolerance obtained in this study is as significant as that reported for the CBF-transgenic Arabidopsis. In addition, NPK1-transgenic maize did not show severe retardation of growth caused by constitutive expression of transcription activators (7, 40).
MAPK cascades play multiple, essential roles in many cell-signal transduction networks, including defense, response to phytohormones, cell-cycle control, induction of programmed cell death, and responses to diverse stress signals (41). As members of the MAPKKK family, NPK1 and its Arabidopsis orthologs, ANP1/2/3, play critical roles in cytokinesis (42), nuclear localization (43), auxin signaling transduction (44), and oxidative stress signaling pathway (45). Whereas suppression of NPK1 results in abnormal cell division (generating multinucleate cells with incomplete cell plates) (41), overexpression of NPK1 at a high level causes detrimental effects on embryogenesis and seed development (44). In the present study, we did not observe significant differences in vegetative or reproductive development between transgenic maize plants and their nontransgenic segregants. It is likely that our transgenic maize expressed NPK1 gene at a relatively low level. However, a slight reduction in transformation efficiency was observed. The Agrobacterium-mediated transformation efficiency of NPK1 construct (pSHX004) was 3.4%, which was lower than a similar construct, pTF102 (5.5%) (18). In a separate experiment that used particle bombardment for transformation, the efficiency for a construct containing NPK1 gene was 7.1%, whereas that of other similar constructs was ≈10%. The reduced transformation efficiency could be caused by the death of the high NPK1 expressers during the transformation process. Thus, the transformation procedure might have provided selection for healthy transgenic plants.
Soluble sugars serve as cryoprotectants and osmolytes to stabilize membranes and prevent dehydration during freezing (30). Gilmour et al. (26) showed that overexpression of CBF3 mimicked the cold-acclimation process and enhanced the levels of total soluble sugar and proline. In this study, we showed that transgenic plants contained higher levels of sugar than did their nontransgenic segregants. However, the increase in TSC level was not tightly correlated with transgene expression level or freezing tolerance performance. It is likely that the active NPK1 induced some cold-acclimation-like biochemical process that elevated sugar content levels, whereas factors other than the transgene also affected the TSC content in these maize seedlings.
From the array analysis, 50% of the stress-induced genes tested showed no significant increase in the NPK1 maize lines. It is possible that expression levels of the NPK1 gene in these transgenic maize events were too low to up-regulate some of these stress-related genes, even though they may be involved in the oxidative signaling pathway. It is also possible that these genes were only transiently induced, hence their changes could not be readily detected. Kasuga et al. (7) showed that the stress-inducible rd29A promoter could minimize negative effects on the growth of the DREB1A transgenic plants because the transgene was not highly expressed until plants were exposed to dehydration conditions. Under stress conditions, the rd29A promoter was induced and resulted in higher expression of the DREB1A protein, compared with the CaMV 35S promoter construct. Transgenic plants in which DREB1A was driven by the rd29A promoter exhibited greater stress tolerance than did those with the 35S promoter. A similar strategy could be used effectively to manipulate NPK1 in transgenic maize plants. A stress-inducible promoter would enable NPK1 gene expression only in the presence of an environmental stress, thereby allowing us to achieve higher NPK1 gene expression than with a constitutive promoter. This strategy, in turn, could lead to the production of transgenic maize, which can more effectively tolerate environmental stress.
Supplementary Material
Acknowledgments
We thank R. Arora, S. Goggi, A. Knapp, M. Westgate, and S. Whitham for helpful discussions; S. Whitham and B. Frame for critical reading of the manuscript; and J. Anhalt, C. Cordes, T. Fonger, B. Frame, Y. Guo, B. Li, J. C. Martinez, J. McMurray, C. Johnson, D. Sundberg, S. Shurney, and E. Wickham for technical support. This work was supported by National Science Foundation Grant DBI-0077692 (to K.W. and J.S.) and U.S. Department of Agriculture Grant 00-35100-9345 (to J.S.).
Abbreviations: DREB, dehydration-responsive element binding; MAPK, mitogen-activated protein kinase; CBF, cold-inducible master transcription factor; NPK1, Nicotiana PK1; PH, plant height, LN, leaf number; EL, electrolyte leakage; TSC, total soluble carbohydrate; NA, nonacclimated; CA, cold-acclimated; HSP, heat shock protein.
References
- 1.Pastori, G. & Foyer, C. H. (2002) Plant Physiol. 129, 460-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guy, C. L. (1990) Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 187-223. [Google Scholar]
- 3.Thomashow, M. F. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571-599. [DOI] [PubMed] [Google Scholar]
- 4.Thomashow, M. F. (2001) Plant Physiol. 125, 89-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artus, N. N., Uemura, M., Steponkus, P. L., Gilmour, S. J., Lin, C. & Thomashow, M. F. (1996) Proc. Natl. Acad. Sci. USA 93, 13404-13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O. & Thomashow, M. F. (1998) Science 280, 104-106. [DOI] [PubMed] [Google Scholar]
- 7.Kasuga, M., Liu, Q., Miura, S., Shinozaki-Yamaguchi, K. & Shinozaki, K. (1999) Nat. Biotechnol. 17, 287-291. [DOI] [PubMed] [Google Scholar]
- 8.Inzé, D. & Van Montagu, M. (1995) Curr. Opin. Biotechnol. 6, 153-158. [Google Scholar]
- 9.Chamnongpol, S., Willekens, H., Moeder, W., Langebartels, C., Sandermann, H., Van Montagu, M., Inze, D. & Van Camp, W. (1998) Proc. Natl. Acad. Sci. USA 95, 5818-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prasad, T., Anderson, M. D. & Stewart, C. R. (1994) Plant Physiol. 105, 619-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerdnaimongkol, K., Bhatia, A., Joly, R. J. & Woodson, W. R. (1997) J. Am. Soc. Hortic. Sci. 122, 485-490. [Google Scholar]
- 12.Li, H. S., Chang, C. S., Lu, L. S., Liu, C. A., Chan, M. T. & Charng, Y. Y. (2003) Bot. Bull. Acad. Sinica 44, 129-140. [Google Scholar]
- 13.Sebehat, A., Weiss, D. & Lurie, S. (1996) Plant Physiol. 110, 531-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovtun, Y., Chiu, W. L., Tena, G. & Sheen J. (2000) Proc. Natl. Acad. Sci. USA 97, 2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banno, H., Hirano, K., Nakamura, T., Irie, K., Nomoto, S., Matsumoto, K. & Machida, Y. (1993) Mol. Cell. Biol. 13, 4745-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miedema, P. (1982) Adv. Agron. 35, 93-128. [Google Scholar]
- 17.Sheen, J. (1993) EMBO J. 12, 3497-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frame, B. R., Shou, H. X., Chikwamba, R. K., Zhang, Z. Y., Xiang, C. B., Fonger, T. M., Pegg, S. E., Li, B. C., Nettleton, D. S., Pei, D. Q., et al. (2002) Plant Physiol. 129, 13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray, M. G. & Thompson, W. F. (1980) Nucleic Acids Res. 8, 4321-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shou, H. X., Palmer, R. G. & Wang, K. (2002) Plant Mol. Biol. Rep. 20, 325-334. [Google Scholar]
- 21.Dubois, M., Guilles, K. A., Hamilton, J. K., Rebers, P. A. & Smith, F. (1956) Anal. Chem. 28, 350-356. [Google Scholar]
- 22.Yeakley, J. M., Fan, J. B., Doucet, D., Luo, L., Wickham, E., Ye, Z., Chee, M. S. & Fu, X. D. (2002) Nat. Biotechnol. 20, 353-358. [DOI] [PubMed] [Google Scholar]
- 23.Oliphant, A., Barker, D. L., Stuelpnagel, J. R. & Chee, M. S. (2002) BioTechniques 32, S56-S61. [PubMed] [Google Scholar]
- 24.Galinsky, V. L. (2003) Bioinformatics 19, 1832-1836. [DOI] [PubMed] [Google Scholar]
- 25.Armstrong, C. L., Green, C. E. & Phillips, R. L. (1991) Maize Genetics Cooperation News Letter 65, 92-93. [Google Scholar]
- 26.Gilmour, S. J., Sebolt, A. M., Salazar, M. P., Everard, J. D. & Thomashow, M. F. (2000) Plant Physiol. 124, 1854-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pontis, H. G. (1989) J. Plant Physiol. 134, 148-150. [Google Scholar]
- 28.Sakai, A. & Yoshida, S. (1968) Cryobiology 5, 160-174. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida, M., Abe, J., Moriyama, M. & Kuwabara, T. (1998) Plant Physiol. 103, 8-16. [Google Scholar]
- 30.Xin, Z. & Browse, J. (2000) Plant Cell Environ. 23, 893-902. [Google Scholar]
- 31.Walt, D. R. (2000) Science 287, 451-452. [DOI] [PubMed] [Google Scholar]
- 32.Brodersen, P., Petersen, M., Pike, H. M., Olszak, B., Skov, S., Ødum, N., JØrgensen, L. B., Brown, R. E. & Mundy, J. (2002) Genes Dev. 16, 490-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheong, Y. H., Chang, H. S., Gupta, R., Wang, X., Zhu, T. & Luan, S. (2002) Plant Physiol. 129, 661-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desikan, R., Mackerness, S. A. H., Hancock, J. T. & Neill, S. J. (2001) Plant Physiol. 127, 159-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang, X., Rad, U. V. & Durner, J. (2002) Planta 215, 914-923. [DOI] [PubMed] [Google Scholar]
- 36.Ishitani, M., Xiong, L. M., Lee, H. J., Stevenson, B. & Zhu, J. K. (1998) Plant Cell 10, 1151-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, J. & Zhu, J. K. (1997) Plant Physiol. 114, 591-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vranova, E., Atichartpongkul, S., Villarroel, R., Van Montagu, M., Inzé,D. & Van Camp, W. (2002) Proc. Natl. Acad. Sci. USA 99, 10870-10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fowler, S. & Thomashow, M. F. (2002) Plant Cell 14, 1675-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinozaaki, K. & Yamaguchi-Shinozaki, K. (2000) Curr. Opin. Plant Biol. 3, 217-223. [PubMed] [Google Scholar]
- 41.Samuel, M. A. & Ellis, B. E. (2002) Plant Cell 14, 2059-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishihama, R., Ishikawa, M., Araki, S., Sayano, T., Asada, T. & Machida, Y. (2001) Genes Dev. 15, 352-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishikawa, M., Soyano, T., Nishihama, R. & Machida, Y. (2002) Plant J. 32, 789-798. [DOI] [PubMed] [Google Scholar]
- 44.Kovtun, Y., Chiu, W. L., Zeng, W. & Sheen, J. (1998) Nature 395, 716-720. [DOI] [PubMed] [Google Scholar]
- 45.Odell, J. T., Nagy, F. & Chua, N. H. (1985) Nature 6, 810-812. [DOI] [PubMed] [Google Scholar]
- 46.Carrington, J. C. & Freed, D. D. (1990) J. Virol. 64, 1590-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Depicker, A., Stachel, S., Dhaese, P., Zambryski, P. & Goodman, H. M. (1982) J. Mol. Appl. Genet. 1, 561-573. [PubMed] [Google Scholar]
- 48.Mason, H. S., DeWald, D. & Mullet, J. E. (1993) Plant Cell 5, 241-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.