Abstract
Light is one of the most important environmental factors regulating expression of photosynthesis genes. The plastid psbD gene encoding the photosystem II reaction center protein D2 is under the control of a unique blue light responsive promoter (BLRP) that is transcribed by a bacterial-type plastid RNA polymerase (PEP). Promoter recognition of PEP is mediated by one of the six nuclear-encoded σ factors in Arabidopsis. The replacement of the plastid σ factor associated with PEP may be the major mechanism for switching of plastid transcription pattern in response to environmental and developmental signals. This study demonstrates that AtSig5 is a unique σ factor that is essential for psbD BLRP activity. A T-DNA insertional mutant with reduced AtSIG5 expression resulted in loss of primary transcripts from the psbD BLRP. Furthermore, transient overexpression of AtSig5 in dark-adapted protoplasts specifically elevated psbD and psbA transcription activities. On the other hand, overproduction of AtSig2 enhanced the transcription of psbA gene and trnE operon, but not psbD transcription. The AtSIG5 gene is phylogenetically distinct from other plastid σ factors, and its expression is induced exclusively by blue light. We propose that AtSig5 acts as a mediator of blue light signaling that specifically activates the psbD BLRP in response to blue light in Arabidopsis.
Chloroplasts use light not only as an energy source for photosynthesis, but also as an environmental signal to regulate biogenesis and environmental adaptation of photosynthesis apparatus. The photosynthesis apparatus of higher plants comprises plastome and nuclear-encoded components. The chloroplast genome encodes ≈40 photosynthesis-related genes and ≈60 housekeeping genes essential for protein synthesis (1, 2). Transcription activity of the majority of chloroplast-encoded photosynthesis genes increases to support rapid construction of the photosynthesis apparatus at an early stage of light-induced chloroplast development, followed by a decline of overall transcription activity in mature chloroplasts. However, even in illuminated mature chloroplasts, psbA and psbD genes, which, respectively, encode D1 and D2 proteins of the photosystem II reaction center core complex, maintain high transcriptional activity differentially (3, 4). It is inferred that light-dependent transcription of psbA and psbD genes supports the light-induced rapid turnover of D1 and D2 proteins in mature chloroplasts to avoid photo inhibition.
A unique blue light responsive promoter (psbD BLRP) mediates light-dependent psbD transcription in mature chloroplasts. The psbD BLRP is activated specifically by high-fluence blue and UV-A light (3, 5). It is also regulated by an endogenous circadian rhythm (6). It is assumed that nuclear-encoded transcription regulators control chloroplast transcription, because no genes encoded by higher plant chloroplasts are presumed to be related to transcriptional regulation. Although it has been demonstrated that light-dependent psbD transcription depends on nuclear-localized cryptochromes (7), little is known about transcription regulators that mediate the blue light signaling.
Chloroplasts contain two types of RNA polymerase. Photosynthesis genes including psbD are transcribed mainly by a bacterial-type chloroplast RNA polymerase named PEP (plastid-encoded plastid RNA polymerase) (8, 9). Another phage-type RNA polymerase named NEP (nuclear-encoded plastid RNA polymerase) is responsible for transcription of housekeeping genes (9, 10). Subunit composition of PEP is almost identical to that of Escherichia coli RNA polymerase (α2ββ′σ) (11). Promoter-specific transcription initiation is conferred by exchangeable σ factors in bacteria. Although PEP core subunits (α, β, β′, β″) are encoded by chloroplast genes, plastid σ factor genes are encoded by the nuclear genome (12-14). In Arabidopsis thaliana, six distinct plastid sigma genes have been cloned and designated as AtSIG1—AtSIG6 (15-17). These proteins are homologous to bacterial group 1 and 2 σ factors and assumed to be targeted to chloroplasts by N-terminal transit peptides (16-18). In general, the bacterial genome encodes multiple σ factors, and heterogeneity of factors is responsible for transcriptional activation of different sets of genes in response to environmental clues (19). In analogy with bacteria, it has been presumed that each plastid σ factor may recognize a specific set of promoters. Replacement of the σ factor may be the major mechanism for switching of the transcription pattern of plastids in response to developmental and environmental signals (20, 21).
Most chloroplast promoters recognized by PEP contain a variant of the -10 (TATAAT) and -35 (TTGACA) consensus sequences of canonical σ70-type E. coli promoters (2, 13, 22). On the other hand, the psbD BLRP is an unusual PEP promoter that lacks a functional -35 element but contains a well-conserved upstream cis element, termed the AAG box (9). The unique promoter structure of the psbD BLRP is responsible for light-dependent switching of its activity. Thus, we inferred that the psbD BLRP must be recognized by a special σ factor. In Arabidopsis, AtSIG5 is structurally distinct from other factors (17). Previously, we found that expression of AtSIG5 transcripts was specifically induced by blue light in a similar manner as the blue light-induced activation of psbD BLRP (23). Therefore, we speculated that AtSig5 might be a specialized σ factor that specifically recognizes the psbD BLRP. The present study identifies a homozygous T-DNA insertional mutant for AtSIG5. In the mutants, the AtSIG5 transcript level was greatly declined, and primary transcripts from the psbD BLRP were completely diminished. Furthermore, transient overproduction of AtSig5 in dark-adapted protoplasts resulted in specific activation of psbD and psbA transcription. These results demonstrate that Sig5 is a unique σ factor that recognizes psbD BLRP specifically and mediates blue light signaling.
Materials and Methods
Plant Materials and Growth Conditions. A. thaliana ecotype Columbia was used as a control in all experiments. The sig5.1 mutant of Arabidopsis ecotype Columbia was identified in a collection of T-DNA insertion lines generated at The Salk Institute (San Diego). Surface sterilized seeds were sown on 0.5× Murashige and Skoog medium (24) and placed at 4°C for 2-3 days followed by germination under long-day conditions (16 h light, 8 h dark) at 22°C. Seedlings were then transferred to Jiffy-7 (Sakata, Yokohama, Japan) and grown under the same conditions. For the selection of T-DNA-induced kanamycin resistance, kanamycin was added to the medium to a final concentration of 50 mg/liter. WT plants used for the protoplast transient assay were grown under continuous white light (10-20 μmol·m-2·s-1) at 22°C for 4 wk. Photon fluence rates were measured with a quantum photometer (LI-250; Li-Cor, Lincoln, NE).
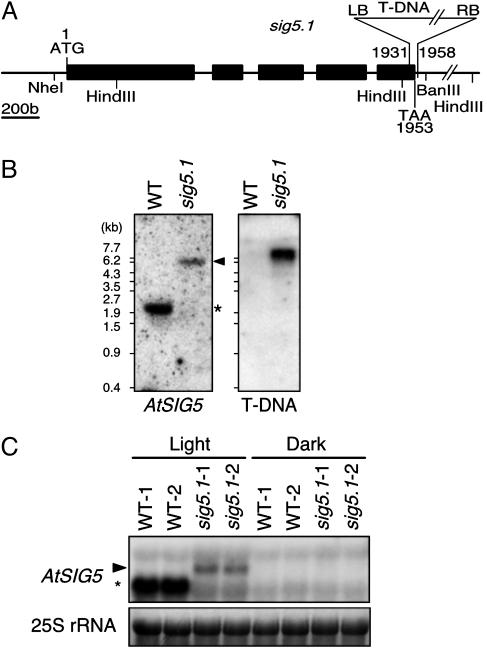
Identification of the T-DNA Insertion Allele of the AtSIG5 Gene. Putative mutants at the T3 generation were subjected to PCR amplification of the NPTII gene using the primers PROK2-1 (5′-GATATCGGCACCTTCGACCGCCTG-3′) and PROK2-2 (5′-GATGGATTGCACGCAGGTTCTCCGGG-3′). The presence of the insertional allele was confirmed by screening the PCR products generated by AtSIG5-specific primers SE-1 (5′-ATGGGAGTTGTGTCTATTTCAAGTTC-3′) and SE-1081 (5′-TCTCCACTCTAACCGATTCAAGTCC-3′). Identified lines were homozygous for the T-DNA insertion; the mutant allele was named sig5.1. The copy number of the insertion allele was determined by restriction digestion with NheI/BanIII or HindIII followed by DNA gel blot analysis.
Northern Blot Analysis. Total RNAs were extracted from seedlings or rosette leaves of Arabidopsis with a RNeasy Plant Mini Kit (Qiagen, Chatsworth, CA). Northern blots containing 10 or 2 μg of total RNA from WT and sig5.1 plants were hybridized in Rapid-Hyb buffer (Amersham Pharmacia Biosciences) at 65°C to random primed [32P] cDNA probes encoding AtSIG5, AtSIG2, psbA, rbcL, trnE-Y-D, and 25S rRNA. The CDR (coding region) probe (+78 to + 1002 of the psbD translational start codon) was designed to detect transcripts produced from all multiple promoters in the psbD/C operon, and the UTR probe (-1085 to -726) to detect specifically the transcripts from the psbD BLRP. Final wash conditions were 0.2× SSC, 0.1% SDS at 65°C for 30 min.
Transient Expression of σ Factors in Mesophyll Protoplasts. cDNAs of SIG genes were amplified by PCR from the Arabidopsis cDNA library based on their nucleotide sequence data taken from GenBank (accession nos. AB019943 and AB021120 for AtSIG2 and AtSIG5, respectively) (15, 17). Primers used were: S2-SalI (5′-AGAGTCGACATGTCTTCTTGTCTTCTTCCTCAG-3′) and S2-NotI (5′-AGAGCGGCCGCATTATGATTGTGCAACCAAGTAT-3′) for AtSIG2; S5-NcoI (5′-AATCCATGGGAGTTGTGTCTATTTCAAGTTCAGC-3′) and S5-NotI (5′-AAAGCGGCCGCTTAGACGATGTATTGACGAAGGTA-3′) for AtSIG5 amplifications. The amplified fragments were subcloned into the pTM vector, which is a pUC18-based plasmid that contains the 35S cauliflower mosaic virus promoter and the nopaline synthase polyadenylation region.
Protoplasts were prepared basically according to Abel and Theologis' method (25): 10 g of rosette leaves was vacuum-infiltrated in 200 ml of protoplasting solution containing 1% Cellulase Onozuka RS (Yakult, Tokyo), 0.25% Macerozyme R-10 (Yakult), 400 mM mannitol, 8 mM CaCl2, and 5 mM Mes-KOH (pH 5.6) for 2 min at 15 mmHg and incubated in the dark at 22°C for 3 h under gentle agitation (40 rpm). The resulting protoplast suspension was filtrated through a 140-μm nylon mesh, washed several times, and finally suspended in W5 solution containing 154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, and 1.5 mM Mes-KOH (pH 5.6). After counting in a hematocytometer, protoplasts were suspended in MaMg solution containing 400 mM mannitol, 15 mM MgCl2, and 5 mM Mes-KOH (pH 5.6) to a density of 5 × 106 protoplasts per ml just before transfection. Then 250 μg of plasmid DNAs and 250 μg of salmon sperm carrier DNA were added to 1.5-ml suspension (7.5 × 106 protoplasts) and mixed well, followed by addition of 1.75 ml of PEG-CMS solution containing 400 mM mannitol, 100 mM Ca(NO3)2, and 40% PEG 4000. The transfection mixture was incubated at room temperature for 30 min. The transfection mixture was diluted with 40 ml of W5 solution, washed with 400 mM mannitol/W5 solution (4:1), and resuspended in 15 ml of protoplast culture medium (pH 5.8) containing 135 g·liter-1 sucrose, 4.3 g·liter-1 Murashige and Skoog salts (GIBCO), 100 mg·liter-1 inositol, 250 mg·liter-1 xylose, 460 mg·liter-1 CaCl2, 10 mg·liter-1 thiamin, 1 mg·liter-1 pyridoxine, and 1 mg·liter-1 nicotinic acid. After incubation in dark conditions at 22°C for 16 h, the protoplasts were collected and resuspended into a suspension buffer containing 300 mM sorbitol, 2 mM EDTA, and 50 mM Hepes-NaOH (pH 7.6) to a density of 2.5 mg chlorophyll per ml.
Run-On Transcription Assay. Twenty-five microliters of the transformed protoplast suspension was added to 160 μl of the reaction mixture containing 10 mM MgCl2, 40 mM KCl, 13.8 mM Hepes-NaOH (pH 7.9), 5 mM ATP, 5 mM CTP, 5 mM GTP, 0.5 mM UTP, 100 μCi [α-32P] UTP, and 1 mg·ml-1 heparin. The protoplasts were disrupted by pipeting the reaction mixture gently up and down >20 times. Then, the solution was incubated for 10 min at 25°C, and the reaction was stopped by adding 20 μl of a lysis buffer containing 50 mg·ml-1 SDS, 50 mM Tris·HCl (pH 8.0), and 25 mM EDTA. In some experiments, 500 μg·ml-1 α-Amanitin (Wako Biochemicals, Osaka) was added to the reaction mixture. Run-on transcripts were extracted twice with phenol/chloroform/isoamyl alcohol (25:25:1) and hybridized with 300 ng of probes blotted onto a Hybond N+ membrane (Amersham Pharmacia Biosciences). PCR fragments, with an average size of 900 nt, covering psbA, psbB, psbD, psbE-F-L-J, ndhF, rpoA, rpoB, rbcL, atpB, trnE-Y-D, rrn16S, and AtSIG1 of Arabidopsis; and MspI-digested pBR322, pUC18, and λ DNA were used as run-on probes. Blots were hybridized with purified run-on transcripts and autoradiographed after washing. The images obtained by BAS2000 (Fuji Photo Film) were processed by using the science lab image-analyzing program (Fuji Photo Film).
Results
Identification of a T-DNA Insertional Mutant Line Deficient for AtSIG5. To elucidate Sig5 function, an insertional mutant was identified in the T-DNA insertion collection generated at The Salk Institute (26). The sig5.1 mutant (SALK049021) contains a T-DNA insertion in the last exon of AtSIG5 (Fig. 1A). DNA gel blot analyses revealed the presence of only one T-DNA insertion site in the sig5.1 line (Fig. 1B). It was possible to identify homozygous sig5 mutant lines based on the genomic PCR (data not shown). Nucleotide sequencing of the T-DNA::genomic borders revealed that the T-DNA insertion site is 1,931 bp downstream of the translation start site and adjacent to the conserved region 4.2, which is required for recognition of the -35 promoter sequence in bacterial group 1 and 2 σ factors (27, 28). To ensure that the T-DNA insertion reduced AtSIG5 gene expression, RNA gel blot analyses were carried out with the AtSIG5 cDNA as a probe. As shown in Fig. 1C, a 1.6-kb AtSIG5 transcript, which is likely to be the mature mRNA, accumulated in a light-dependent manner in WT plants. In contrast, the endogenous AtSIG5 transcript was undetectable in the illuminated homozygous sig5 mutants, although a slight accumulation (≈5%) of an aberrant transcript (2.9 kb) was observed. It is supposed that the 2.9-kb aberrant transcript was destabilized as a result of the loss of endogenous 3′ UTR. Thus, it is expected that the functional AtSig5 protein was decreased greatly and truncated protein, if any, was accumulated faintly in the sig5.1 mutants.
Fig. 1.
Characterization of a T-DNA insertion mutant of AtSIG5. A homozygous AtSIG5::T-DNA (sig5.1) line was established. (A) The exon-intron structure of the AtSIG5 gene and the position of T-DNA insertion in sig5.1 allele. (B) Southern blot analysis of genomic DNA of WT and sig5.1 plants digested with NheI/BanIII or HindIII and fractionated by electrophoresis in a 1.0% agarose gel; blots were hybridized with AtSIG5 gene probe or NPTII gene probe (T-DNA), respectively. DNA marker sizes are shown at left. * marks the WT AtSIG5 band, and the arrowhead indicates the hybridizing band corresponding to the sig5.1 insertional allele. (C) Northern blot analysis of AtSIG5 transcript levels in total RNA (10 μg per lane) isolated from dark-adapted (Dark) and reilluminated (Light) leaves of two independent plants of WT (WT-1 and WT-2) and two of the sig5.1 line (sig5.1-1 and sig5.1-2). The ethidium bromide-stained 25S rRNAs are gel-loading references. * indicates the endogenous AtSIG5 transcript.
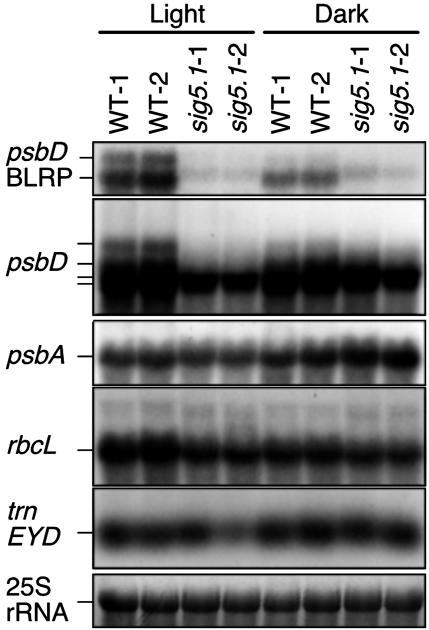
AtSIG5 Deficiency Results in Loss of the psbD BLRP Activity. The homozygous sig5.1 mutant showed no phenotypic alterations in standard growth conditions. To elucidate a role of Sig5 on light-dependent activation of the psbD BLRP, we analyzed the effect of AtSIG5 deficiency on expression levels of several chloroplast-encoded transcripts in response to light irradiation. WT and homozygous sig5.1 mutant plants were grown under white light with a 16-h light, 8-h dark cycle for 21 d and dark-adapted for 24 h followed by 3-h white light illumination. Total RNAs were prepared from rosette leaves, and RNA blot analyses were carried out. Fig. 2 shows that the psbD coding region probe (see Materials and Methods) detected four bands (4.5, 3.7, 2.8, and 2.6 kb) in illuminated leaves of WT plants. Among them, the 4.5- and 3.7-kb transcripts were detected by the psbD UTR probe. They were induced by light in WT plants, indicating that these two transcripts (psbD BLRP transcripts) originated from the psbD BLRP. In sig5.1 mutant plants, we found that AtSIG5 deficiency caused a severe reduction of psbD BLRP transcript accumulation (8% of the WT level) in response to 3-h light irradiation (Fig. 2). On the other hand, accumulation of 2.8- and 2.6-kb psbD transcripts were somewhat decreased but not greatly in sig5.1 mutants, suggesting that these transcripts might arise from additional promoters or might be produced by RNA processing that generates the extraordinary stable transcripts. The transcript levels of psbA and rbcL were decreased faintly in reilluminated sig5.1 mutant plants. Previously, it was suggested that AtSIG2 recognizes certain plastid tRNA promoters (29). Contrary to sig2 mutant, the expression of trnE operon was unchanged in the sig5.1 mutant. These results indicate that Sig5 is specifically required for activation of psbD BLRP in illuminated plants.
Fig. 2.
Northern blot analysis of transcript levels of various genes in total RNA (2 μg per lane) isolated from leaves of dark-adapted (Dark) and reilluminated (Light) plants. RNA samples were obtained from leaves of two independent plants of WT (WT-1 and WT-2) and two of the mutant sig5.1 line (sig5.1-1 and sig5.1-2). Transcript of trnE operon was reduced in sig5.1-2 sample because of unequal loading.
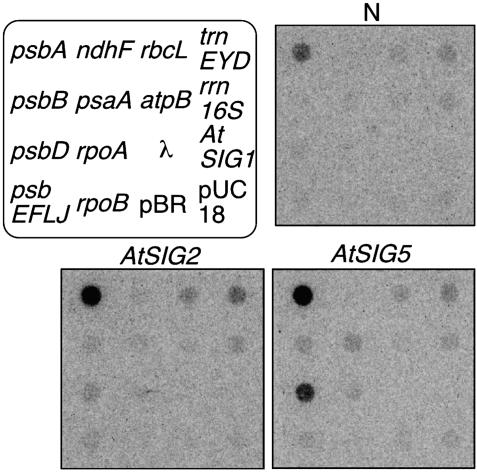
Transient Expression of AtSig5 Results in Activation of psbD BLRP Activity in Dark-Adapted Protoplasts. It is expected that overexpression of Sig5 would result in light-independent activation of psbD BLRP because psbD BLRP activity requires AtSIG5 in illuminated plants. To confirm this possibility, we transformed the dark-adapted protoplasts prepared from Arabidopsis with the expression vector pTM-AtSIG2 and AtSIG5 to transiently express each σ factor under the control of cauliflower mosaic virus 35S promoter. After transformation, cells were incubated for 16 h in the dark at 22°C and submitted to transcriptional run-on assays. Run-on transcripts in each of the protoplast preparations were hybridized to dot blots containing DNA fragments specific for plastid-encoded genes including psbD. Nontransformed protoplasts (N) and protoplasts transformed with the plasmid expressing GFP instead of σ factors, pTM-gfp (data not shown), were used for control experiments. Fig. 3 shows a typical result comparing transcription patterns obtained with two differently treated protoplasts and nontransformed protoplasts. Table 1 summarizes average values of several independent experiments. In nontransformed dark-adapted protoplasts (N), psbA was actively transcribed compared to other plastid-encoded genes examined. Transient expression of GFP (pTM-gfp) in the dark-adapted protoplasts did not enhance the transcription activities of any genes (data not shown). On the other hand, as expected, the transient expression of AtSig5 drastically activated the psbD transcription in the dark-adapted protoplasts. Transient expression of AtSig5 also caused significant activation of psbA transcription and slight enhancement of psaA and psbB transcription. Transcription of other genes including a nuclear-encoded control gene (AtSig1) was not significantly activated by AtSig5. Because the selective activation of psbA and psbD transcription was observed in the presence of α-Amanitin that specifically inhibits transcription elongation of nuclear RNA polymerase II (data not shown), it is unlikely that the nuclear RNA polymerase II is directly involved in the transcription of psbD and psbA genes in the lysed protoplasts. On the other hand, the transient expression of AtSig2 specifically activated transcription of trnEYD operon and psbA in the dark-adapted protoplasts. These results concur with previous findings that AtSIG2 deficiency reduced the accumulation of some plastid-encoded tRNAs (29) and psbA transcripts (30).
Fig. 3.
Autoradiograms representing run-on transcription activities of plastid-encoded genes in dark-adapted protoplasts to overexpress AtSig2 or AtSig5. α-32P-labeled run-on transcripts derived from 25 μl of protoplast suspension (a density of 2.5 mg chlorophyll per ml) were hybridized to dot blots containing DNA fragments specific for plastid-encoded genes including psaA, psbA, psbD, psbE-F-L-J, ndhF, rbcL, atpB, rpoA, rpoB, trnE-Y-D, and rrn16S genes that encode P700 apoprotein A1 of PSI, D1, D2, CP47, small subunit proteins of PSII, NADH dehydrogenase NDS, the large subunit of riblose-1,5-bisphosphate carboxylase/oxygenase, α-subunit of ATPase, α,β-subunit of PEP, the set of transfer RNAs, and 16S ribosomal RNA, respectively. To detect transformed vector sequences, pUC18 vector DNA (pUC) was used as a control. MspI-digested pBR322 DNA fragments (pBR) and lambda DNA (λ) probes indicated that there were no nonspecific transcripts. Because no transcription signal was detected from AtSig1 probe, nuclear RNA polymerase is unlikely involved in the protoplast run-on transcription activity.
Table 1. Quantitation of plastid gene transcription in AtSig2 and AtSig5-expressing protoplasts.
| Gene | N | AtSIG2 | AtSIG5 |
|---|---|---|---|
| psbA | 100.0 [5] | 299.9 ± 31.1 [4] | 760.8 ± 448.6 [4] |
| psbB | 38.9 ± 12.3 [5] | 55.2 ± 13.6 [4] | 133.9 ± 47.8 [4] |
| psbD | 54.4 ± 8.9 [5] | 73.4 ± 9.4 [4] | 498.0 ± 292.0 [4] |
| psbEFLJ | 43.7 ± 14.0 [4] | 60.4 ± 15.1 [3] | 53.0 ± 16.1 [3] |
| psaA | 40.4 ± 12.2 [5] | 49.5 ± 14.2 [4] | 117.5 ± 42.0 [4] |
| rbcL | 46.3 ± 10.9 [5] | 64.8 ± 13.2 [4] | 65.4 ± 10.0 [4] |
| atpB | 39.1 ± 14.5 [4] | 54.8 ± 15.1 [3] | 50.2 ± 15.8 [3] |
| ndhF | 39.4 ± 14.6 [4] | 49.6 ± 18.7 [3] | 49.8 ± 15.9 [3] |
| rpoA | 39.9 ± 14.8 [4] | 54.4 ± 17.3 [3] | 59.3 ± 13.8 [3] |
| rpoB | 36.0 ± 13.3 [5] | 41.6 ± 15.4 [4] | 44.5 ± 13.8 [4] |
| trnEYD | 50.1 ± 11.7 [4] | 109.2 ± 11.0 [3] | 71.3 ± 13.4 [3] |
| rrn16S | 39.7 ± 12.7 [5] | 51.5 ± 15.5 [4] | 49.4 ± 14.2 [4] |
| AtSIG1 | 34.6 ± 13.6 [5] | 37.6 ± 16.7 [4] | 41.8 ± 14.4 [4] |
The amount of radioactivity hybridized was quantified by using the Science Lab Image Gauge and given as photo-stimulated luminescence per square millimeter (psl/mm2). Each value represents the transcription activity of genes relative to that of psbA in nontransformed protoplast lysates (%). The data represent mean values of three to five independent experiments (mean ± SD). Numbers in brackets represent the number of assay repetitions.
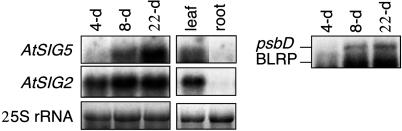
Tissue-Specific and Developmental Regulation of AtSIG5 Gene Expression. Expression of AtSIG5 gene was examined in WT plants at different ages. As shown in Fig. 4, transcripts of AtSIG5 were highly accumulated in leaves of 8-d-old and 22-d-old plants, but not in 4-d-old plants. Similarly, the accumulation of psbD BLRP transcripts also depended on seedling growth. On the other hand, AtSIG2 transcript was expressed constitutively during seedling growth. Furthermore, AtSIG5 and AtSIG2 were expressed highly in rosetta leaves and faintly in roots. These results indicate that AtSIG5 expression correlates with development of functional chloroplasts.
Fig. 4.
Northern blot analysis of the transcript levels of AtSIG2 and AtSIG5 and in total RNA (10 μg per lane) isolated from leaves of WT plants at different ages and roots. RNA samples were isolated from leaves of 4-d-, 8-d-, and 22-d-old plants. Primary transcripts from the psbD BLRP were also shown. Ethidium bromide-stained 25S rRNAs are gel-loading references.
Discussion
AtSig5 Is Required for psbD BLRP Activity. We took a reverse genetic approach using a T-DNA insertional mutant of Arabidopsis to test directly the possibility that nuclear-encoded Sig5 mediates blue light signaling that regulates chloroplast psbD BLRP activity. We identified a homozygous allele of sig5.1 that contains a T-DNA insertion 22 bp upstream of the AtSIG5 stop codon. This mutant was expected to produce a truncated AtSig5 protein that lacks 7 aa of its C terminus. However, the homozygous sig5.1 mutants showed no detectable accumulation of AtSIG5 endogenous transcript. Although the 2.9-kb aberrant transcript was slightly induced by light, its level was <5% of the endogenous AtSIG5 transcript level in WT plants, suggesting that the 2.9-kb aberrant transcript was destabilized by the loss of endogenous 3′ UTR. Presumably, the functional AtSig5 protein is greatly deficient in the homozygous sig5.1 mutants. As was expected, the light-induced accumulation of 4.5- and 3.7-kb psbD transcripts completely disappeared in homozygous sig5.1 plants, presenting unambiguous evidence supporting our hypothesis that AtSig5 is involved in light-induced activation of psbD BLRP. On the other hand, transcripts of other plastid-encoded genes accumulated nearly equivalent to WT levels in the mutant plants, although psbA and rbcL transcripts were slightly decreased. In addition, transient expression of the AtSIG5 gene in dark-adapted protoplasts resulted in exclusive enhancement of psbD and psbA transcription activities and slight elevation of psaA and psbB transcription activities. Expression of other σ factor genes could not activate psbD transcription in the dark-adapted protoplasts (data not shown). Furthermore, AtSIG5 expression depended on chloroplast development similarly to psbD BLRP activity. Taken together, we concluded that AtSig5 represents a unique σ factor that specifically recognizes psbD BLRP.
Recently, Yao et al. (31) reported on two other AtSIG5 mutant alleles generated by T-DNA insertion and designated them sig5-1 and sig5-2. These mutants showed embryonic lethality. Because T-DNA insertions are located at exons 5 and 2 of the AtSIG5 gene in these mutants, respectively, it is presumed that these mutants would produce truncated proteins lacking functionally important domains. The authors speculate that Sig5 may play an important role in plant reproduction in addition to transcriptional regulation in chloroplasts. On the other hand, we have identified a homozygous progeny line with T-DNA insertion at the last exon of AtSIG5 gene (sig5.1) and demonstrated that the psbD BLRP was completely inactivated in this mutant plant. We speculate that AtSig5 serves dual functions: one as a specific transcription initiation factor for the psbD BLRP, and the other as an essential factor for plant reproduction. The sig5.1 mutants may synthesize a faint amount of functional Sig5 protein that retains important domains required for plant reproduction, but sig5-1 and sig5-2 could not. In fact, AtSIG5 signal with altered size was detectable in Northern blots (Fig. 1C), suggesting that our sig5.1 line produced a trace of the truncated AtSig5 protein. Crossing between sig5-1/-2 and our sig5.1 lines would give further insight into additional function of Sig5.
AtSig5 Acts as a Specialized σ Factor in Plastids. In bacteria, the σ70 family is classified into three subgroups including primary, primary-like, and alternative σ factors. It is presumed that plastid σ factors also would be classified into functionally distinct subgroups. The most characterized plastid σ factor to date is AtSig2. Deficiency of AtSIG2 resulted in reduced accumulation of several tRNAs including trnE in Arabidopsis (29, 30). Furthermore, we demonstrated that overproduction of AtSig2 enhanced transcription of psbA and trnE operon. Because these genes are preceded by typical σ70-type promoters, we cannot exclude the possibility that AtSig2 also recognizes other σ70-type PEP promoters. In fact, hybrid protein of E. coli σ70 with a C-terminal fragment of AtSig2 containing regions 1.2 through 4.2 could complement E. coli rpoD mutants (32), indicating that AtSig2 could recognize σ70-type standard promoters in E. coli.
On the other hand, this work demonstrated that AtSig5 is specifically required for psbD BLRP activity. AtSig5 is also involved in the transcription of psbA, and possibly psaA and psbB genes. It should be noted that other σ factors including AtSig2 and AtSig6 likely recognizes these PEP promoters redundantly except for psbD BLRP. In vitro analysis of AtSig5 transcription would provide further insight into the promoter specificity of AtSig5.
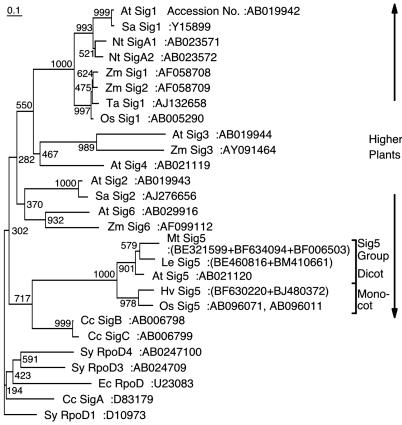
Taken together, it was suggested that AtSig5 might act as a specialized σ factor in chloroplasts. Multiple cDNA clones show close similarity to AtSIG5 sequences in various angiosperms including Lycopersicon esculentum, Medicago truncatula, and Hordeum vulgare. We also determined the complete cDNA sequence of SIG5 homolog in Oryza sativa (GenBank accession no. AB096071). Phylogenetic analyses revealed that AtSIG5 homologs form their own branch (Fig. 5) (14, 17). Furthermore, intron sites of AtSIG5 are distinct among plastid σ factor genes in Arabidopsis (17). It has been inferred that multiple plastid σ factor genes were the product of gene duplication events from one or a few origins during plant evolution. It is likely that the Sig5 family has evolved from ancestral plastid σ factor(s) at an early stage of plant evolution. It should be noted that the psbD BLRP is conserved in all plastid genomes of higher plants including monocots, dicots, and black pine (33), but not in liverwort Marchantia polymorpha (34). Detailed analysis of σ factor genes in moss may provide a novel insight into plastid σ factor evolution.
Fig. 5.
Phylogenetic analysis of two σ factors from E. coli (Ec), three from cyanobacteria Synechococcus PCC7942 (Sy), three from red algae Cyanidium caldarium (Cc), six from A. thaliana (At), two from O. sativa (Os), two from Sinapis alba (Sa), two from Nicotiana tabacum (Nt), four from Zea mays (Za) and putative Sig5 proteins of M. truncatula (Mt), L. esculentum (Le), and H. vulgare (Hv). Only the highly conserved amino acid residues containing region 1.2-2.3 were chosen for calculation of evolutionary distances by the clustal-w multiple sequence alignment program (version 1.8) using the blosum matrix method. The unrooted tree was constructed from the distance data by the neighbor-joining method. The scale bar represents 10% substitutions per amino acid position. Bootstrap analysis was performed by resampling the data set 1,000 times. Numbers at nodes indicate bootstrap values.
AtSig5 Mediates Blue Light Signaling. The psbD BLRP is an unusual promoter in plastids in terms of specific activation by high-fluence blue and UV-A light (3, 5). Distinct from standard σ70-type PEP promoters, psbD BLRP activity basically depends on transcription-enhancing sequences, termed the AAG-box, that are located immediately upstream of the -35 sequence (9). Several proteins are known to bind to the AAG-box and enhance the psbD transcription; they are named AGF (AAG-box factor) (6, 35). Considerable debate has arisen regarding molecular mechanisms mediating light regulation of the psbD BLRP. Nuclear/cytoplasm-localized cryptochromes are involved in blue light-specific activation of the psbD BLRP (7). Thus, it has been inferred that blue light signaling to activate specifically the psbD BLRP is mediated by a nuclear-encoded transcription factor, such as AGF and σ factors. Recently, PTF-1 was cloned as an AAG-box binding factor in Arabidopsis (36). However, PTF-1 is unlikely to be involved in blue light signaling because light-dependent psbD transcription was not diminished in PTF-1-deficient mutants. Another candidate would be the specific σ factor that recognizes the psbD BLRP. The psbD BLRP contains the less conserved -35 element. Extensive in vivo and in vitro mutation analyses revealed that light-dependent transcription from the psbD BLRP depends on the -10, but not the -35, element (6, 37, 38). The present report demonstrates that AtSig5 is a specialized σ factor that is required for light-dependent activation of the psbD BLRP. It should be noted that region 4.2 of AtSig5 that is involved in the recognition of -35 element is most poorly conserved with that of E. coli σ70 among plastid σ factors (17). We demonstrated previously that tip-type PEP in mature chloroplasts could recognize an extended -10 promoter and did not require -35 element for transcription initiation at the psbA promoter in wheat (39). Interestingly, psbA transcription was also enhanced in AtSig5-overexpressing protoplasts.
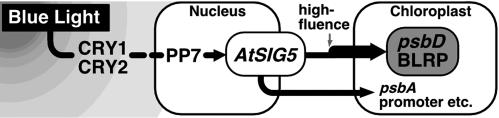
AtSig5 is the only σ factor of plastids that is specifically induced by blue light illumination (23). It was demonstrated previously that cryptochromes mediate blue light-dependent activation of psbD BLRP (7). Furthermore, we found that cryptochromes are responsible for the blue light-dependent expression of AtSig5 transcripts (T. Mochizuki, Y. Onda, Y. Tsunoyama, T.S., M. Wada, and Y. Toyoshima, unpublished data). Recently a novel Ser/Thr protein phosphatase (PP7) was shown to mediate cryptochrome signaling in nuclei (40). The blue light-dependent accumulation of AtSIG5 transcripts was diminished in PP7-deficient mutants, suggesting that PP7 functions upstream of the blue light-induced expression of AtSIG5. Taken together, we speculate that blue light signals absorbed by cryptochromes induce expression of nuclear-encoded Sig5 via PP7 as a positive regulator. Subsequently, Sig5 transported to chloroplasts specifically recognizes the psbD BLRP and the psbA promoter to initiate light-dependent transcription (Fig. 6). Thus, Sig5 likely functions as a unique σ factor, which communicates blue light signaling between nuclei and chloroplasts. Interestingly, overexpression of AtSig5 simultaneously elevated the transcription activities of psbA and psbD genes encoding D1 and D2 proteins of photosystem II, respectively. They are known to undergo light-dependent rapid turnover.
Fig. 6.
A model for the action of σ factor Sig5 in the light-signaling pathway regulating the psbD BLRP. High-fluence light enhances psbD transcription (23).
Acknowledgments
We thank Dr. Y. Isozumi for providing the facilities of the Radioisotope Research Center. This work was funded by the New Energy and Industrial Technology Development Organization. This work was also supported by Grants-in-Aids for Scientific Research (Grant 14540598 to T.S.) and for Young Scientists (Grant 200203019 to Y.N.) and the Foundation for Bio-venture Research Center from the Japanese Ministry of Education, Culture, Sports, Science, and Technology. Y.N. is a Research Fellow of the Japan Society for the Promotion of Science.
Abbreviations: BLRP, blue light responsive promoter; PEP, plastid-encoded plastid RNA polymerase.
References
- 1.Palmer, J. D. (1991) in The Photosynthetic Apparatus: Molecular Biology and Operation, ed. Vasil, I. K. (Academic, San Diego), Vol. 7A, pp. 5-53. [Google Scholar]
- 2.Sugiura, M. (1992) Plant Mol. Biol. 19, 149-168. [DOI] [PubMed] [Google Scholar]
- 3.Christopher, D. A. & Mullet, J. E. (1994) Plant Physiol. 104, 1119-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg, B. M., Gaba, V., Canaani, O., Malkin, S., Mattoo, A. K. & Edelman, M. (1989) Proc. Natl. Acad. Sci. USA 86, 6617-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gamble, P. E. & Mullet, J. E. (1989) EMBO J. 8, 2785-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakahira, Y., Baba, K., Yoneda, A., Shiina, T. & Toyoshima, Y. (1998) Plant Physiol. 118, 1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thum, K. E., Kim, M., Christopher, D. A. & Mullet, J. E. (2001) Plant Cell 13, 2747-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu, J. & Bogorad, L. (1990) Proc. Natl. Acad. Sci. USA 87, 1531-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liere, K. & Maliga, P. (2001) in Regulation of Photosynthesis, ed. Aro, E. M. (Kluwer, Dordrecht, The Netherlands), pp. 29-49.
- 10.Hedtke, B., Börner, T. & Weihe, A. (1997) Science 277, 809-811. [DOI] [PubMed] [Google Scholar]
- 11.Igloi, G. L. & Kössel, H. (1992) Crit. Rev. Plant Sci. 10, 525-558. [Google Scholar]
- 12.Troxler, R. F., Zhang, F., Hu, J. & Bogorad, L. (1994) Plant Physiol. 104, 753-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Link, G. (1996) BioEssays 18, 465-471. [Google Scholar]
- 14.Allison, L. A. (2000) Biochimie 82, 537-548. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka, K., Tozawa, Y., Mochizuki, N., Shinozaki, K., Nagatani, A., Wakasa, K. & Takahashi, H. (1997) FEBS Lett. 413, 309-313. [DOI] [PubMed] [Google Scholar]
- 16.Isono, K., Shimizu, M., Yoshimoto, K., Niwa, Y., Satoh, K., Yokota, A. & Kobayashi, H. (1997) Proc. Natl. Acad. Sci. USA 94, 14948-14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiwara, M., Nagashima, A., Kanamaru, K., Tanaka, K. & Takahashi, H. (2000) FEBS Lett. 481, 47-52. [DOI] [PubMed] [Google Scholar]
- 18.Kanamaru, K., Fujiwara, M., Seki, M., Katagiri, T., Nakamura, M., Mochizuki, N., Nagatani, A., Shinozaki, K., Tanaka, K. & Takahashi, H. (1999) Plant Cell Physiol. 40, 832-842. [DOI] [PubMed] [Google Scholar]
- 19.Ishihama, A. (2000) Annu. Rev. Microbiol. 54, 499-518. [DOI] [PubMed] [Google Scholar]
- 20.Liu, B. & Troxler, R. F. (1996) Proc. Natl. Acad. Sci. USA 93, 3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan, S. & Troxler, R. F. (1999) Proc. Natl. Acad. Sci. USA 96, 5316-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruissem, W. & Tonkyn, J. C. (1993) Crit. Rev. Plant Sci. 12, 19-55. [Google Scholar]
- 23.Tsunoyama, Y., Morikawa, K., Shiina, T. & Toyoshima, Y. (2002) FEBS Lett. 516, 225-228. [DOI] [PubMed] [Google Scholar]
- 24.Murashige, T. & Skoog, F. (1962) Physiol. Plant 15, 473-497. [Google Scholar]
- 25.Abel, S. & Theologis, A. (1994) Plant J. 5, 421-427. [DOI] [PubMed] [Google Scholar]
- 26.Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- 27.Gardella, T., Moyle, H. & Susskind, M. M. (1989) J. Mol. Biol. 206, 579-590. [DOI] [PubMed] [Google Scholar]
- 28.Dombroski, A. J., Walter, W. A., Record, M. T., Jr., Siegele, D. A. & Gross, C. A. (1992) Cell 70, 501-512. [DOI] [PubMed] [Google Scholar]
- 29.Kanamaru, K., Nagashima, A., Fujiwara, M., Shimada, H., Shirano, Y., Nakabayashi, K., Shibata, D., Tanaka, K. & Takahashi, H. (2001) Plant Cell Physiol. 42, 1034-1043. [DOI] [PubMed] [Google Scholar]
- 30.Privat, I., Hakimi, M. A., Buhot, L., Favory, J. J. & Mache-Lerbs, S. (2003) Plant Mol. Biol. 51, 385-399. [DOI] [PubMed] [Google Scholar]
- 31.Yao, J., Roy-Chowdhury, S. & Allison, L. A. (2003) Plant Physiol. 132, 739-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hakimi, M. A., Privat, I., Valay, J. G. & Lerbs-Mache, S. (2000) J. Biol. Chem. 275, 9215-9221. [DOI] [PubMed] [Google Scholar]
- 33.Christopher, D. A., Kim, M. & Mullet, J. E. (1992) Plant Cell 4, 785-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohyama, K., Fukuzawa, H., Kohchi, T., Shirai, H., Sano, T., Sano, S., Umesono, K., Shiki, Y., Takeuchi, M., Chang, Z., et al. (1986) Nature 322, 572-574. [Google Scholar]
- 35.Kim, M. & Mullet, J. E. (1995) Plant Cell 7, 1445-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baba, K., Nakano, T., Yamagishi, K. & Yoshida, S. (2001) Plant Physiol. 125, 595-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, M., Thum, K. E., Morishige, D. T. & Mullet, J. E. (1999) J. Biol. Chem. 274, 4684-4692. [DOI] [PubMed] [Google Scholar]
- 38.Thum, E. T., Kim, M., Morishige, D. T. Eibl, C., Koop, H.-U. & Mullet, J. E. (2001) Plant Mol. Biol. 47, 353-366. [DOI] [PubMed] [Google Scholar]
- 39.Satoh, J., Baba, K., Nakahira, Y., Tsunoyama, Y., Shiina, T. & Toyoshima, Y. (1999) Plant J. 18, 407-415. [DOI] [PubMed] [Google Scholar]
- 40.Moller, S. G., Kim, Y. S., Kunkel, T. & Chua, N. H. (2003) Plant Cell 15, 1111-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]