Abstract
Background & objectives:
Bartonella henselae is a fastidious gram-negative bacterium usually causing self limiting infections in immunocompetent individuals but often causes potentially life threatening infection, such as bacillary angiomatosis in immunocompromised patients. Both diagnosis of infections and research into molecular mechanisms of pathogenesis have been hindered by lack of appropriate and reliable diagnostic techniques. We undertook this study to standardize methods to characterize B. henselae in clinical samples to diagnose Bartonella infection correctly.
Methods:
B. henselae ATCC 49882 strain was procured from American type culture collection, USA. This strain was revived and maintained in the laboratory, and identification and characterization of this strain was done by conventional and molecular techniques, which included culture on various media, staining by different methods including electron microscopy, biochemical analysis by conventional methods and API, polymerase chain reaction (PCR) for amplification of citrate synthase gene followed by restriction fragment length polymorphism (RFLP).
Results:
This organism was biochemically inert due to slow growth and generated unique identification code with API. The amplification of the citrate-synthase gene with primers yielded a 381 bp product followed by specific RFLP profile for B. henselae.
Interpretation & conclusions:
Bartonella is fastidious and fragile organism and should be handled carefully. Extra effort and careful observation are required to isolate and characterize this organism.
Keywords: API, Bartonella, culture, electron microscopy, PCR-RFLP
Human infections due to Bartonella species (formerly Rochalimaea species) are widely considered emerging diseases. Bartonella genus belongs to the family Bartonellaceae1. Of the 22 species described, about half are known to infect healthy or immunocompromised humans. Most of our current knowledge on Bartonella infections is restricted to B. bacilliformis, B. henselae, B. quintana, and B. tribocorum. Genomes of these four species have been sequenced and significant knowledge about the pathogenicity and infection biology exists2. Although, at least one mammalian reservoir host is known for each of the described Bartonella species, but the predominant arthropod vector(s) and the mode of transmission for many novel Bartonella species remain elusive as only three Bartonella species are known to infect and replicate in the digestive tract of their respective vectors: B. quintana in body lice3, B. henselae in cat fleas4,6, B. bacilliformis in sandflies (Lutzomyia). Recent studies have shown that fleas play an important role in the maintenance and transmission of many Bartonella species5.
Bartonella genus causes a variety of manifestations in humans. The common theme in Bartonella infection in mammalian hosts is a prolonged period of intra-erythrocytic bacteremia, which represents a specific adaptation to the mode of transmission by blood-sucking arthropods. The establishment of a chronic intra-erythrocytic bacteremia takes place exclusively in the mammalian reservoir host(s) e.g., B. quintana and B. bacilliformis in humans, B. henselae in cats 6. Clinical presentation depends on the immune status of the host. In immunocompetent individuals, the response is granulomatous and suppurative, while in immunocompromised patients the response is mainly vasoproliferative7. These include long-recognized diseases such as Carrion's disease due to B. bacilliformis, trench fever due to B. quintana, and cat scratch disease (CSD) due to B. henselae and B. clarridgeiae8. Newer clinical manifestations such as bacillary angiomatosis (BA) and peliosis hepatitis, endocarditis, and Parinaud oculoglandular syndrome caused by both B. henselae and B. quintana have been identified. Rarely these organisms can lead to neurological, hematologic and renal manifestations 9. The clinical features of bartonellosis are surprisingly variable; at times the clinical illness is so indistinctive from other organisms that Bartonella infection would not be at or near the top of differential diagnosis due to lack of knowledge of its clinical spectrum.
Bartonellae are small (0.3 to 0.6 by 1.0 to 1.7 μm), pleomorphic, non capsulated, non sporing, Gram-negative rods. On electron microscopy, B. henselae does not reveal flagella, but the organism may exhibit twitching motility in wet mounts due to the presence of pili 10. However, presence of flagella has been shown in B. bacilliformis, B. clarridgeiae, and B.schoenbuchii11. Bartonellae are often cultivable, take generally 5-15 days and up to 45 days on primary culture to form visible colonies on enriched blood containing media as these are highly hemin-dependent. Repeated subcultures, however, reduces this time to only 3 to 5 days, although colonial morphology may differ 12. Isolation is more difficult from clinical samples because of prior antibiotic use and intracellular presence of the organism 13. Because of slow growth of these bacteria, standard biochemical methods for identification may not be applicable. Measurement of preformed enzymes has revealed minimal difference between Bartonella spp.; most species are biochemically inert except for production of peptidase 14. Recently, a novel, chemically modified, insect-based, liquid culture medium (Bartonella alpha-Proteobacteria growth medium, BAPGM) has been developed for isolation of at least seven species of Bartonella and other non-cultured bacteria from blood and tissue samples 15,16.
Diagnosis of Bartonella infection is based on either serology or PCR-based methods, or observation of distinctive pathology and organisms. Serological tests confirm current or prior infection and are 90-95 per cent sensitive but less specific 17. PCR offers a rapid, sensitive, and specific means to detect the organism directly from clinical samples. Two approaches can be used (i) sequencing or restriction fragment length polymorphism (RFLP) analysis of PCR-amplified genes, such as the 16S rRNA gene, the 16S-23S rRNA ITS region 18, the citrate synthase (gltA) gene19, the riboflavin synthase (ribC) gene20, the groEL gene, or the RNA polymerase beta subunit gene; (ii) species-specific amplification of the cell division protein (ftsZ) gene 21. PCR can be done directly from specimens or pure culture, and has been widely used for detecting and characterizing Bartonella. Whole-cell fatty acid (CFA) analysis has proved to be useful for identification because Bartonella species as a group have a unique and characteristic CFA profile22.
Diagnosis of bartonellosis is hindered by variable clinical manifestations and due to non-availability of appropriate, simple, and reliable technology for both detection as well as characterization of Bartonella spp. to the level necessary to permit clear differentiation from already recognized pathogens with similar clinical presentation. Bacterial isolation, serology, and PCR amplification of Bartonella species DNA directly from patient samples have diagnostic limitations. The purpose of our study was, therefore, standardization and optimization of methods to characterize B. henselae (ATCC 49882™ strain), to implement these techniques on clinical samples to diagnose infection in suspected cases of bartonellosis.
Material & Methods
The study was conducted in the Department of Microbiology, All India Institute of Medical Sciences, New Delhi, Inidia.
B. henselae ATCC® 49882™ strain which was obtained from American type culture collection (ATCC), USA, was examined. The strain was characterized by Gram stain, colony morphology, biochemical substrate degradation by conventional and commercial methods and PCR-RFLP.
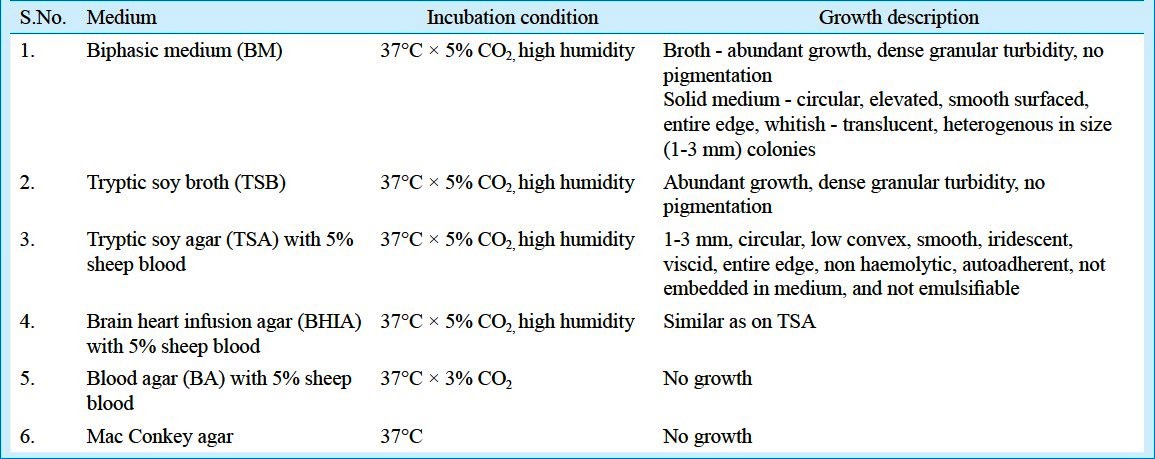
Culture and growth conditions: Representative strain was recevied in lyophilzed form and was propagated on biphasic medium consisting of tryptic soy agar (Becton Dickinson, NJ, USA) with 5 per cent defibrinated sheep blood and tryptic soy broth (Becton Dickinson, NJ, USA) and tryptic soy agar with 5 per cent defibrinated sheep blood plates, as recommended by ATCC, USA. The medium was sealed and incubated in a 5 per cent CO2 atmosphere for up to 6 wk at 37°C. Humidity of 85 per cent was obtained with sterile cotton moistened with sterile saline. All the procedures were done under Bio-Saftey Level (BSL)-223. Biphasic media and plates were examined on every 3rd day until growth becomes evident. Subculture was done using broth pool as the inoculum source and was done on various media (Table).
Table.
List of media used, growth conditions and growth description of B. henselae
Colony morphology on various media, wet mount to check motility, different types of staining including Gram's staining, acridine orange, Albert's, modified Ziehl-Neelsen staining, and negative staining for electron microscopy were used for identification and characterization of B. henselae as per standard protocol. For electron microscopy, one to two colonies were taken and fixed in fixative (2% paraformaldehyde + 2.5% gluteraldehyde) for 30 min, then washed once in PBS, followed by three washings with distilled water. After washing, homogenous suspension of pellet was made in 50 μl distilled water. One drop of homogenous suspension was put on parafilm and covered with coated copper grid (TAAB laboratories, Aldermaston, UK) with coated side below for 1 min. The grid containing the adherent bacteria was placed on a drop of 2 per cent phosphotungstinic acid (Qualigen, Carlsbad, CA, USA) for 20-30 seconds. Excess of fluid was drained on blotting paper and the grid was examined under the electron microscope (Philips, Netherlands). Biochemical characterization was performed with the conventional and three commercially available systems designed for detection of preformed enzymes: API Staph, API 20 Strep and API 20E (identification system for Staphylococci and Micrococci, Streptococci, and Enterobacteriaceae and other Gram-negative bacilli, respectively) (bioMerieux, Inc. USA). Each system was used according to manufacturers’ instructions for preparation, incubation and reading of results.
Antimicrobial sensitivity: The Clinical Laboratory Standard Institute (CLSI) has not laid down any guidelines to perform antibiotic susceptibly testing for Bartonella species. Antibiotic susceptibility was performed testing by disk diffusion as recommended by CLSI guidelines, 2006 for Acinetobacter species24. Our hypothesis of following these guidelines was that as Acinetobacter species, B. henselae is also a Gram-negative, non-fermenter organism (according to results of API Staph, API 20 Strep and API 20E). Organisms were harvested from growth on TSA with 5 per cent defibrinated sheep blood (5-7 days), resuspended in 3 ml of sterile 0.9 per cent PBS and adjusted to a McFarland standard of 0.5. The suspension (1.5 ml) was poured onto the TSA with 5 per cent defibrinated sheep blood and then aspirated off. Plates were allowed to dry before the application of disks. The tests were run in duplicate. The plates were incubated at 35 °C in 5 per cent CO2 and observed daily for growth and zone size over a period of 7 days; zones of inhibition were recorded on day 7. A growth control plate was inoculated with each test run. Antibiotic (in μg) impregnated disks (HiMedia, Mumbai, India) of ampicillin (10), ceftriaxone (30), gentamicin (10), meropenem (10) erythromycin (15), tetracycline (30), doxycycline (30), rifampin (10), ciprofloxacin (5), trimethoprim- sulphamethoxazole (1.25/23.75), and vancomycin (30) were used for susceptibility testing.
PCR-RFLP: For PCR, DNA extraction from colony growth was done by boiling method, phenol-chloroform-ethanol method, and extraction by commercial kit (Quiagen, Hidlden, Germany). Extracted DNA was stored at -80°C for further use in PCR. Primers described by Joblet M et al25 were used for amplification of citrate synthase gene of Bartonella species. PCR amplification was carried out in 25 μl reaction volumes with AMP PCR system 9700 thermocycler (MJ Research Inc., USA). The standard PCR reaction mixture consisted of the following: 5 μl of appropriate DNA template, 1.5 units of Taq DNA polymerase (Bangalore Genei, India), 20 pmol of forward and reverse primers, 200 μM each of the dNTPs and sterile distilled water to a final volume of 25 μl. Amplification of template with citrate synthase gene primers included 30 cycles of denaturation at 95°C for 20 sec, annealing of primers at 48°C for 30 sec, and primer extension at 60°C for 90 sec. Amplified products were examined by agarose gel electrophoresis. Products were electrophoresed at 110v on horizontal agarose gel, stained with ethidium bromide and visualized under UV transilluminator GelDoc system (Alpha Innotech Corp. St. San Leandro, CA, USA) for the presence of 381 base pair band. For RFLP analysis, PCR-amplified DNA, was digested with restriction enzymes (Taq I, Aci I), incubated at 65°C for Taq I and at 37°C for Aci I for 4 h. Restriction profiles were analyzed by using 3 per cent agarose gel stained with ethidium bromide, and examined under ultra violet light.
Results
On primary subculture, there was no growth in broth as well as in media on day 3, which was followed by slight turbidity in broth without visible growth on day 5. On day 7, growth appeared in both broth and solid media. Broth showed abundant growth with dense granular turbidity, but no pigmentation. On solid medium circular, elevated, smooth surfaced, entire edge, whitish- translucent colonies grew which were heterogenous in size (1-3 mm) (Fig. 1). Later on, relatively rapid growth (4-5 days between subcultures) was achieved. The organism showed twitching motility in wet mount. In Gram's staining, Gram negative, slightly curved small bacilli in clumps (Fig. 2) were visible, while in acridine orange staining, pleomorphic, apple green coloured bacilli in clumps were seen. Green colour bacilli without metachromatic granules were seen in Albert's staining. Bacilli were non acid fast. In electron microscopy, the organism appeared to form cohesive aggregates, with relatively few organisms existing freely. The average size of the organism was approximately 2μm in length by 0.5 to 0.6 μm in width (Fig. 3). Flagella were not observed. Biochemical characterization based upon conventional methods showed that organism was non fermentative, catalase, oxidase, urease, and indole negative. It also did not utilize citrate and Triple Sugar Iron (TSI) showed alkaline/no change. Profile code in API Staph -0004001, showed that the organism was positive for alkaline phosphatase, arginine dihydrolase and negative for sugar fermentation, acetoin production, urease and reduction of nitrate to nitrite, API 20 Strep - 0161000 - indicated that the organism positive for pyrrolidonylarylamidase, leucin arylamidase, alkaline phosphatase, arginine dihydrolase and negative for β-glucosidase, α-galactosidase, β-glucuronidase, β-galactosidase, sugar fermentation and in API 20 E - 610000040 - indicated positive reaction for arginine dihydrolyse, lysine decarboxylase, ornithine decarboxylase and negative reaction for β-galactosidase, citrate utilization, H2S production, urease, indole production, tryptophan deaminase, acetoin production, gelatinase, sugar production, NO2 production. All these profile codes were not provided in database of these identification kits.
Fig. 1.
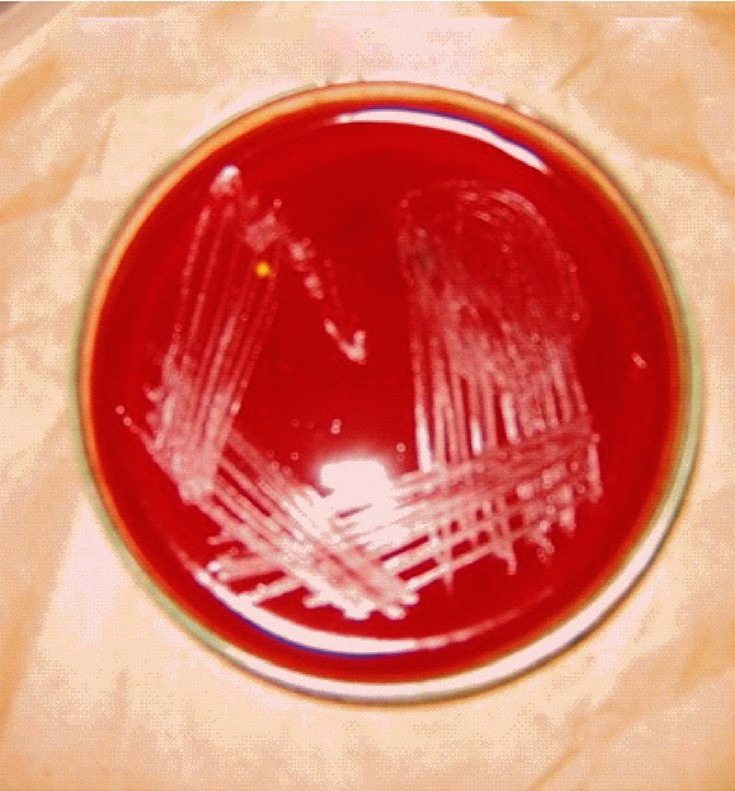
Tryptic soy agar showing of growth B. henselae.
Fig. 2.
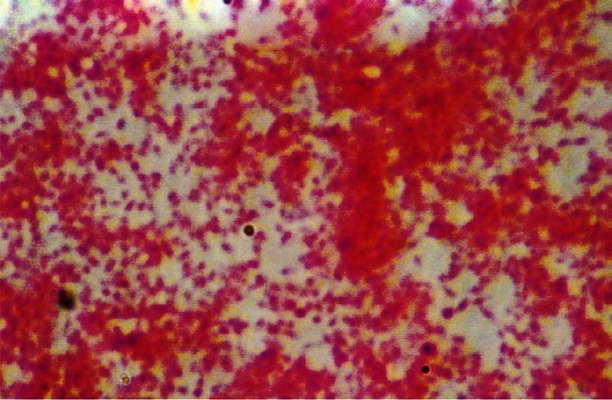
Gram's staining showing gram negative, pleomorphic bacilli in clumps at 100× magnification.
Fig. 3.
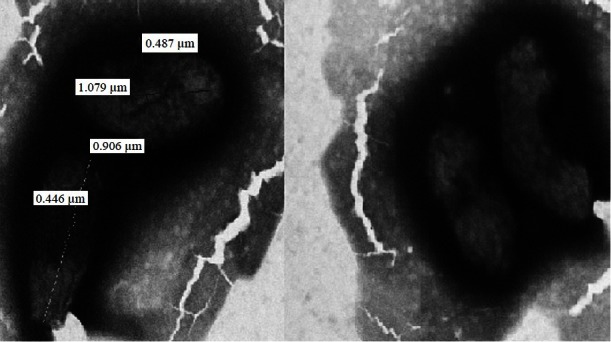
Electron microscopic photograph of B. henselae.
Antimicrobial susceptibility: The organism was sensitive to ampicillin (zone size: >29 mm), ceftriaxone (>21 mm), meropenem (>16 mm), gentamicin (>15 mm), tetracycline (>19 mm), doxycycline (>16 mm), erythromycin (>23 mm), rifampicin (>20 mm), ciprofloxacin (>21 mm), and trimethoprim- sulphamethoxazole (>16 mm), and resistant to vancomycin (<15 mm) only.
PCR-RFLP: For DNA extraction, the boiling method was found to be the easiest, less time consuming, cost effective and resulted good yield of DNA. The product size was approximately 381 bp after amplification in PCR (Fig. 4). The sensitivity of this PCR analysis was evaluated by 10 fold serial dilution of bacterial DNA and found to be 20 pg/μl. Specificity of oligonucleotide primers was evaluated by testing purified DNA from a panel of bacteria having citrate synthase gene. Similar PCR amplification cycle was used. No amplification products were observed for Escherichia coli, Klebisella pneumoniae, Salmonella Typhi, Salmonella Typhimurium, Shigella dysenteriae, Proteus mirabilis, Staphylococcus aureus, coagulase negative Staphylococci, Streptococcus pyogenes, Enterococcus fecalis, Clostridium perfringens, Mycoplasma pneumoniae, Campylobacter jejuni, Legionella pneumophila, and Acinetobacter species. Digestion of PCR products with restriction endonuclease, TaqI yielded three bands of 71, 137, and 177 bp and with AciI two bands of 100, and 260 bp (Fig. 5). The PCR-RFLP profile of this isolate was identical to B. henselae.
Fig. 4.
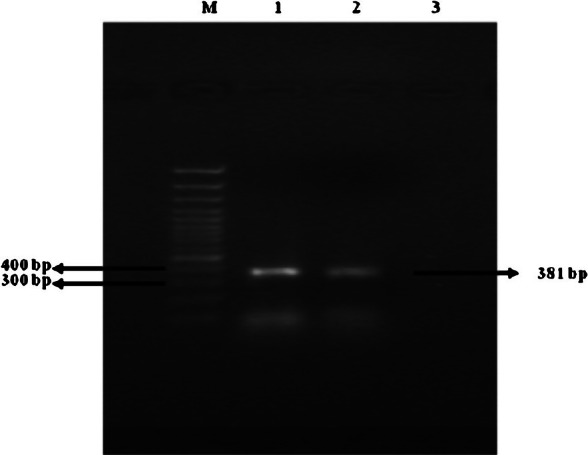
Agarose gel showing bands obtained by electrophoresis of PCR products from B. henselae using primers targeted at conserved bacterial citrate synthase gene sequence. Lane at left extreme is with marker DNA (100bp). Lanes 1 and 2 show B. henselae PCR product (381 bp); lane 3-negative control.
Fig. 5.
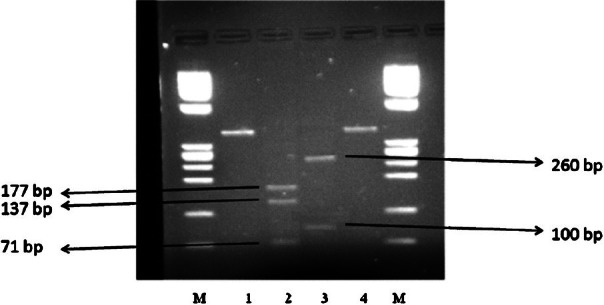
Agarose gel showing bands obtained by electrophoresis of RFLP products of PCR product of B. henselae using restriction enzymes Taql and Acil. Lanes at right and left extreme are with marker DNA. Lanes 1 and 4 - positive control (B. henselae PCR product), lane 2 Taql and lane 3 Acil restriction profile.
Discussion
Although the predisposing factors for Bartonella infection, i.e. vector, reservoir, susceptible population, and similar geographical conditions are present in India, but only a couple of reports available about presence of infection by bartonellae. Chaudhry et al26 reported association of Bartonella sp. with optic neuritis. In another recent report from Chennai, India 27, IgG antibodies against Bartonella quintana spp. were found in 14 per cent of blood culture negative cases and in 8 per cent of all infective endocarditis cases by microimmunofluorescence. Clinicians are often unaware of the various clinical presentations of Bartonella infection and do not even consider it in the differential diagnosis of the diseases of similar clinical features. Further, non availability of the appropriate technology for both the detection of bartonellae and its characterization to the level necessary to permit clear differentiation from already recognized pathogens with similar clinical presentation is also responsible for underdiagnosis of the infection.
Several studies have concluded that Bartonella species grow on axenic medium at 35-37°C in 5 per cent CO2 atmosphere with high humidity. Almost all studies conclude that growth in axenic medium is hemin dependent and agar should be enriched with blood. The recently developed BAPGM followed by PCR might provide a complimentary diagnostic approach for the isolation and identification of potentially fastidious bacteria from blood and tissue samples. As Bartonella is highly fastidious bacteria and that these organisms are unlikely to be detected with conventional microbiological approaches in many patients, enrichment culture combined with molecular detection methods using the potential target genes such as ITS, gltA, ribC, etc. can replace conventional culture techniques for the identification of Bartonella in clinical specimens 28.
We used tryptic soy agar with 5 per cent sheep blood, as recommended by ATCC. These colonies were, shiny, smooth, circular, entire edge, low convex, non haemolytic, auto-adherent, not embedded in agar; a similar description has also been given by Regnery et al29. On staining, the organism was non acid fast, Gram-negative bacilli, showing no metachromatic granules. On electron microscopy, the organism appeared non- flagellated and formed cohesive aggregates, with relatively a few organisms existing freely; similar to the description given by Welch et al30. As the organism is biochemically inert, conventional biochemical tests can rule out other organisms but cannot characterize Bartonella. Commercial tests helps in identification, as these are the only tests available to detect the enzymatic and peptidase activity of the organism. In our study API 20E and API 20 Strep, with the exception of the API Staph, generated profile codes unique to their data bases. The profile codes in these tests indicated positive reaction for arginine dihydrolase, lysine decarboxylase, pyrrolidonylarylamidase, leucine arylamidase, and alkaline phosphatase, as shown previously 29,30.
The boiling method was found to be the easiest and cost-effective method for DNA extraction but for long term storage of DNA, extraction by commercial DNA kits has shown better results31. The gltA coding sequence of B. henselae is 1293 nuclotides in length, encoding a protein of 431 amino residues. PCR analysis under stringent conditions with primer specific for Bartonella spp. yielded amplified products for only Bartonella spp., similar specificity was noted in previous studies32. Joblet et al25 proved that citrate synthase gene PCR amplification was restricted only to Bartonella species; there was no amplification with other members of α2 proteobacteria. Further PCR-RFLP is able to identify Bartonella species using TaqI and AciI enzyme. With TaqI - two bands, 63 and 316 bp for B. quintana, three bands, 58, 71, 250 bp for B. vinsonii, three bands, 71, 137, 177 bp for B. henselae and two bands, 72 and 245 bp for B. bacilliformis were observed. With AciI - three bands 15, 155, 209 bp for B. quintana, two bands 15 and 361 bp for B. vinsonii, two bands 100 and 260 bp for B. henselae and three bands 15, 115, 160 bp for B. bacilliformis were appeared. No divergence was noted between the RFLP profiles obtained with different isolates belonging to the same species. We also tried to standardize the PCR-RFLP technique with amplification of partial citrate sythase gene followed by restriction with TaqI and AciI. Our observations were different from that of Jablet et al25, we were able to visualize <200 bp bands (up to around 70 bp) in agarose gel elecrophorosis after RFLP. The reason for this difference may be the use of 3 per cent agarose gel in contrast to the 1.8 per cent used in previous study, to visualize digestion fragments after RFLP. Limitation of this study was that the primer specificity was not checked with other members of α-proteobacteria phylum due to the non availability of these strains.
In conclusion, Bartonella is a fastidious and fragile organism and should be handled carefully. This organism is slow growing and grows only on special media; hence routine blood culture methods are not able to isolate it. As shown earlier, the citrate synthase gene was found to be specific for Bartonella genus and RFLP followed by PCR may be able to differentiate between species of Bartonella genus.
In this study isolation and characterization of B. henselae was done on reference ATCC strain. These techniques need to be tested for isolation and characterization of Bartonella from clinical samples. Clinical samples should be taken from patients having diseases with similar presentation to Bartonella infection, especially patients with lymphadenopathy, optic neuritis, endocarditis, pyrexia of unknown origin and also from diseases with unknown aetiology.
References
- 1.Anna Sander. Bartonella and Afipia. In: Collier L, Balows A, Sussman M, Duerden BI, editors. Topley & Wilson's microbiology and microbial infections. 9th ed. II. New York: Hodder Arnold; 1998. p. 133. [Google Scholar]
- 2.Kaiser PO, Riess T, O’Rourke F, Linke D, Kempf VAJ. Bartonella spp.: throwing light on uncommon human infections. Int J Med Microbiol. 2011;301:7–15. doi: 10.1016/j.ijmm.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Fournier PE, Minnick MF, Lepidi H, Salvo E, Raoult D. Experimental model of human body louse infection using green fluorescent protein-expressing Bartonella quintana. Infect Immun. 2001;69:1876–79. doi: 10.1128/IAI.69.3.1876-1879.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higgins JA, Radulovic S, Jaworski DC, Azad AF. Acquisition of the cat scratch disease agent Bartonella henselae by cat fleas (Siphonaptera: Pulicidae) J Med Entomol. 1996;33:490–5. doi: 10.1093/jmedent/33.3.490. [DOI] [PubMed] [Google Scholar]
- 5.Marié JL, Fournier PE, Rolain JM, Briolant S, Davoust B, Raoult D. Molecular detection of Bartonella quintana. B. Elizabethae, B. Koehlerae, B. Doshiae, B. Taylorii, and Rickettsia Felis in rodent fleas collected in Kabul, Afghanistan. Am J Trop Med Hyg. 2006;74:436–9. [PubMed] [Google Scholar]
- 6.Rolain JM, La SB, Liang Z, Davoust B, Raoult D. Immunofluorescent detection of intraerythrocytic Bartonella henselae in naturally infected cats. J Clin Microbiol. 2001;39:2978–80. doi: 10.1128/JCM.39.8.2978-2980.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bass JW, Vincent JM, Person DA. The expanding spectrum of Bartonella infections: II. Cat-scratch disease. Pediatr Infect Dis J. 1997;16:163–79. doi: 10.1097/00006454-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Jerris RC, Regnery RL. Will the real agent of cat-scratch disease please stand up? Annu Rev Microbiol. 1996;50:707–25. doi: 10.1146/annurev.micro.50.1.707. [DOI] [PubMed] [Google Scholar]
- 9.Kalogeropoulos C, Koumpoulis I, Mentis A, Pappa C, Zafeiropoulos P, Aspiotis M. Bartonella and intraocular inflammation: a series of cases and review of literature. Clin Ophthalmol. 2011;5:817–29. doi: 10.2147/OPTH.S20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benson LA, Kar S, McLaughlin G, Ihler GM. Entry of Bartonella bacilliformis into erythrocytes. Infect Immun. 1986;54:347–53. doi: 10.1128/iai.54.2.347-353.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dehio C, Lanz C, Pohl R, Behrens P, Bermond D, Piémont Y, et al. Bartonella schoenbuchii sp. nov., isolated from the blood of wild roe deer. Int J Syst Evol Microbiol. 2001;51:1557–65. doi: 10.1099/00207713-51-4-1557. [DOI] [PubMed] [Google Scholar]
- 12.Maurin M, Roux V, Stein A, Ferrier F, Viraben R, Raoult D. Isolation and characterization by immunofluorescence, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, western blot, restriction fragment length polymorphism-PCR, 16S rRNA gene sequencing, and pulsed-field gel electrophoresis of Rochalimaea quintana from a patient with bacillary angiomatosis. J Clin Microbiol. 1994;32:1166–71. doi: 10.1128/jcm.32.5.1166-1171.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.La Scola B, Raoult D. Culture of Bartonella quintana and Bartonella henselae from human samples: a 5-year experience (1993 to 1998) J Clin Microbiol. 1999;37:1899–905. doi: 10.1128/jcm.37.6.1899-1905.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breitschwerdt EB, Kordick DL. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin Microbiol Rev. 2000;13:428–38. doi: 10.1128/cmr.13.3.428-438.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maggi RG, Duncan AW, Breitschwerdt EB. Novel chemically modified liquid medium that will support the growth of seven Bartonella species. J Clin MIcrobiol. 2005;43:2651–5. doi: 10.1128/JCM.43.6.2651-2655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cadenas MB, Maggi RG, Diniz PP, Breitschwerdt KT, Sontakke S, Breithschwerdt EB. Identification of bacteria from clinical samples using Bartonella alpha-Proteobacteria growth medium. J Microbiol Methods. 2007;71:147–55. doi: 10.1016/j.mimet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 17.La Scola B, Raoult D. Serological cross-reactions between Bartonella quintana, Bartonella henselae, and Coxiella burnetii. J Clin Microbiol. 1996;34:2270–4. doi: 10.1128/jcm.34.9.2270-2274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maggi RG, Breitschwerdt EB. Potential limitations of the 16S-23S rRNA intergenic region for molecular detection of Bartonella species. J Clin Microbiol. 2005;43:1171–6. doi: 10.1128/JCM.43.3.1171-1176.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Birtles RJ, Rauolt D. Comparison of partial citrate synthase gene (glt A) sequence for phylogenetic analysis of Bartonella species. J Syst Bacteriol. 1996;46:891–7. doi: 10.1099/00207713-46-4-891. [DOI] [PubMed] [Google Scholar]
- 20.Bereswill S, Hinkelmann S, Kist M, Sander A. Molecular analysis of riboflavin synthesis gene in Bartonella henselae and use of the ribC gene for differentiation of Bartonella species by PCR. J Clin Microbiol. 1999;37:3159–66. doi: 10.1128/jcm.37.10.3159-3166.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeaiter Z, Liang Z, Raoult D. Genetic classification and differentiation of Bartonella species based on comparison of partial ftsZ gene sequences. J Clin Microbiol. 2002;40:3641–7. doi: 10.1128/JCM.40.10.3641-3647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarridge JE, 3rd, Raich TJ, Pirwani D, Simon B, Tsai L, Rodriguez-Barradas MC, et al. Strategy to detect and identify Bartonella species in routine clinical laboratory yields Bartonella henselae from human immunodeficiency virus-positive patient and unique Bartonella strain from his cat. J Clin Microbiol. 1995;33:2107–13. doi: 10.1128/jcm.33.8.2107-2113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Available from: https://www- envirinfo.llnl.gov/contant/enviroRecent/livermoreSite/BSL_ 3_EA_Appendix_A_Final_R1_Revised_25Jan08.pdf .
- 24.Performance standards for antimicrobial disk susceptibility tests M100-S16. 9th ed. Wayne, PA: CLSI; 2006. Clinical Laboratory Standard Institute (CLSI) [Google Scholar]
- 25.Joblet C, Roux V, Drancourt M, Gouvernet J, Raoult D. Identification of Bartonella (Rochalimaea) species among fastidious gram-negative bacteria on the basis of the partial sequence of the citrate-synthase gene. J Clin Microbiol. 1995;33:1879–83. doi: 10.1128/jcm.33.7.1879-1883.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudhry R, Mukherjee A, Menon V. Isolation and characterisation of Bartonella sp. from potic neuritis patients. Indian J Med Res. 2012;136:985–90. [PMC free article] [PubMed] [Google Scholar]
- 27.Balakrishnan N, Menon T, Fournier PE, Raoult D. Bartonella quintana and Coxiella burnetii as causes of endocarditis, India. Emerg Infect Dis. 2008;14:1168–9. doi: 10.3201/eid1407.071374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maggi RG, Kempf VA, Chomel BB, Breitschwerdt EB. Bartonella. In: Versalovic J, Carroll KC, Funke G, Jorgensen JH, Landry ML, Warnock DW, editors. Manual of clinical microbiology. 10th ed. I. Washington DC: ASM Press; 2011. p. 791. [Google Scholar]
- 29.Regnery RL, Anderson BE, Clarridge JE, 3rd, Rodriguez-Barradas MC, Jones DC, Carr JH. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol. 1992;30:265–74. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch DF, Pickett DA, Slater LN, Steigerwalt AG, Brenner DJ. Rochalimaea henselae sp. nov., a cause of septicemia, bacillary angiomatosis, and parenchymal bacillary peliosis. J Clin Microbiol. 1992;30:275–80. doi: 10.1128/jcm.30.2.275-280.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhry R, Chandel DS, Verma N, Singh N, Singh P, Dey AB. Rapid diagnosis of typhoid fever by an in-house flagellin PCR. J Med Microbiol. 2010;59:1391–3. doi: 10.1099/jmm.0.020982-0. [DOI] [PubMed] [Google Scholar]
- 32.Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


