Abstract
One measure of the severity of a pandemic influenza outbreak at the individual level is the risk of death among people infected by the new virus. However, there are complications in estimating both the numerator and denominator. Regarding the numerator, statistical estimates of the excess deaths associated with influenza virus infections tend to exceed the number of deaths associated with laboratory-confirmed infection. Regarding the denominator, few infections are laboratory confirmed, while differences in case definitions and approaches to case ascertainment can lead to wide variation in case fatality risk estimates. Serological surveillance can be used to estimate the cumulative incidence of infection as a denominator that is more comparable across studies. We estimated that the first wave of the influenza A(H1N1)pdm09 virus in 2009 was associated with approximately 232 (95% confidence interval: 136, 328) excess deaths of all ages in Hong Kong, mainly among the elderly. The point estimates of the risk of death on a per-infection basis increased substantially with age, from below 1 per 100,000 infections in children to 1,099 per 100,000 infections in those 60–69 years of age. Substantial variation in the age-specific infection fatality risk complicates comparison of the severity of different influenza strains.
Keywords: death, human influenza, severity
The severity profile of a pandemic influenza virus, in combination with its transmissibility, determines the impact it will have in a population (1). One commonly reported measure of severity at the individual level is the risk of death among people infected by the virus, and this conditional measure is referred to as the “case fatality risk” or, sometimes, as the “case fatality rate” or “ratio.” There are well-known complications in quantifying both the numerator and the denominator of the case fatality risk (2). Regarding the numerator, deaths of individuals with confirmed infection would underascertain all deaths associated with infection (3–6). Instead of directly counting “confirmed” deaths, it is also possible to statistically estimate excess deaths (5–9). Regarding the denominator, most influenza infections are mild; laboratory testing has limited capacity, is expensive, and is often unnecessary for clinical management; and therefore few infections would be laboratory confirmed (10, 11).
It is feasible to estimate the incidence rates of infections in a population if relevant surveillance data are available (12, 13), and the estimated cumulative incidence of infection may provide more unbiased denominators of case fatality risk that address the unobservability of infection and minimize ascertainment bias. In the present study, we propose such a severity measure referred to as the “infection fatality risk” and define it as the number of influenza-associated deaths divided by the number of infections in a population or subgroup. The infection fatality risk is expected to permit comparisons (e.g., across age groups and risk groups), and we investigate the infection fatality risk based on deaths of confirmed cases (IFRc) as well as the infection fatality risk based on excess influenza-associated deaths (IFRe). IFRc involves ascertainment and underreporting biases, while IFRe assumes full description of observed data by statistical modeling.
The first objective of our study was to estimate the number of excess deaths associated with the first wave of pandemic influenza A(H1N1)pdm09 (pH1N1) in Hong Kong. The second objective was to estimate the age-specific severity profiles of IFRc and IFRe and to investigate the differences between them.
MATERIALS AND METHODS
Sources of data
Information on age-specific all-cause deaths and the corresponding annual midyear populations from 2001 through 2009 was obtained from the Hong Kong Government Census and Statistics Department (14, 15). Information on age-specific hospitalizations and deaths associated with laboratory-confirmed pH1N1 infection from May 1, 2009, through December 31, 2009, was provided by the Hong Kong Hospital Authority. Surveillance data on influenza-like illness (ILI) from around 50 sentinel general practitioners were available as the weekly proportion of outpatients reporting a fever of >37.8°C plus a cough or sore throat (denoted “ILI data” hereafter), along with local laboratory (LAB) data on the weekly proportion of specimens from sentinel outpatient clinics and local hospitals that tested positive for influenza (denoted “LAB data” hereafter) (16). Surveillance data stratified by age were not available. Data on temperature and humidity were obtained from the Hong Kong Observatory (17). Age-specific estimates of the cumulative incidence of pH1N1 infection in the first wave were determined in separate serological surveillance studies (18, 19) and used as the denominators for estimation of IFRc and IFRe.
Statistical analysis
We assume that all excess deaths are truly associated with pH1N1. To address the uncertainty, 4 statistical models were used to estimate the excess deaths, namely, time-series regression, linear regression, and Poisson regression with log links and identity links. In each approach, we compared excess death estimates based on 4 different proxy measures of local influenza activity including the following: 1) weekly incidence rates of pH1N1 infection, 2) weekly ILI data, 3) weekly LAB data, and 4) the product of weekly ILI and LAB data. Incidence rates of pH1N1 infection were estimated by deconvoluting the time series of hospitalizations associated with pH1N1, allowing for the delay from infection to hospitalization and scaling to serial cross-sectional serological data (Web Appendix available at http://aje.oxfordjournals.org/) (18).
We applied each regression model to the time series of weekly all-cause mortality rates from 2001 through 2009, excluding February–September of 2003, which was affected by the Severe Acute Respiratory Syndrome epidemic. The data were stratified into 8 age groups: 0–4 years, 5–14 years, 15–29 years, 30–39 years, 40–49 years, 50–59 years, 60–69 years, and ≥70 years. In each regression model, we included one of the measures of influenza activity as a covariate, lagged by 1 week to allow for a delay between infection and death, and also adjusted for covariates including seasonal influenza activity, respiratory syncytial virus activity, mean temperature, and absolute humidity (Web Appendix). Trigonometric components were included to allow for cyclic annual seasonality. The influenza-associated excess mortality rates were calculated by subtracting the predicted mortality rate estimated from each fitted regression model setting influenza activity as zero from the predicted mortality rate from the same model based on the observed weekly influenza activity. Further details of the statistical methods are described in the Web Appendix. All analyses were conducted in R, version 2.13.1, language and environment (20).
RESULTS
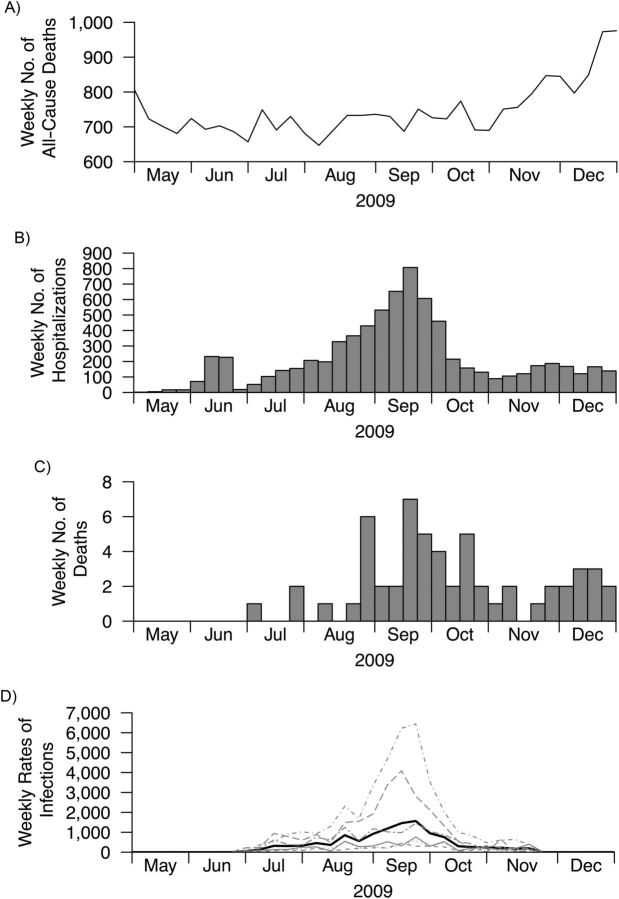
The first wave of pH1N1 in Hong Kong began in the summer and peaked in September 2009 before activity declined to low levels (Figure 1). Local all-cause mortality rates generally increased in the winter, and there was no obvious increase in all-cause deaths during the peak of the pandemic (Web Appendix). Because the patterns of age-specific pH1N1 incidence rates were similar in each age group (Figure 1), we directly standardized these age-specific pH1N1 incidence rates using the Hong Kong population. The resulting age-standardized incidence rates were then used as a single proxy measure of influenza activity.
Figure 1.
All-cause deaths, pandemic A(H1N1)2009 virus (pH1N1) hospitalizations and deaths, and estimated pH1N1 incidence rates in Hong Kong, 2009. A) Weekly number of all-cause deaths; B) weekly number of hospitalizations of patients with confirmed pH1N1; C) weekly number of deaths of patients with confirmed pH1N1; D) age-specific estimated incidence rates of pH1N1 infection, estimated by deconvoluting hospital admission rates and scaling to serological surveillance data. Incidence rates by age group: 5–14 years (dot-dash-dot); 15–19 years (long dash); 20–29 years (double dash); 30–39 years (thin solid); 40–49 years (short dash); and 50–59 years (dotted), respectively, and the age-standardized incidence rates (thick solid), expressed as rates per 100,000 population per week. Incidence rates were standardized to the local Hong Kong population. Three-letter names and abbreviations on the inner label of the x-axis represent months of the year.
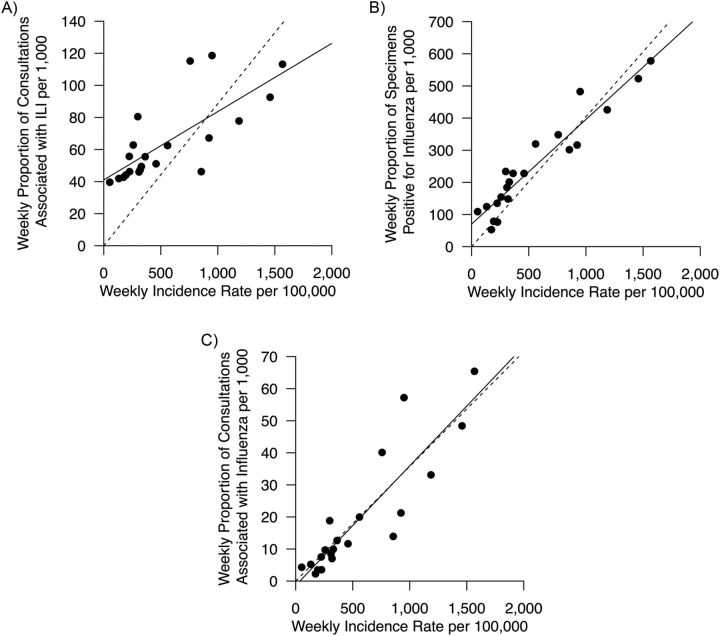
We compared the correlation between the ILI data, the LAB data, and the product of ILI and LAB data versus age-standardized incidence rates (Figure 2). ILI data tended to overestimate the lower levels of pH1N1 incidence rates, as did the LAB data to a lesser extent, while the product of ILI data and LAB data had the strongest correlation with pH1N1 incidence rates.
Figure 2.
Correlation between influenza-like illness (ILI) surveillance data, laboratory (LAB) detection data, and age-standardized incidence rate estimates (per 100,000 population per week) based on hospital admissions and serological surveillance data in Hong Kong, 2009. A) Correlation between ILI data based on general practitioners and incidence rates; B) correlation between LAB data and incidence rates; C) correlation between ILI × LAB and incidence rates. In each panel, the solid line indicates the least-squares regression line, and the dashed line indicates the constrained ordinary least-squares regression line through the origin. The dashed line in each panel has the intercept coefficient fixed at zero. The estimated intercept coefficients for the solid line in each panel were estimated as follows: A) 41 (95% confidence interval (CI): 29, 53); B) 70 (95% CI: 37, 100); C) −1.3 (95% CI: −7.4, 4.7).
Using age-standardized pH1N1 incidence rates as the proxy of influenza activity, we estimated that the overall number of excess deaths associated with the first wave of pH1N1 was 232 (95% confidence interval (CI): 136, 328) under the time-series regression model with most of the excess deaths in the elderly (Table 1, Web Table 1). Estimates of excess deaths were similar in each of the 4 regression models (Table 1, Web Table 1). Estimates of excess mortality based on proxy measures of influenza activity gave estimates similar to those based on estimated pH1N1 incidence rates (Table 1, Web Table 1). In comparison, there were 54 deaths in patients with laboratory-confirmed pH1N1 before December 31, 2009 (7). In sensitivity analyses, point estimates of the excess deaths were lower when influenza activity was lagged by 2 weeks and when it was not lagged (Web Table 2).
Table 1.
Estimated Excess All-Cause Deaths by 4 Statistical Methods and 4 Measures of Influenza Activity, Assuming a 1-Week Lag Between Influenza Incidence and Death, in Hong Kong, 2009
| Age and Statistical Model | Influenza Incidence Proxy |
|||||||
|---|---|---|---|---|---|---|---|---|
| Age-standardized Incidence Proxy |
ILI × LAB |
ILI |
LAB |
|||||
| No. | 95% CI | No. | 95% CI | No. | 95% CI | No. | 95% CI | |
| <60 years | ||||||||
| Time-series regression | 2 | −57, 61 | −15 | −72, 42 | −19 | −79, 42 | −1 | −62, 59 |
| Linear regression | 19 | −18, 55 | −1 | −37, 36 | 9 | −27, 45 | 21 | −16, 57 |
| Poisson regression with log link | 9 | −23, 42 | −8 | −41, 25 | −7 | −39, 26 | 8 | −25, 40 |
| Poisson regression with identity link | 16 | −18, 50 | −3 | −37, 31 | 7 | −26, 41 | 18 | −16, 52 |
| ≥60 years | ||||||||
| Time-series regression | 231 | 154, 307 | 223 | 149, 298 | 162 | 84, 240 | 201 | 123, 279 |
| Linear regression | 230 | 128, 333 | 223 | 120, 326 | 160 | 58, 263 | 200 | 98, 302 |
| Poisson regression with log link | 230 | 154, 307 | 221 | 144, 298 | 161 | 85, 237 | 197 | 121, 273 |
| Poisson regression with identity link | 248 | 169, 326 | 236 | 158, 315 | 192 | 115, 270 | 224 | 146, 302 |
Abbreviations: CI, confidence interval; ILI, influenza-like illness based on consultations with general practitioners; ILI × LAB, general practitioner consultations associated with influenza; LAB, laboratory specimens positive for influenza.
Based on deaths of patients with laboratory-confirmed pH1N1, point estimates of the IFRc increased with age from 0.4 to 164 deaths per 100,000 infections for individuals from 5–14 years to 60–69 years of age (Table 2). Similarly, based on estimated excess deaths by the time-series regression model using age-standardized incidence rates as the proxy measure of influenza activity, point estimates of the IFRe increased from approximately zero (point estimate −1.1, 95% CI: −6.1, 4.2) deaths per 100,000 infections in those 5–14 years of age to 1,100 (95% CI: 180, 4,700) deaths per 100,000 infections in those 60–69 years of age.
Table 2.
Severity Profile of Pandemic A(H1N1)2009 Virus During the First Wave in Hong Kong, 2009
| Age, years | Population, no. | CII, % | 95% CI | Confirmed Deaths, no. | IFRca | 95% CI | Estimated Deaths, no. | IFRea | 95% CI |
|---|---|---|---|---|---|---|---|---|---|
| 0–4 | 229,200 | NA | 0 | NA | −8 | NA | |||
| 5–14 | 644,200 | 43.5 | 39.6, 48.3 | 1 | 0.4 | 0, 1.1 | −3 | −1.1 | −6.1, 4.2 |
| 15–29 | 1,430,500 | 16.9 | 12.4, 21.3 | 4 | 1.7 | 0.3, 3.8 | 2 | 0.8 | −12.3, 14.6 |
| 30–39 | 1,114,500 | 5.8 | 3.1, 9.7 | 7 | 10.8 | 3.6, 25.5 | 2 | 3.1 | −40.1, 43.7 |
| 40–49 | 1,273,000 | 3.8 | 1.1, 7.5 | 6 | 12.5 | 3.4, 51.4 | 8 | 16.7 | −51.2, 119 |
| 50–59 | 1,085,400 | 5.0 | 2.4, 8.3 | 15 | 27.9 | 14.6, 61.7 | 1 | 1.9 | −59.2, 49.9 |
| 60–69 | 555,500 | 0.8 | 0.2, 4.2 | 7 | 164 | 18, 741 | 47 | 1,099 | 176, 4,657 |
| ≥70 | 671,400 | NA | 14 | NA | 184 | NA |
Abbreviations: CI, confidence interval; CII, cumulative incidence of infection based on serological surveillance studies (18, 19); IFRc, infection fatality risk based on deaths of confirmed cases; IFRe, infection fatality risk based on excess influenza-associated deaths; NA, not available; pH1N1, pandemic influenza A(H1N1)pdm09.
a Infection fatality risks are expressed as number of deaths per 100,000 infections. The denominators for IFRc and IFRe were the numbers of estimated pH1N1 infections in each age group. A bootstrap method was used to compute 95% confidence intervals. IFRc = deaths in confirmed cases/(CII × population) × 100,000. IFRe = estimated excess deaths/(CII × population) × 100,000.
DISCUSSION
We estimated that, during the first wave of the pandemic, pH1N1 was associated with 232 excess deaths among individuals at all ages, mostly among the elderly (Table 2). Among the elderly, we estimated that there were around 10 times as many excess deaths as deaths in patients with confirmed pH1N1 (21). Although the population-wide estimate involves a large standard error due to overall small age-specific estimates and large variations in the estimates by age group, previous estimates of the excess mortality associated with pH1N1 in Hong Kong were similar to those presented here (7). Excess deaths typically exceed confirmed deaths because some deaths occur in individuals who do not present to the health-care system, while others occur in patients who are tested for pH1N1 only after cessation of detectable viral shedding or who never receive a laboratory test (3–6). In particular, excess influenza deaths in the elderly tend to exceed confirmed deaths because of nonspecific presentation of influenza infections and the association with nonrespiratory causes of deaths in this age group (22, 23).
Although caution must be exercised in attempting causal interpretation of statistical estimates of excess deaths due to their ecological nature, 4 different statistical methods each gave similar estimates of the excess deaths associated with pH1N1 (Table 1). The ILI × LAB proxy, a measure of the proportion of ILI due to pH1N1 among outpatients, was highly correlated with the estimated incidence rates of pH1N1 infection, suggesting that it may be a better proxy of influenza activity than either the ILI data or LAB data alone (Figure 2) (24, 25).
Using estimates of the excess deaths and the cumulative incidence of infection, we found that the risk of death on a per-infection basis increased substantially with age, with the IFRe varying from below 1 per 100,000 infections in children to the order of 1,100 per 100,000 infections in those 60–69 years of age. Combining information from a serological study in the Netherlands (26) with excess death estimates (21), we found that it is possible to obtain very similar estimates of the IFRe, varying from 2.1 to 2,900 deaths per 100,000 infections for individuals from 5–24 years to 65–74 years of age. Although the incidence of infection was very low among the elderly, probably due to preexisting immunity associated with historical exposures to similar viruses (27), we found that the high severity in this age group led to a substantial impact on mortality (Table 2). We did not identify substantial differences between IFRc and IFRe for individuals below the age of 60 years.
Other studies provided a wide range of estimates of the case fatality risk of pandemic influenza (3, 11, 19, 28–31). The earliest study of the severity of pH1N1 did not account for age and estimated that the case fatality risk was 400 deaths per 100,000 infections (29). Another study from Mexico estimated that the fatality risk varied from 3 to 30 deaths per 100,000 cases with ILI in individuals aged 1–9 years and ≥70 years, respectively (30). In the United Kingdom, the infection fatality risk was estimated to range from 5 to 9 deaths per 100,000 infections for all ages (28). Studies on previous influenza pandemics in 1918, 1957, and 1968 estimated the fatality risk among clinically apparent illnesses to be 100–2,500 per 100,000 patients (31). However, estimates of the cumulative incidence of symptomatic infection are likely to vary depending on the case definition as well as health-care–seeking behaviors and surveillance systems. The IFRe, measured by using the cumulative incidence of infection derived from the serological data as the denominator and statistically estimated deaths as the numerator, may provide a more consistent and less biased approach for comparatively assessing the severity of infection than case fatality risk estimates.
Our study has a few limitations. First, estimates of IFRe may not be comparable between populations because the severity of infection could be affected by virus mutations, environmental conditions, or host factors that may vary in different countries (32). In addition, estimates of the number of excess deaths may be zero or even negative because of harvesting or virus interference (33, 34) and, in the presence of stronger such effects, the IFRe may not fully capture the risk of mortality associated with influenza infection. Second, the ILI and laboratory surveillance systems in Hong Kong are not population based, while the laboratory specimens are mostly diagnostic specimens submitted by hospitals rather than routinely collected through the ILI network. Nevertheless, ILI × LAB was highly correlated with pH1N1 incidence rates (Figure 2). Third, it is unclear whether serological surveillance accurately measures the actual cumulative incidence in the population, because serum samples may not be collected from a representative sample of the population, and also because the validity and reliability of using seropositivity or seroconversion as indicators of infection have yet to be explored in detail. Finally, individuals who died would not have had a chance to seroconvert and would not be reflected in the infection fatality risk denominator. This would be a relevant consideration for studies of diseases with much greater severity than pH1N1.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong Special Administrative Region, People's Republic of China (Jessica Y. Wong, Peng Wu, Hiroshi Nishiura, Eric H. Y. Lau, Lin Yang, J. S. Malik Peiris, Joseph T. Wu, Benjamin J. Cowling); Precursory Research for Embryonic Science and Technology, Japan Science and Technology Agency, Saitama, Japan (Hiroshi Nishiura); Center for Communicable Disease Dynamics, Department of Epidemiology, Harvard School of Public Health, Boston, Massachusetts (Edward Goldstein); Centre for Health Protection, Department of Health, Government of the Hong Kong Special Administrative Region, Hong Kong Special Administrative Region, People's Republic of China (S. K. Chuang, Thomas Tsang); and Centre for Influenza Research, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong Special Administrative Region, People's Republic of China (J. S. Malik Peiris).
J.T.W. and B.J.C. are joint senior authors.
This project was supported by the Harvard Center for Communicable Disease Dynamics from the National Institute of General Medical Sciences (grant U54 GM088558), the Area of Excellence Scheme of The University of Hong Kong Grants Committee (grant AoE/M-12/06), and the US National Institutes of Health K01 award 1K01AI101010-01. H.N. received funding support from the Japan Science and Technology Corporation (JST) Precursory Research for Embryonic Science and Technology (PRESTO) Program.
We thank Horace Choi, Vicky Fang, Dr. Andrew Ho, Kathy Leung, and Dr. Irene Wong for technical support. We thank Dr. Dennis Ip and Dr. Heath Kelly for helpful discussions.
The funding bodies had no role in the study design, data collection and analysis, preparation of the manuscript, or the decision to publish.
Conflict of interest: J.S.M.P. receives research funding from Crucell N.V. J.T.W. is an associate editor of the American Journal of Epidemiology. B.J.C. has received research funding from MedImmune, Inc., and consults for Crucell N.V. The authors report no other potential conflicts of interest.
REFERENCES
- 1.Van Kerkhove MD, Asikainen T, Becker NG, et al. Studies needed to address public health challenges of the 2009 H1N1 influenza pandemic: insights from modeling. PLoS Med. 2010;7(6):e1000275. doi: 10.1371/journal.pmed.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishiura H. Case fatality ratio of pandemic influenza. Lancet Infect Dis. 2010;10(7):443–444. doi: 10.1016/S1473-3099(10)70120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu JT, Ma ES, Lee CK, et al. The infection attack rate and severity of 2009 pandemic H1N1 influenza in Hong Kong. Clin Infect Dis. 2010;51(10):1184–1191. doi: 10.1086/656740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warren-Gash C, Bhaskaran K, Hayward A, et al. Circulating influenza virus, climatic factors, and acute myocardial infarction: a time series study in England and Wales and Hong Kong. J Infect Dis. 2011;203(12):1710–1718. doi: 10.1093/infdis/jir171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson WW, Moore MR, Weintraub E, et al. Estimating influenza-associated deaths in the United States. Am J Public Health. 2009;99(suppl 2):S225–S230. doi: 10.2105/AJPH.2008.151944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charu V, Chowell G, Palacio Mejia LS, et al. Mortality burden of the A/H1N1 pandemic in Mexico: a comparison of deaths and years of life lost to seasonal influenza. Clin Infect Dis. 2011;53(10):985–993. doi: 10.1093/cid/cir644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang L, Chan KP, Cowling BJ, et al. Excess mortality associated with the 2009 pandemic of influenza A(H1N1) in Hong Kong. Epidemiol Infect. 2012;140(9):1542–1550. doi: 10.1017/S0950268811002238. [DOI] [PubMed] [Google Scholar]
- 8.Hardelid P, Pebody R, Andrews N. Mortality caused by influenza and respiratory syncytial virus by age group in England and Wales 1999–2010. Influenza Other Respi Viruses. 2013;7(1):35–45. doi: 10.1111/j.1750-2659.2012.00345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson WW, Shay DK, Weintraub E, et al. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA. 2003;289(2):179–186. doi: 10.1001/jama.289.2.179. [DOI] [PubMed] [Google Scholar]
- 10.Lipsitch M, Hayden FG, Cowling BJ, et al. How to maintain surveillance for novel influenza A H1N1 when there are too many cases to count. Lancet. 2009;374(9696):1209–1211. doi: 10.1016/S0140-6736(09)61377-5. [DOI] [PubMed] [Google Scholar]
- 11.Presanis AM, De Angelis D, Hagy A, et al. The severity of pandemic H1N1 influenza in the United States, from April to July 2009: a Bayesian analysis. PLoS Med. 2009;6(12):e1000207. doi: 10.1371/journal.pmed.1000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baguelin M, Hoek AJ, Jit M, et al. Vaccination against pandemic influenza A/H1N1v in England: a real-time economic evaluation. Vaccine. 2010;28(12):2370–2384. doi: 10.1016/j.vaccine.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Lipsitch M, Riley S, Cauchemez S, et al. Managing and reducing uncertainty in an emerging influenza pandemic. N Engl J Med. 2009;361(2):112–115. doi: 10.1056/NEJMp0904380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Census and Statistics Department. Population and vital events. Hong Kong Special Administrative Region, China: Census and Statistics Department; 2011. http://www.censtatd.gov.hk/hong_kong_statistics/statistics_by_subject/ (Accessed January 1, 2012)
- 15.Department of Health. Vital statistics. Hong Kong Special Administration Region, China: Department of Health; 2011. http://www.healthyhk.gov.hk/phisweb/en/enquiry/index.html. (Accessed January 1, 2012) [Google Scholar]
- 16.Centre for Health Protection; 2011. Sentinel surveillance. Hong Kong Special Administrative Region, China: Centre for Health Protection. http://www.chp.gov.hk/en/dns_submenu/10/26/44.html. (Accessed January 1, 2012) [Google Scholar]
- 17.Hong Kong Observatory. Climatological information services. Hong Kong Special Administrative Region, China: Hong Kong Observatory; 2011. http://www.hko.gov.hk/cis/climat_e.htm#. (Accessed January 1, 2012) [Google Scholar]
- 18.Wu JT, Ho A, Ma ES, et al. Estimating infection attack rates and severity in real time during an influenza pandemic: analysis of serial cross-sectional serologic surveillance data. PLoS Med. 2011;8(10):e1001103. doi: 10.1371/journal.pmed.1001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riley S, Kwok KO, Wu KM, et al. Epidemiological characteristics of 2009 (H1N1) pandemic influenza based on paired sera from a longitudinal community cohort study. PLoS Med. 2011;8(6):e1000442. doi: 10.1371/journal.pmed.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.R Development Core Team. Vienna, Austria: R Foundation for Statistical Computing; 2005. R: a language and environment for statistical computing. http://www.R-project.org. (Accessed January 1, 2012) [Google Scholar]
- 21.van den Wijngaard CC, van Asten L, Koopmans MPG, et al. Comparing pandemic to seasonal influenza mortality: moderate impact overall but high mortality in young children. PLoS One. 2012;7(2):e31197. doi: 10.1371/journal.pone.0031197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finelli L, Chaves SS. Influenza and acute myocardial infarction. J Infect Dis. 2011;203(12):1701–1704. doi: 10.1093/infdis/jir175. [DOI] [PubMed] [Google Scholar]
- 23.Simonsen L, Clarke MJ, Williamson GD, et al. The impact of influenza epidemics on mortality: introducing a severity index. Am J Public Health. 1997;87(12):1944–1950. doi: 10.2105/ajph.87.12.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstein E, Cobey S, Takahashi S, et al. Predicting the epidemic sizes of influenza A/H1N1, A/H3N2, and B: a statistical method. PLoS Med. 2011;8(7):e1001051. doi: 10.1371/journal.pmed.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstein E, Viboud C, Charu V, et al. Improving the estimation of influenza-related mortality over a seasonal baseline. Epidemiology. 2012;23(6):829–838. doi: 10.1097/EDE.0b013e31826c2dda. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steens A, Waaijenborg S, Teunis PF, et al. Age-dependent patterns of infection and severity explaining the low impact of 2009 influenza A (H1N1): evidence from serial serologic surveys in the Netherlands. Am J Epidemiol. 2011;174(11):1307–1315. doi: 10.1093/aje/kwr245. [DOI] [PubMed] [Google Scholar]
- 27.Hancock K, Veguilla V, Lu X, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361(20):1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 28.Presanis AM, Pebody RG, Paterson BJ, et al. Changes in severity of 2009 pandemic A/H1N1 influenza in England: a Bayesian evidence synthesis. BMJ. 2011;343:d5408. doi: 10.1136/bmj.d5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraser C, Donnelly CA, Cauchemez S, et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science. 2009;324(5934):1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Echevarria-Zuno S, Mejia-Arangure JM, Mar-Obeso AJ, et al. Infection and death from influenza A H1N1 virus in Mexico: a retrospective analysis. Lancet. 2009;374(9707):2072–2079. doi: 10.1016/S0140-6736(09)61638-X. [DOI] [PubMed] [Google Scholar]
- 31.Taubenberger JK, Morens DM. 1918 influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12(1):15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Kerkhove MD, Vandemaele KA, Shinde V, et al. Risk factors for severe outcomes following 2009 influenza A (H1N1) infection: a global pooled analysis. PLoS Med. 2011;8(7):e1001053. doi: 10.1371/journal.pmed.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muscatello DJ, Cretikos MA, Macintyre CR. All-cause mortality during first wave of pandemic (H1N1) 2009, New South Wales, Australia, 2009. Emerg Infect Dis. 2010;16(9):1396–1402. doi: 10.3201/eid1609.091723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cowling BJ, Fang VJ, Nishiura H, et al. Increased risk of noninfluenza respiratory virus infections associated with receipt of inactivated influenza vaccine. Clin Infect Dis. 2012;54(12):1778–1783. doi: 10.1093/cid/cis307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.