Abstract
This study reports the preparation and characterization of cysteine-terminated B-domain (Bd-cys) of Staphylococcus aureus protein A, in combination with IgG antibodies directed against the ρ1 and α1 subunits of GABAA receptors, for localizing reagents of interest to the target receptor. A cysteine residue was inserted at the C-terminus of the cysteine-lacking B-domain (Bd) and used for conjugating maleimide-containing compounds. As determined by ELISA, binding of a Bd-cys-S-fluorescein conjugate to polyclonal guinea pig anti-GABAA-ρ1 and rabbit anti-GABAA-α1 IgG was similar to that exhibited by full-length protein A. Surface plasmon resonance analysis of the interaction of Bd-cys-S-PEG3400-biotin conjugate with anti-GABAA-ρ1 and anti-GABAA-α1 yielded KD values of 6.4 ± 1.9 and 0.4 ± 0.1 nM, respectively. Fluorescence anisotropy analysis of the binding of Bd-cys-S-fluorescein to the two antibodies yielded respective EC50 values of 65 and 18 nM. As determined with biotin-reactive fluorescent reagents, Bd-cys-S-PEG3400-biotin specifically bound to the plasma membrane of Xenopus laevis oocytes that expressed α1β2γ2 or homomeric ρ1 GABAA receptors and were pre-treated with the corresponding anti-GABAA IgG. The IgG binding specificity and high affinity of Bd-cys conjugates illustrate the potential of these conjugates, in combination with a selected IgG, to localize compounds of interest at specific cell surface proteins.
Keywords: Protein A, B-domain scaffold, GABA receptor
Protein A (SpA), a cell wall-associated protein of Staphylococcus aureus bacteria [1], protects S. aureus against the host’s immune system by binding to host antibodies [2–5]. SpA consists of a tandem sequence of five homologous repeats termed (sequentially, from the N-terminal) domains E, D, A, B, and C [6–8]. Each of these domains is arranged as three helices [5,9], and binds to the Fc fragment of IgG [5,10,11]. The B-domain, a 58 amino acid segment (about 7 kDa) that lacks cysteine residues, has been studied extensively as a representative domain of SpA. The similar IgG-binding affinity of B-domain and parent SpA [5,11–13], together with B-domain’s ease of bacterial expression and high solubility, have motivated interest in designing affinity reagents based on the B-domain structure [2,12,14–17].
The robust IgG-binding activity of B-domain (henceforth abbreviated Bd), along with its small size and defined structure, make Bd, in combination with an IgG directed against the ectodomain of a cell surface protein, a potentially versatile scaffold for localizing reagents at the target protein. Our motivation for investigating such an approach comes, in particular, from recent studies demonstrating the light-controlled activation or blockage of ion channels by photosensitive analogs of channel ligands that are anchored to a mutant (cysteine-substituted) form of the channel [18–20]. That is, conjugating Bd with a physiologically active analog of the channel ligand, and interfacing the Bd/ligand conjugate with an IgG that binds to the channel’s ectodomain, might enable modulation of the activity of the native ion channel, avoiding the need to express a mutant (i.e., anchoring) form of the channel in the target cell. Mazzucchelli et al. [21], in a recent study addressing the targeting of HER2 membrane receptors with magnetic beads, conjugated the beads with a form of Bd that had been modified to contain multiple cysteine residues (that served as the bead anchoring site), interfaced the Bd/bead conjugate with a monoclonal anti-HER2 antibody, and, with use of FITC-labeled secondary antibody directed against the monoclonal antibody, demonstrated HER2 receptor labeling. To our knowledge, however, no one has studied the workability of engineered Bd as an IgG-associated scaffold for targeting ion channel proteins.
GABAA receptors, an abundant and widely distributed class of heteropentameric ion channels that are gated by the neurotransmitter γ-aminobutyric acid (GABA) and mediate neural signaling at numerous locations in the CNS including the retina, are a logical target for physiological modulation by a ligand localized at the receptor’s ectodomain (cf. [18–20]). Among the 19 known subunits of GABAA receptors are α1, a component of the α1β2γ2 GABAA subtype found in many neural tissues [22,23]; and ρ1, a component of GABAA-ρ receptors (also known as GABAC receptors) whose distribution includes multiple locations within the retina [23–26]. As a step toward developing a ligand-anchoring system for GABA receptors, we have prepared a cysteine-terminated form of Bd (here termed Bd-cys) and conjugated Bd-cys with maleimide-terminated test reagents. The experiments have employed a single cysteine inserted at the end of Bd’s third α-helix, a site distinct from the Fc binding positions on the first and second α-helices [2,27]. The absence of native cysteine residues in Bd [12] avoids the possibility of unwanted intra-molecular disulfide linkage to the introduced cysteine [21]. The introduced cysteine is, however, capable of forming inter-molecular disulfide linkages with another cysteine-terminated Bd, forming a Bd-cys dimer. With the use of appropriate IgG antibodies, we have tested the ability of complexes consisting of IgG and Bd-cys-tethered reagents to interact with GABAA ρ1 and α1 subunits.
MATERIALS and METHODS
Preparation of SpA, Bd and Bd-cys
Plasmid DNA from E. coli with the pGX2907 plasmid containing the S. aureus genome, including the full SpA sequence (American Type Culture Collection, ATCC 39344, Manassas, VA; SpA gene NCBI accession number J01786), was used as a template for PCR preparation of DNA fragments coding for wild-type SpA and wild-type Bd. PCR was performed using Pfu polymerase (Promega, Madison, WI) and primers designed to allow cloning of the PCR products into the Champion pET-100 TOPO-cloning and expression system (Invitrogen by Life Technologies, Carlsbad, CA), which features addition of a hexa-histidine fusion tag at the N-terminal followed by an enterokinase (EK) cleavage site to the protein of interest. Primers were: SpA (forward: 5′-CAC CGC GCA ACA CGA TGA A-3′; reverse: 5′-TTA TAG TTC GCG ACG ACG TCC A-3′); Bd (forward: 5′-CAC CGC TGA TAA CAA ATT C-3′; reverse: 5′-CTA TTT TGG TGC TTG TGC ATC-3′). PCR products were ligated to a TOPO plasmid and transformed into XL-1 One Shot TOPO competent cells, sequenced, and subcloned. In addition, the TOPO plasmid containing the coding sequence for Bd was used as a template for preparation of Bd containing cysteine at the C-terminus. The procedure for cysteine addition was similar to that described above, with the following primers (cys-codon underlined): forward: 5′-GCA CAA GCA CCA AAA TGT TAG AAG GGC GAG CTC-3′; reverse: 5′-GAG CTC GCC CTT CTA ACA TTT TGG TGC TTG TGC-3′.
Plasmid DNA of the SpA, Bd and Bd-cys clones was transformed into BL-21 star E. coli for protein expression. Lysis of the cell pellets was performed using the BugBuster with Benzonase protein extraction kit (Novagen/EMD Biosciences, Madison, WI), with lysozyme and protease inhibitor cocktail added. Proteins were purified from the supernatant by affinity chromatography using HiTrap affinity columns (GE Healthcare, Piscataway, NJ). Prior to analysis by SDS-PAGE (4–20% acrylamide gradient; BioRad, Carlsbad, CA), sample aliquots were prepared in buffer that either contained 5% (v/v) β-mercaptoethanol (β-ME) (reducing buffer) or lacked β-ME (non-reducing buffer). Gels were then run and stained by Coomassie blue.
Preparation of Bd-cys conjugates
Because affinity-purified Bd-cys consisted largely of dimers linked by a disulfide bond (see Results), linkage of Bd-cys to the investigated ligands fluorescein-5-maleimide (Fisher Scientific, Pittsburgh, PA) and maleimide-PEG3400-biotin (Laysan Bio, Arab, AL) was carried out under reducing conditions. The fluorescein-5-maleimide and maleimide-PEG3400-biotin conjugates were prepared by adding 200 µL of 6 µM (largely dimerized) Bd-cys to each of several wells of a HIS-Select high capacity (HC) nickel-coated plate (Sigma-Aldrich, St. Louis, MO), and incubation overnight at 4°C. Following three washes with TBST buffer (Tris-buffered saline with Tween20; 10 mM Tris, 150 mM NaCl, 0.1% Tween20), 200 µL of freshly prepared 100 mM dithiothreitol reducing agent (DTT) in water was added to the empty wells, and the plate was incubated for 40 min at 37°C. Immediately following five washes with TBST (~5 min total), 120 µM of thiol-reactive fluorescein-5-maleimide or maleimide-PEG3400-biotin was added, forming the thioether products Bd-cys-S-fluorescein (Bd-cys-S-FL) or Bd-cys-S-PEG3400-biotin, respectively. The plate was incubated for 2 hr at room temperature and then washed 3 times with TBST. The conjugate was removed from the plate by incubation with 500 mM imidazole buffer for 20 min at room temperature. Bd-cys conjugate solutions were then pooled, concentrated to 1–2 mg/mL, and dialyzed against PBS; aliquots were then prepared in reducing (5% β-ME) or non-reducing buffer, and analyzed by SDS-PAGE. Experiments were conducted also on preparations that, by design, were dominated by Bd-cys dimer. FL-labeled Bd-cys dimer conjugate was prepared by FL-labeling of HiTrap Ni-column purified Bd-cys dimer (4–6 FLs/protein) and purification according to manufacturer’s instructions, using the FluoroTag FL conjugation kit (FITC1-1KT; Sigma-Aldrich). The FL-labeled product was concentrated to 1–2 mg/mL and dialyzed against PBS. This FL-labeled Bd-cys dimer was then analyzed by SDS-PAGE under reducing or non-reducing conditions.
ELISA
The interaction of Bd-cys-S-FL monomer and of FL-labeled Bd-cys dimer with IgG was tested using an ELISA similar to that described [12]. Wells of HIS-Select Ni plates were incubated overnight at 4°C with 200 µL of 100 nM affinity-purified Bd-cys dimer, FL-labeled Bd-cys dimer, Bd-cys-S-FL monomer, SpA protein, or TBST buffer control (nine wells for each preparation). After three washes with TBST, three wells containing each preparation were supplemented with 200 µL of 0.5 µg/mL HRP-conjugated guinea pig IgG (Rockland Immunochemicals, Gilbertsville, PA) in binding buffer [TBST + 1% (w/v) bovine serum albumin] and incubated for 2 hr at room temperature. An additional three wells of each preparation were incubated with 200 µL of 0.5 µg/mL HRP-conjugated rabbit IgG (Rockland Immunochemicals) in binding buffer, and three wells of each preparation were similarly incubated with binding buffer alone (TBST control wells). Wells were washed three times with TBST buffer; 200 µL of 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) liquid substrate (Sigma-Aldrich) was then added, and the plate was incubated at room temperature for 30–40 min. Three separate wells received the ABTS substrate only (i.e., no protein preparation and no antibody), and termed blank wells. Raw absorbance values at λ = 405 nm (A405) were determined (GENios Pro plate reader; Tecan Group Ltd., Männedorf, Switzerland). For each well, net absorbance was defined as [A405(sample well) – A405(blank well)].
Surface plasmon resonance (SPR)
The binding of Bd-cys-S-PEG3400-biotin to anti-GABAA-ρ1 and anti-GABAA-α1 antibodies was analyzed by SPR (Biacore T100; GE Healthcare). All SPR experiments were performed at room temperature. In the first type of experiment, which was designed to establish an antibody-binding profile, Bd-cys-S-PEG3400-biotin was immobilized on a flow cell of a precoated streptavidin biosensor chip (0.75–1.5 µg/mL) for 60 s. This immobilization step led to an increase of ~2350 response units (RU). Guinea pig anti-GABAA-ρ1 IgG [28], rabbit anti-GABAA α1 IgG (Abcam, Cambridge, MA), or control antibodies [rat IgG2a (Abcam), chicken IgY (Abcam) or anti-insulin chicken IgY (Sigma-Aldrich)] were loaded into the flow cell (0.03 mg/mL) at a rate of 10 µL/min. The chip was then washed with HBS-EP+ buffer (GE Healthcare) for 20–30 min, and 10 mM glycine buffer, pH 2.0, was added for 60 s to dissociate bound antibody prior to addition of the next antibody. The second type of experiment was designed to determine dissociation constants (KD), association rate constants (ka) and dissociation rate constants (kd) for the interaction of Bd-cys-S-PEG3400-biotin with antibody. Here, Bd-cys-S-PEG3400-biotin was immobilized on the precoated streptavidin biosensor chip with the application of 40 nM solution at 10 µL/min for 120 s; this produced a response of about 50–60 RU. Then, guinea pig anti-GABAA-ρ1, rabbit anti-GABAA-α1, or (as controls) rat IgG2a or chicken IgY, was added to the flow cell in triplicate at concentrations of 20, 10, 5, 2.5, 1.25, and 0.625 nM for 240 s, which yielded a maximum response of ~100 and ~200 RU for anti GABAA-ρ1 and anti-GABAA-α1, respectively. A wash with 10 mM glycine buffer (pH 2.0) was performed after each run to dissociate bound antibody. Raw sensograms within each experiment (triplicate samples for each of the six antibody concentrations) were analyzed by Langmuir 1:1-binding kinetics using the Biacore T100 evaluation software to obtain fitted values of KD, ka and kd. For each parameter, values obtained in three separate experiments of a given type were analyzed to yield the mean ± SD.
Fluorescence anisotropy (FA)
The binding of Bd-cys-S-FL and FL-labeled Bd-cys dimer to anti-GABAA-ρ1 and anti-GABAA-α1 antibodies was also analyzed by FA (POLAR-star OPTIMA Microplate Reader; BMG Labtech Inc., Cary, NC). Each experiment testing guinea pig anti-GABAA-ρ1 IgG and rabbit GABAA-α1 IgG, as well as rat IgG2a and chicken IgY controls, involved study of a group of 42 mixtures. Each mixture, 50 µL in volume, contained 15 nM of FL-labeled Bd-cys conjugate (Bd-cys-S-FL or FL-labeled Bd-cys dimer), and antibody at a concentration ranging from 0.078 to 200 nM (14 different concentrations; triplicate samples for each concentration). In addition, as control experiments, groups of samples (30 mixtures, each 50 µL in volume, per group) containing 15 nM of ubiquitin-FL and anti-GABAA-α1, rat IgG2a, or chicken IgY at a concentrations ranging from 1.25 to 200 nM (triplicates at each concentration) were prepared. The mixtures were incubated overnight at 4°C, added to a 96-well microplate and analyzed by FA. Using MARS data analysis software (BMG Labtech, Inc.), a 4-parameter curve was fitted to the data set. For both Bd-cys-S-FL and FL-labeled Bd-cys dimer, binding to the anti-GABAA-ρ1 and anti-GABAA-α1 antibodies was analyzed in two FA experiments, each of the type just described. The excursions of the fitted 4-parameter curves in these experiments, i.e., the vertical (ordinate) distances between the lower and upper asymptotes of the fitted curves, were, in units of milli-anisotropy (mA): 43 and 49 for (Bd-cys-S-FL, anti-GABAA-ρ1), 40 and 42 for (Bd-cys-S-FL, anti-GABAA-α1), 78 and 83 (FL-labeled Bd-cys dimer, anti-GABAA-ρ1), and 12 and 11 for (FL-labeled Bd-cys dimer, anti-GABAA-α1). For both Bd-cys-S-FL monomer and FL-labeled Bd-cys dimer, there was no change in mA with varying antibody concentration when the Bd-cys preparation was incubated with either rat IgG2a or with chicken IgY. Similarly, there was no change in mA with antibody concentration for the preparations consisting of (ubiquitin-FL, anti-GABAA-α1), (ubiquitin-FL, rat IgG2a), or (ubiquitin-FL, chicken IgY). Each data point from the experiment was referenced to the minimum (i.e., low-concentration asymptotic) value of the fitted curve and normalized to the excursion of this curve. The referenced, normalized data obtained in each experiment were averaged. Using OriginPro software (OriginLab Corporation, Northampton, MA), the averaged data were then analyzed through the equation
| (1) |
where y/y0 is the normalized anisotropy value at a given concentration of antibody, x is the antibody concentration, and EC50 is a fitted parameter.
Oocyte preparation and fluorescence imaging
All animal procedures conformed with institutional policies and with the Statement for the Use of Animals in Ophthalmic Research adopted by the Association for Research in Vision and Ophthalmology. Xenopus laevis oocytes expressing human ρ1 GABAA, or α1β2γ2 GABAA receptors (α1, β2 and γ2 subunits of rat, rat and human. respectively) were prepared as described previously [25, 29, 30]. Briefly, mRNA, obtained for each selected sequence from in vitro transcription (mMessage mMachine, Ambion Inc./Applied Biosystems by Life Technologies, Austin, TX) from linearized cDNAs, was injected into the oocytes (Drummond Nanoject II; Drummond Scientific Co., Broomall, PA). Oocytes were analyzed for GABA receptor expression after 24–72 hr incubation in physiological saline (100 mM NaCl, 2 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 5 mM HEPES, and 10 mM glucose, pH 7.2–7.4) containing 0.1 mg/mL gentamycin at 16°C. Immunofluorescence labeling of oocytes expressing either GABAA ρ1 or GABAA α1β2γ2 receptors and non-expressing control oocytes involved, sequentially, a blocking step with 10% fetal bovine serum and 1% bovine serum albumin diluted in physiological saline (1 hr, room temperature); 3–4 washes with physiological saline; and a 1-hr incubation at room temperature with 20 µg/mL of primary antibody (guinea pig anti-ρ1, rabbit anti-α1), or in the absence of antibody as a control. Following 3–4 washes with physiological saline, oocytes were incubated for 1 hr at room temperature either with 100 nM Bd-cys-S-PEG3400-biotin with streptavidin-DyLight 488 (Fisher Scientific, Pittsburgh, PA), or with 100 nM FL-conjugated Bd-cys dimer. Following six washes, oocytes were visualized and imaged using a Leica DM-IRE2 confocal microscope (Leica Microsystems Inc., Buffalo Grove, IL) at 20X magnification [28,29].
RESULTS
Preparation and cysteine reactivity of Bd-cys monomer
The Bd component of the desired conjugate consisted of monomeric Bd containing a terminating cysteine, to which the test ligand was attached. Although the absence of other cysteines in the Bd monomer precluded intra-molecular disulfide bond formation, Bd-cys monomers were capable of intermolecular disulfide linkage (i.e., dimerization). The preparation of monomeric Bd-cys thus necessitated the reduction of dimerizing disulfide bonds that formed upon the expression and affinity purification of the protein. To determine the extent of contamination of conjugated Bd-cys preparations (Bd-cys-S-FL, FL-labeled Bd-cys dimer, and Bd-cys-S-PEG3400-biotin) by Bd-cys dimers, aliquots of these preparations were analyzed by SDS-PAGE in reducing conditions (i.e., dilution in buffer containing β-ME prior to loading onto the gel) or non-reducing conditions (buffer lacking β-ME). Figure 1A shows the electrophoretic profile, in non-reducing conditions, for affinity-purified Bd-cys (lane 1). Lanes 2–4 show samples resulting from conjugation of the affinity-purified Bd-cys with maleimide-FL (lane 2), with FITC, (lane 3), and with maleimide-PEG3400-biotin (lane 4). These four samples contained varying contribution from the dimer form of Bd-cys. Specifically, as normalized to the combined density of the monomer and dimer bands in a given lane, the dimer contribution in lanes 1–4 was 0.63, 0.25, 0.65 and 0.51, respectively. When the samples were treated with β-ME prior to loading onto the gel (Fig. 1B), Bd-cys was efficiently converted to monomer (normalized dimer contributions of ≤0.1 for all lanes), consistent with the reduction of disulfide bonds under the reducing conditions. Together, the Figure 1A–B results indicated that, following the standard treatment with DTT as part of the preparation of the conjugates, contaminating dimers remained in the final product. This contamination presumably reflected incomplete reduction by DTT, and/or the re-formation of dimerizing disulfide linkages during subsequent washes (that removed DTT) or during the conjugation reaction (which was carried out in the absence of DTT). As a measure of the possible effects of dimer in the conjugate preparations, the examination of Bd-cys binding kinetics and of oocyte surface interaction (see below) also included tests of preparations that by design were dominated by dimer, i.e., not subjected to the reducing preparative step.
Fig. 1. SDS-PAGE analysis.
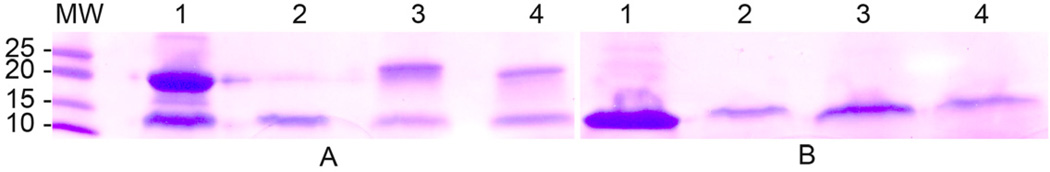
Profiles obtained for (1) affinity-purified Bd-cys, (2) Bd-cys-S-FL, (3) FL-labeled Bd-cys dimer, and (4) Bd-cys-S-PEG3400-biotin. Samples were prepared in buffer lacking β-ME (A) or containing 5% (v/v) β-ME (B) immediately prior to electrophoresis (4–20% acrylamide Tris-HCl gel). MW: molecular weight (kDa) of standards. Dimer bands were present in the profiles of all three conjugates (lanes 2–4) in non-reducing conditions (A), but not when the loading buffer contained β-ME (B).
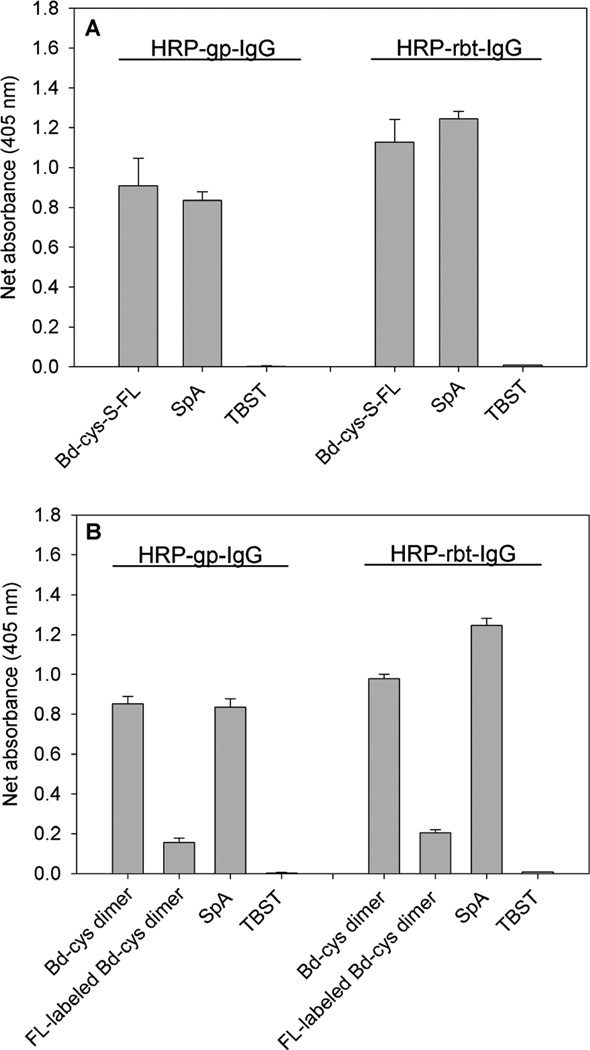
To compare the IgG binding affinity of the Bd-cys construct with that of SpA, and because guinea pig and rabbit IgGs were used as the two investigated anti-GABAA subunit antibodies, we employed ELISA to assess the binding of Bd-cys monomer and dimer to HRP-conjugated guinea pig IgG and rabbit IgG. Figure 2A shows net absorbance data (sample minus blank; see Experimental Procedures) obtained for the binding of Bd-cys-S-FL monomer and SpA to the HRP-conjugated IgG preparations. There were no significant differences in binding of Bd-cys-S-FL and SpA to HRP-guinea pig IgG (0.91 ± 0.14 and 0.84 ± 0.04, respectively; p=0.43) or to HRP-rabbit IgG (1.13 ± 0.11 and 1.25 ± 0.04, respectively; p=0.16). Furthermore, net absorbance data obtained for the binding of both Bd-cys-S-FL and SpA to both IgG preparations significantly exceeded those obtained from TBST controls from which Bd-cys-S-FL or SpA was omitted [p= 7.2 × 10−5 and 2.45 × 10−5 (HRP-guinea pig IgG), 2.42 × 10−4 and 1.28 × 10−5 (HRP-rabbit IgG), respectively]. Figure 2B shows that, as with monomer Bd-cys-S-FL in panel A, net absorbance for the binding of unlabeled Bd-cys dimer and FL-labeled Bd-cys dimer to the IgG preparations also significantly exceeded those of TBST controls [p= 2.0 × 10−6 and 2.9 × 10−4 (HRP-guinea pig IgG), 1.8 × 10−7 and 1.8 × 10−5 (HRP-rabbit IgG), respectively]. While higher than the net absorbance for TBST controls, net absorbances for the binding of FL-labeled Bd-cys dimer (0.16 ± 0.02 for HRP-guinea pig IgG, and 0.21 ± 0.01 for HRP-rabbit IgG) were significantly less than those for unlabeled Bd-cys dimer (0.85 ± 0.04 for HRP-guinea pig IgG, and 0.98 ± 0.02 for HRP-rabbit IgG) and for SpA (0.84 ± 0.04 for HRP-guinea pig IgG, and 1.25 ± 0.04 for HRP-rabbit IgG) (p < 0.001).
Fig. 2. ELISA for binding of Bd-cys preparations and SpA to HRP-conjugated guinea pig IgG (HRP-gp-IgG) and HRP-conjugated rabbit IgG (HRP-rbt-IgG).
Net absorbance (sample minus blank) data obtained at 405 nm, indicated as the mean ± SD of triplicate values. A: Bd-cys-S-FL and SpA binding vs. TBST control. B: Bd-cys dimer, FL-labeled Bd-cys dimer and SpA vs. TBST control. For both of the investigated IgGs, net absorbance obtained with the Bd-cys and SpA preparations in all cases exceeded that exhibited by the corresponding TBST control (p < 9.0 × 10−4).
Affinity and kinetics of binding to anti-GABAA-ρ1 and anti-GABAA-α1 antibodies
Surface plasmon resonance (SPR)
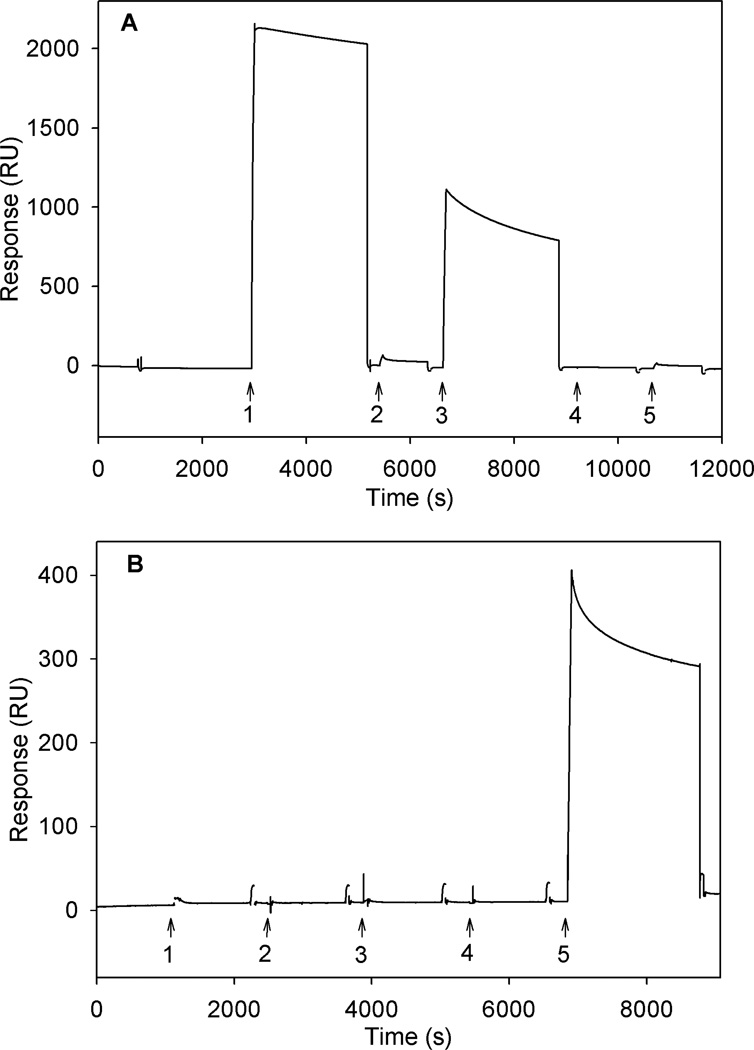
Using a streptavidin-coated chip, we examined binding of the anti-GABAA-ρ1 and anti-GABAA-α1 antibodies to a surface immobilized with Bd-cys-S-PEG3400-biotin. Figure 3 (top) shows results obtained when anti-GABAA-α1 (arrow 1) and anti-GABAA-ρ1 (arrow 3) were introduced to the flow cells containing immobilized Bd-cys-S-PEG3400-biotin. The response produced by antibody binding for anti-GABAA-α1 and anti-GABAA-ρ1 amounted to about 2000 and about 800 RU, respectively. The larger size of the response to anti-GABAA-α1 is consistent with the somewhat stronger affinity of Bd-cys-S-PEG3400-biotin to the anti-GABAA-α1 described in Figure 2A. Also shown in the Figure 3 top panel are responses to rat IgG2a (arrow 2) and chicken IgY (arrow 4), two types of immunoglobulin known not to be reactive with SpA [3,31]; no substantial signal was produced with either of these immunoglobulins, or with chicken anti-insulin IgY (arrow 5; see below). Figure 3 (bottom) shows results from an experiment in which insulin-biotin was used as a control capture agent. Here, chicken anti-insulin IgY (positive control) showed a substantial signal, but neither anti-GABAA-α1 (arrow 1), anti-GABAA-ρ1 (arrow 3), rat IgG2a (control; arrow 2) nor chicken IgY (control; arrow 4) produced a significant signal. Overall, the Figure 3 data indicate a specificity of interaction of the Bd-cys monomer with the two investigated anti-GABAA-subunit antibodies.
Fig. 3. SPR sensograms (responses in RU) for binding of IgG and IgY antibodies.
A: Bd-cys-S-PEG3400-biotin capture agent. Here and in Panel B, arrows indicate the times of introduction of (1) anti-GABAA-α1 rabbit IgG, (2) rat IgG2a, (3) anti-GABAA-ρ1 guinea pig IgG, (4) chicken IgY, and (5) anti-insulin chicken IgY. B: Insulin-biotin capture agent. In both A and B, sensogram electronic spikes of ≤ 4 s duration have been removed. Surface-bound Bd-cys-S-PEG3400-biotin thus specifically binds anti-GABAA-α1 IgG and anti-GABAA-ρ1 IgG.
Binding affinities and kinetics of the interactions of anti-GABAA-ρ1 and anti-GABAA-α1 antibodies with Bd-cys-S-PEG3400-biotin were investigated over the concentration range of 0.625 – 20 nM (10 µL/min flow rate and 240-s incubation time). To test for the possible influence of a mass transfer effect (i.e., of antibody flow rate) [32] on the kinetics of interaction of Bd-cys-S-PEG3400-biotin with the anti-GABAA-subunit antibodies, data were obtained with 2 nM antibody at varying flow rate (5, 15 and 75 µL/min). Binding profiles obtained with the three flow rates produced responses within the range 43.5–45.4 RU, indicating an essential independence of binding kinetics on flow rate and thus a minimal effect of mass transport. Global χ2 analysis of the data yielded 0.27 ± 0.18 and 1.9 ± 0.56 for the interaction of Bd-cys-S-PEG3400-biotin with anti-GABAA-ρ1 and anti-GABAA-α1, respectively (mean ± SD of χ2 determinations in three experiments for each antibody). Analysis through the Langmuir model also yielded standard errors (SEs) for the investigated parameter; the SE determinations in all cases represented less than 10% of the average value.
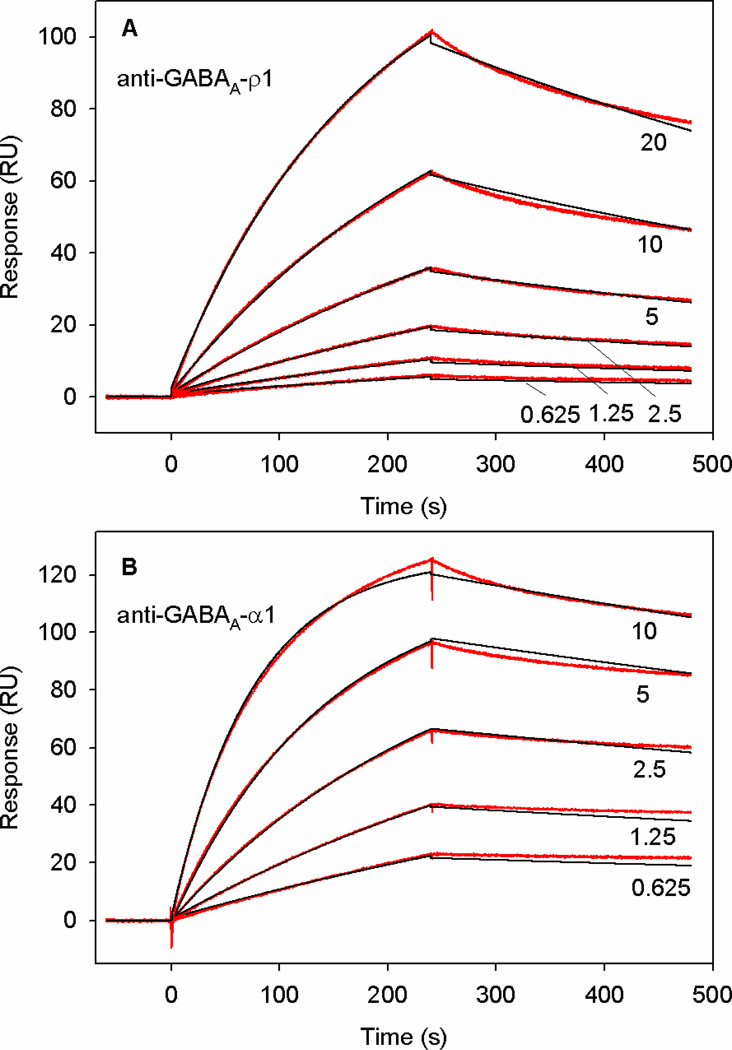
Figure 4 shows two representative plots for the binding of anti-GABAA-ρ1 (upper plot) and anti-GABAA-α1 (bottom plot) to B-domain-cys-S-PEG3400-biotin that was immobilized on a SA-coated flow cell. Each plot shows the response (RU) versus time (seconds) at 5–6 different concentrations (triplicate samples at each concentration) of the anti-GABAA-ρ1 antibody (upper plot) and anti-GABAA-α1 antibody (bottom plot), 0.625, 1.25, 2.5, 5.0, 10.0, and 20.0 nM. The plots also show the fitting of the binding curves to a 1:1 ratio binding fit using the Biacore evaluation software. The plots show the close fit to the binding curves for anti-GABAA-ρ1 and anti-GABAA-α1 antibodies. Table 1 shows KD values of 6.4 ± 1.9 nM and 0.4 ± 0.1 nM determined for the interaction of Bd-cys-S-PEG3400-biotin with anti-GABAA-ρ1 and anti-GABAA-α1, respectively. These KD values, in addition to ka and kd data in Table 1, indicate high affinity of this Bd-cys monomer conjugate for anti-GABAA-ρ1 guinea pig IgG and anti-GABAA-α1 rabbit IgG. The ka and KD values for the binding of Bd-cys-S-PEG3400-biotin to anti-GABAA-α1 rabbit IgG indicate slightly higher affinity for this IgG than for the guinea pig IgG. Data obtained with the addition of rat IgG2a and chicken IgY negative controls to immobilized Bd-cys-S-PEG3400-biotin and insulin-biotin, as well as the addition of the rabbit anti-GABAA-α1 IgG and guinea pig anti-GABAA-ρ1 IgG to immobilized insulin-biotin, showed no change in the response and thus indicated the absence of binding.
Fig. 4. SPR sensograms (RU) for kinetics of binding of anti-GABAA-ρ1 and anti-GABAA-α1 to immobilized Bd-cys-S-PEG3400-biotin.
A: Red: sensograms obtained with anti-GABAA-ρ1 at 20, 10, 5, 2.5, 1.25, and 0.625 nM. Black: fitted curves obtained using Biacore software in a 1:1 binding analysis. B: Sensograms (red) and fitted curves (black) obtained with the addition of anti-GABAA- α1 at 10, 5, 2.5, 1.25 and 0.625. nM. Binding kinetics of both antibodies to Bd-cys-S-PEG3400-biotin are well described by Langmuir 1:1 binding analysis (see Methods).
Table 1. Kinetic parameters.
Rows 1–3: SPR determinations of binding constant (KD), association rate constant (ka), and dissociation rate constant (kd), for the binding of anti-GABAA-ρ1 and anti-GABAA-α1 to Bd-cys-S-PEG3400-biotin capture agent. Data for a given antibody were obtained in 3 experiments, each involving triplicate samples at each investigated antibody concentration. Rows 4–5: FA determinations of EC50 for binding of Bd-cys-S-FL monomer and FL-labeled Bd-cys dimer to anti-GABAA-ρ1 and anti-GABAA-α1. EC50 values derived from eq. 1 fitting to data obtained in 1 (anti-GABAA-ρ1) or 2 (anti-GABAA-α1) experiments, each involving triplicate samples at each of 14 concentrations of antibody.
| Row | Method | Preparation | Parameter | Anti-GABAA-ρ1 | Anti-GABAA-α1 |
|---|---|---|---|---|---|
| 1 | SPR | Bd-cys-S-PEG3400-biotin | KD (nM) | 6.4 ± 1.9 | 0.4 ± 0.1 |
| 2 | SPR | Bd-cys-S-PEG3400-biotin | ka (M−1 s−1) | 2.1 × 105 ± 0.3 × 105 | 1.3 × 106 ± 5.0 × 104 |
| 3 | SPR | Bd-cys-S-PEG3400-biotin | kd (s−1) | 1.3 × 10−3 ± 2.3 × 10−4 | 4.8 × 10−4 ± 1.0 × 10−4 |
| 4 | FA | Bd-cys-S-FL | EC50 (nM) | 65 | 18 |
| 5 | FA | FL-labeled Bd-cys dimer | EC50 (nM) | 93 | 25 |
Fluorescence Anisotropy
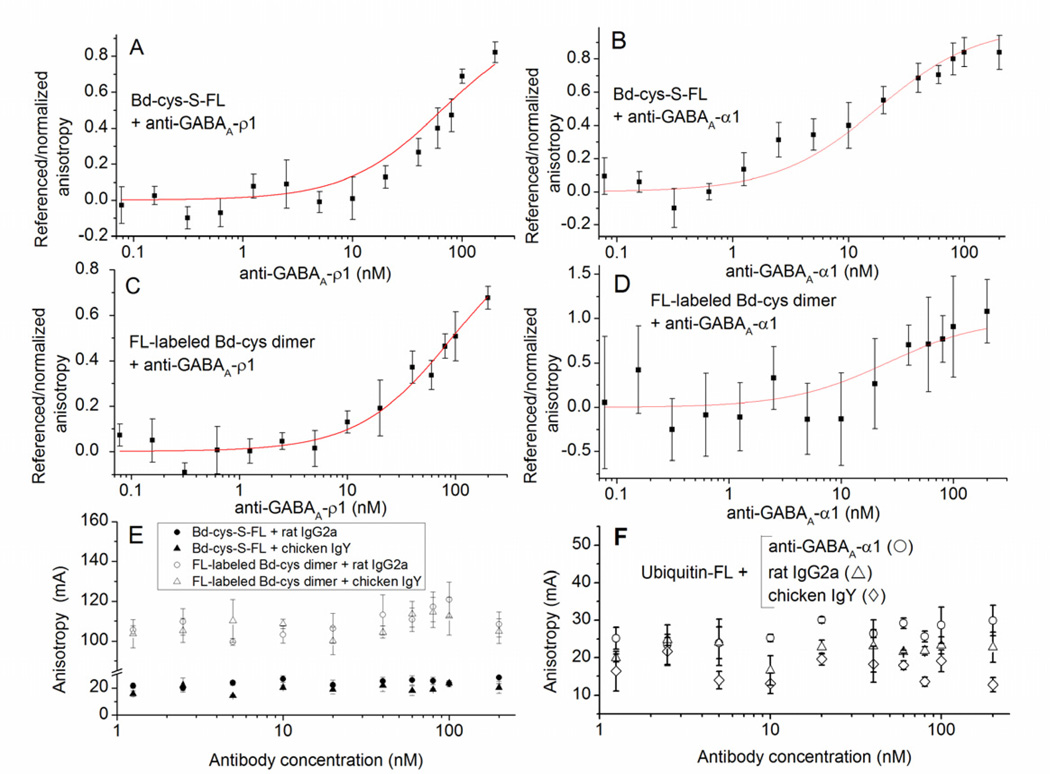
In FA experiments, we investigated the binding of (monomeric) Bd-cys-S-FL, FL-labeled Bd-cys dimer, and (as a negative control) FL-labeled ubiquitin to anti-GABAA-ρ1 and anti-GABAA-α1, as well as to (negative controls) rat IgG2a and chicken IgY. Figure 5A–B shows data for referenced/normalized anisotropy as a function of antibody concentration (nM) for the binding of Bd-cys-S-FL to anti-GABAA-ρ1 (A) and anti-GABAA-α1 (B). Analysis of these data through eq. 1 yielded EC50 values of 65 nM and 18 nM, respectively (Table 1). Similarly, Figure 5C–D illustrate plots of referenced/normalized anisotropy vs. antibody concentration for the binding of FL-labeled Bd-cys dimer to anti-GABAA-ρ1 (C) and anti-GABAA-α1 (D); analysis of these plots yielded EC50 values of 93 nM and 25 nM, respectively (Table 1). (The Fig. 5D and Table 1 results describing the interaction of FL-labeled Bd-cys dimer with anti-GABAA-α1 should be taken as only a relatively rough estimate, as the absolute range of FA signal amplitudes obtained for this interaction were relatively small (see Methods text noting excursions of fitted curves in the FA analysis). We furthermore examined, by FA, the binding of Bd-cys-S-FL and FL-labeled-Bd-cys dimer to control immunoglobulins that do not bind to SpA (rat IgG2a and chicken IgY) [3]. Shown in Figure 5E are raw data for anisotropy (mA) vs. antibody concentration (nM) obtained for the binding of Bd-cys-S-FL (closed symbols) and FL-labeled Bd-cys dimer (open symbols) to rat IgG2a (circles) and chicken IgY (triangles). By contrast with results obtained for binding to anti-GABAA-ρ1 and anti-GABAA-α1, none of the four conditions examined in Figure 4E yielded a systematic increase in anisotropy with increasing antibody concentration; analysis of the data failed to yield a smooth function of substantial ordinate excursion, and thus the data could not be normalized as in Fig. 4A–D. Control experiments (Fig. 5F) testing the binding of FL-labeled ubiquitin (8.5 kDa) to anti-GABAA-α1, rat IgG2a and chicken IgY also failed to yield a systematic increase in anisotropy with increasing antibody concentration.
Fig. 5. Binding of Bd-cys preparations to anti-GABAA-subunit antibodies determined by FA.
The horizontal axis in each panel represents concentration of the tested antibody. (A–D) Referenced/normalized anisotropy in relation to antibody concentration (mean ± SD of values obtained in ≥ 2 experiments, each involving duplicate/triplicate samples). A: Bd-cys-S-FL with anti-GABAA-ρ1. Here and in (B–D), curves illustrate equation (1) fitted to the data. B: Bd-cys-S-FL with anti-GABAA-α1. C: FL-labeled Bd-cys dimer with anti-GABAA-ρ1. D: FL-labeled Bd-cys dimer with anti-GABAA-α1. (E–F) Anisotropy (mA) in relation to antibody concentration (mean ± SD of values obtained in one experiment, each involving triplicate samples). E: Anisotropy (mA) for Bd-cys preparations with control antibodies. Mixtures were: Bd-cys-S-FL with rat IgG2a (filled circles); Bd-cys-S-FL with chicken IgY (filled triangles); FL-labeled Bd-cys dimer with rat IgG2a (open circles); and FL-labeled Bd-cys dimer with chicken IgY (open triangles). F: Anisotropy (mA) for ubiquitin-FL with anti-GABAA-α1 (open circles), rat IgG2a (open triangles) and chicken IgY (open diamonds). An increase in anisotropy with antibody concentration occurred only when the antibody incubated with the Bd preparation was anti-GABAA-ρ1 or anti-GABAA-α1.
Binding of B-domain/IgG to oocytes expressing GABAA receptors
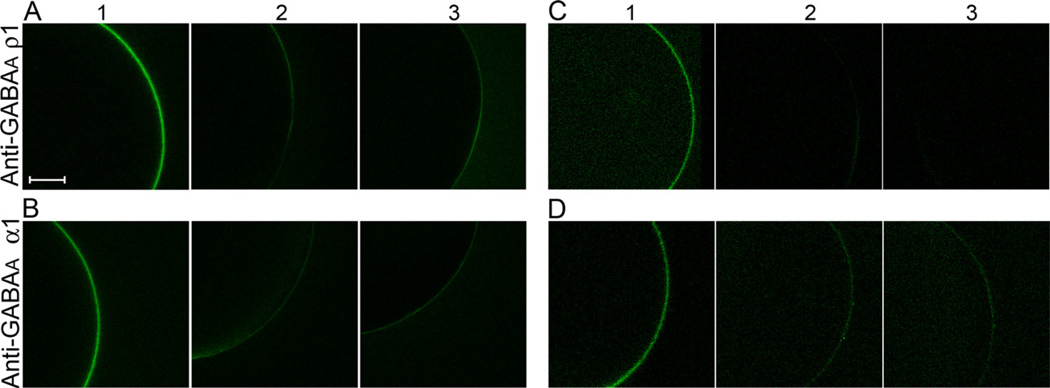
By fluorescence microscopy we investigated the interaction of Bd-cys monomer conjugates with anti-GABAA-subunit antibodies that were pre-bound to the surface membrane of Xenopus oocytes expressing homomeric GABAA-ρ1 and α1β2γ2 GABAA receptors (Fig. 6). In the experiment described by column 1 of Figure 6A, a GABAA-ρ1-expressing oocyte was treated, sequentially, with anti-GABAA-ρ1 guinea pig IgG, Bd-cys-S-PEG3400-biotin, and streptavidin-DyLight 488 (see Experimental Procedures). This treatment led to the appearance of a fluorescence halo at the surface membrane of the (approximately spherical) oocyte. Figure 6A columns 2–3 show results obtained from two control oocytes: a GABAA-ρ1-expressing oocyte that was treated only with Bd-cys-S-PEG3400-biotin and streptavidin-DyLight 488 (i.e., primary antibody omitted) (column 2), and a non-expressing oocyte that received the same treatment as the oocyte of column 1. As illustrated in the columns 2–3 images, both control preparations exhibited little, if any, fluorescence halo. In summary, the Figure 6A data indicate the binding of anti-GABAA-ρ1 antibody to the GABAA-ρ1-expressing oocyte, and the binding of Bd-cys-S-PEG3400-biotin to the receptor-bound primary antibody. As shown in Figure 6B, treatment of α1β2γ2 GABAA-expressing oocytes with Bd-cys-S-PEG3400-biotin similarly indicated specific binding of the Bd-cys-S-PEG3400-biotin to receptor-bound anti-GABAA-α1 antibody. In addition, FL-labeled Bd-cys dimer exhibited specific binding to receptor-bound anti-GABAA-ρ1 and anti-GABAA-α1 IgG (Figs. 6C–D).
Fig. 6. Binding of Bd-cys preparations and anti-GABAA-subunit antibodies to GABAA-expressing oocytes.
A: GABAA-ρ1-expressing oocyte with anti-GABAA-ρ1, Bd-cys-S-PEG3400-biotin and streptavidin-DyLight 488 (column 1) or with Bd-cys-S-PEG3400-biotin and streptavidin-DyLight 488 only (column 2). Column 3: non-expressing oocyte with same treatment as in column 1. B: α1β2γ2 GABAA-expressing oocyte with anti-GABAA-α1, Bd-cys-S-PEG3400-biotin and streptavidin-DyLight 488 (column 1) or with Bd-cys-S-PEG3400-biotin and streptavidin-DyLight 488 only (column 2). Column 3: non-expressing oocyte with same treatment as in column 1. C: GABAA-ρ1-expressing oocyte with anti-GABAA-ρ1 and FL-labeled Bd-cys dimer (column 1) or with FL-labeled Bd-cys dimer only (column 2). Column 3: non-expressing oocyte with same treatment as in column 1. D: α1β2γ2 GABAA-expressing oocyte with anti-GABAA-α1 and FL-labeled Bd-cys dimer (column 1) or with FL-labeled Bd-cys dimer only (column 2). Column 3: non-expressing oocyte with same treatment as in column 1. Similar microscope acquisition settings were used for all conditions. For visual clarity, a fixed adjustment of brightness and contrast was applied to all images of C and D. Scale bar = 150 µm. The data show that the binding of Bd-cys-S-PEG3400-biotin (A-B) and of FL-labeled Bd-cys dimer (C-D) to the oocyte surface membrane depends on both the expression of GABAA receptor (GABAA-ρ1 in A and C; α1β2γ2 GABAA in B and D) and the presence of the cognate antibody.
DISCUSSION
The present study describes the preparation of cysteine-terminated B-domain of SpA, and, in combination with IgG antibodies, the investigation of Bd-cys as a scaffold for localizing compounds of interest to oocyte-expressed ρ1 and α1β2γ2 GABAA receptors. The results show that when Bd-cys is conjugated with either maleimide-FL or maleimide-PEG3400-biotin, binding affinities of the conjugate for anti-GABAA-ρ1 and anti-GABAA-α1 IgG are similar to those exhibited by full-length SpA (Figs. 2A–B). This finding, and the observed binding of Bd-cys-S-PEG3400-biotin/IgG complex to oocyte-expressed ρ1 and α1β2γ2 GABAA receptors (Fig. 5), provide direct evidence that C-terminal cysteine insertion and ligand conjugation preserve a robust affinity of Bd for IgG in vitro and in a receptor-containing cell membrane environment.
The SPR and FA results provide quantitative information on the Bd-cys/IgG interaction. For the binding of Bd-cys-S-PEG3400-biotin to anti-GABAA-ρ1 and anti-GABAA-α1, SPR data yielded respective KD values of 6.4 ± 1.9 and 0.4 ± 0.1 nM, and ka values of 2.1 × 105 ± 0.3 × 105 and 1.3 × 106 ± 5.0 × 104 M−1 s−1 (Table 1), consistent with previously reported values for SpA and its domains [5,11,33–35]. The FA experiments yielded EC50 values of 65 and 18 nM, respectively, for the binding of the Bd-cys-S-FL monomer to guinea pig anti-GABAA-ρ1 and rabbit anti-GABAA-α1 antibodies, respectively (Table 1). These nM-range values of KD and EC50 indicate strong affinity of the Bd-cys conjugate for the two antibodies, and are in agreement with the range of reported values for the unlabeled and cysteine free SpA, B-domain and other single (and double) domains (about 2–70 nM) [5,33,36,37]
Previous studies employing radioimmunoassay have found that the affinities of SpA for guinea pig and rabbit IgG are similar [3,31,38]. However, the KD and EC50 determinations from the SPR and FA experiments, respectively, indicate that the affinity of Bd-cys conjugates for the anti-GABAA-α1 antibody exceeds that for the anti-GABAA-ρ1 antibody (Fig. 3A, Fig. 5A–D and Table 1). This relationship is consistent also with the results obtained by ELISA (Fig. 2). The higher affinity of Bd-cys for the anti-GABAA-α1 could reflect a tighter binding of SpA (and its domains) to rabbit IgG as compared to guinea pig IgG. That is, because SpA and its domains bind to the Fc fragment and not the Fab fragment of IgG, the binding of SpA and B-domain to IgG is species- and isotype-dependent. Alternatively, the evident difference in relative affinity for anti-GABAA-α1 vs. anti-GABAA-ρ1 determined by SPR and FA could reflect a differential sensitivity of the two analytical techniques to signals arising from complexes in which anti-GABAA-α1 vs. anti-GABAA-ρ1 participate.
The present study included tests of the activity of Bd-cys dimer, a moiety present in Bd-cys monomer preparations (Fig. 1). The FA data indicate that FL-labeled Bd-cys dimer has substantial affinity for the investigated anti-GABAA-ρ1 and anti-GABAA-α1 antibodies, although the affinity of the dimer for a given antibody appears less than that exhibited by the Bd-cys monomer preparation (Fig. 5). This somewhat lower antibody-binding affinity of FL-labeled dimer might be a consequence of masking, by the FL label, of residues that are within the IgG binding region of the dimer [35]. In addition, the ELISA results of Fig. 2 indicate significantly lower net absorbance values for the FL-labeled Bd-cys dimer than for unlabeled Bd-cys dimer and the FL-labeled monomer, indicating relatively weak antibody binding for the FL-labeled Bd-cys dimer. The larger difference observed in the ELISA assay for the antibody binding affinities of FL-labeled Bd-cys dimer vs. FL-labeled monomer could be due to the use of different antibodies in the ELISA vs. FA experiments. Because the SA-coated surface of the SPR flow cell captured only (monomeric) Bd-cys-S-PEG3400-biotin, Bd-cys dimer did not contribute to the detected SPR signals. In the FA experiments testing Bd-cys-S-FL, contaminating Bd-cys dimer (which lacked cys-S-conjugated FL) was not a reporting moiety but presumably bound antibodies in solution and rendered them unavailable for binding to the Bd-cys-S-FL. The evident variation among samples in the amount of dimer present (Fig. 1) could be due to differences in preparative conditions, or to the difference in the maleimide compounds added, i.e., maleimide-FL vs. maleimide-PEG3400-biotin. A bulkier maleimide compound, for example, could exhibit a slower reaction with monomeric Bd-cys, allowing more time for reduced sulfhydryl groups to combine and form dimers.
The present findings extend the approach described by Mazzucchelli et al. [21], and demonstrate specifically the ability of Bd-cys, in combination with a SpA-binding antibody, to localize Bd-cys-bound compounds at transmembrane ion channels. By contrast with direct functionalization of the antibody through technologies that often yield conjugates of multiple and variable valency (e.g., 1-ethyl-3-(3-dimethylaminopropyl) carbo-diimide (EDC) coupling of the ligand to lysine residues), the approach studied here enables localization of the ligand at a single, defined site on the Bd-cys scaffold, and thus (given the specificities of Bd’s attachment to IgG and of IgG’s attachment to the target protein) a uniformity of spatial positioning of the ligand relative to the target protein’s structure. Of particular note is our finding that conjugation of Bd-cys with a long-chain PEG moiety (maleimide-PEG3400-biotin) preserves robust binding of the conjugate/antibody complex to the target membrane protein. That is, both high-affinity interaction of the Bd-cys scaffold for anti-GABAA antibody, and high-affinity interaction of the complex with the investigated GABAA receptors, are maintained in the presence of the long, highly flexible PEG3400. Particularly in cases where the cysteine site of the Bd-cys may be relatively distant from the target protein’s ligand-binding site, Bd-cys/antibody complexes that employ a long-chain linker for ligand tethering to Bd-cys may prove useful for achieving localized physiological control of GABA receptors as well as other transmembrane ion channels [18–20].
ACKNOWLEDGMENTS
We thank Dr. Ambarish Pawar, Ms. Ruth Zelkha and Ms. Feng Feng for their assistance, respectively, with preparation of Bd-cys/antibody complexes, confocal microscopy, and protein and oocyte preparation. This work was supported by NIH grants EY016094 and EY001792; the Daniel F. and Ada L. Rice Foundation (Skokie, IL); Hope for Vision (Washington, DC); the American Health Assistance Foundation (Clarksburg, MD); and Research to Prevent Blindness (New York, NY).
Abbreviations
- SpA
Staphylococcus aureus protein A
- Bd
B-domain
- Bd-cys
cysteine-terminated B-domain
- FL
fluorescein
- SPR
surface plasmon resonance
- FA
fluorescence anisotropy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Graille M, Stura EA, Corper AL, Sutton BJ, Taussig MJ, Charbonnier J-B, Silverman GJ. Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5399–5404. doi: 10.1073/pnas.97.10.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cedergren L, Andersson R, Jansson B, Uhlén M, Nilsson B. Mutational analysis of the interaction between staphylococcal protein A and human IgG1. Protein Eng. 1993;6:441–448. doi: 10.1093/protein/6.4.441. [DOI] [PubMed] [Google Scholar]
- 3.Langone JJ. 125I-Labeled protein A: reactivity with IgG and use as a tracer in radioimmunoassay. Methods Enzymol. 1980;70(A):356–375. doi: 10.1016/s0076-6879(80)70064-2. [DOI] [PubMed] [Google Scholar]
- 4.MacSween JM, Eastwood SL. Recovery of antigen from Staphylococcal protein A-antibody adsorbents. Methods Enzymol. 1981;73:459–471. [PubMed] [Google Scholar]
- 5.Ljungberg UK, Jansson B, Niss U, Nilsson R, Sandberg BE, Nilsson B. The interaction between different domains of staphylococcal protein A and human polyclonal IgG, IgA, IgM, and F(ab’)2: separation of affinity from specificity. Mol. Immunol. 1993;30:1279–1285. doi: 10.1016/0161-5890(93)90044-c. [DOI] [PubMed] [Google Scholar]
- 6.Uhlén M, Guss B, Nilsson B, Gatenbeck S, Philipson L, Lindberg M. Complete sequence of the staphylococcal gene encoding protein A. J Biol. Chem. 1984;259:1695–1702. [PubMed] [Google Scholar]
- 7.Sjödahl J. Structural studies on the four repetitive Fc-binding region in protein A from Staphylococcus aureus . Eur. J. Biochem. 1977;78:471–490. doi: 10.1111/j.1432-1033.1977.tb11760.x. [DOI] [PubMed] [Google Scholar]
- 8.Sjödahl J. Repetitive sequences in protein A from Staphylococcus aureus . Eur. J. Biochem. 1977;73:343–351. doi: 10.1111/j.1432-1033.1977.tb11324.x. [DOI] [PubMed] [Google Scholar]
- 9.Nilsson B, Moks T, Jansson B, Abrahmsén L, Elmblad A, Holmgren E, Henrichson C, Jones TA, Uhlén M. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987;1:107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- 10.Moks T, Abrahmsén L, Nilsson B, Hellman U, Sjöquist J, Uhlén M. Staphylococcal protein A consists of five IgG-binding domains. Eur. J. Biochem. 1986;156:637–643. doi: 10.1111/j.1432-1033.1986.tb09625.x. [DOI] [PubMed] [Google Scholar]
- 11.Oda M, Kozono H, Morii H, Azuma T. Evidence of allosteric conformational changes in the antibody constant region upon antigen binding. Int. Immunology. 2003;15:417–426. doi: 10.1093/intimm/dxg036. [DOI] [PubMed] [Google Scholar]
- 12.Bratkoviĉ T, Berlec A, Popoviĉ T, Lunder M, Kreft S, Urleb U, Ŝtrukelj B. Staphylococcal protein A’s IgG-binding domain with cathepsin L inhibitory activity. Biochem. Biophys. Res. Commun. 2006;349:449–453. doi: 10.1016/j.bbrc.2006.08.078. [DOI] [PubMed] [Google Scholar]
- 13.Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.9-A resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 14.Wikman M, Steffen A-C, Gunneriusson E, Tolmachev V, Adams GP, Carlsson J, Ståhl S. Selection and characterization of HER2/neu-binding affibody ligands. Protein Eng. Des. Sel. 2004;17:455–462. doi: 10.1093/protein/gzh053. [DOI] [PubMed] [Google Scholar]
- 15.Steffen A-C, Wikman M, Tolmachev V, Adams GP, Nilsson FY, Ståhl S, Carlsson J. In vitro characterization of a bivalent anti-HER-2 affibody with potential for radionuclide-based diagnostics. Cancer Biother. Radiopharm. 2005;20:239–248. doi: 10.1089/cbr.2005.20.239. [DOI] [PubMed] [Google Scholar]
- 16.Wikman M, Rowcliffe E, Friedman M, Henning P, Lindholm L, Olofsson S, Ståhl S. Selection and characterization of an HIV-I gp120-binding affibody ligand. Biotechnol. Appl. Biochem. 2006;45:93–105. doi: 10.1042/BA20060016. [DOI] [PubMed] [Google Scholar]
- 17.Mume E, Orlova A, Larsson B, Nilsson A-S, Nilsson FY, Sjöberg S, Tolmachev V. Evaluation of ((4-hydroxyphenyl)ethyl)maleimide for site-specific radiobromination of anti-HER2 affibody. Bioconjug. Chem. 2005;16:1547–1555. doi: 10.1021/bc050056o. [DOI] [PubMed] [Google Scholar]
- 18.Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat. Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, Trauner D. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat. Chem. Biol. 2006;2:47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tochitsky I, Banghart MR, Mourot A, Yao JZ, Gaub B, Kramer RH, Trauner D. Optochemical control of genetically engineered neuronal nicotinic acetylcholine receptors. Nat. Chem. 2012;4:105–111. doi: 10.1038/nchem.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazzucchelli S, Colombo M, De Palma C, Salvadè A, Verderio P, Coghi MD, Clementi E, Tortora P, Corsi F, Prosperi D. Single-domain protein A-engineered magnetic nanoparticles: toward a universal strategy to site-specific labeling of antibodies for targeted detection of tumor cells. ACS Nano. 2010;4:5693–5702. doi: 10.1021/nn101307r. [DOI] [PubMed] [Google Scholar]
- 22.Farrant M, Nusser Z. Variations on an inhibitory theme: phasic and tonic activation of GABAA receptors. Nat. Rev. Neurosci. 2005;6:215–229. doi: 10.1038/nrn1625. [DOI] [PubMed] [Google Scholar]
- 23.Olsen RW, Sieghart W. GABAA receptors: subtypes provide diversity of function and pharmacology. Neuropharmacology. 2009;56:141–148. doi: 10.1016/j.neuropharm.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enz R, Brandstatter JH, Hartveit E, Wässle H, Bormann J. Expression of GABA receptors ρ1 and ρ2 subunits in the retina and brain of the rat. Eur. J. Neurosci. 1995;7:1495–1501. doi: 10.1111/j.1460-9568.1995.tb01144.x. [DOI] [PubMed] [Google Scholar]
- 25.Qian H, Hyatt G, Schanzer A, Hazra R, Hackam AS, Cutting GR, Dowling JE. A comparison of GABAC and ρ subunit receptors from the white perch retina. Vis. Neurosci. 1997;14:843–851. doi: 10.1017/s0952523800011585. [DOI] [PubMed] [Google Scholar]
- 26.Lukasiewicz PD, Eggers ED, Sagdullaev BT, McCall MA. GABAC receptor-mediated inhibition in the retina. Vision Res. 2004;44:3289–3296. doi: 10.1016/j.visres.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 27.Gouda H, Torigoe H, Saito A, Sato M, Arata Y, Shimada I. Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures. Biochemistry. 1992;31:9665–9672. doi: 10.1021/bi00155a020. [DOI] [PubMed] [Google Scholar]
- 28.Gussin HA, Khasawneh FT, Xie A, Feng F, Memic A, Qian H, Le Breton GC, Pepperberg DR. Subunit-specific polyclonal antibody targeting human ρ1 GABAC receptor. Exp. Eye Res. 2011;93:59–64. doi: 10.1016/j.exer.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gussin HA, Tomlinson ID, Little DM, Warnement MR, Qian H, Rosenthal SJ, Pepperberg DR. Binding of muscimol-conjugated quantum-dots to GABAC receptors. J. Am. Chem. Soc. 2006;128:15701–15713. doi: 10.1021/ja064324k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vu TQ, Chowdhury S, Muni NJ, Qian H, Standaert RF, Pepperberg DR. Activation of membrane receptors by a neurotransmitter conjugate designed for surface attachment. Biomaterials. 2005;26:1895–1903. doi: 10.1016/j.biomaterials.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Richman DD, Cleveland PH, Oxman MN, Johnson KM. The binding of staphylococcal protein A by the sera of different animal species. J. Immunol. 1982;128:2300–2305. [PubMed] [Google Scholar]
- 32.Baglia FA, Badellino KO, Ho DH, Dasari VR, Walsh PN. A binding site for the kringle II domain of prothrombin in the apple 1 domain of factor XI. J. Biol. Chem. 2000;275:31954–31962. doi: 10.1074/jbc.M005465200. [DOI] [PubMed] [Google Scholar]
- 33.Arouri A, Garidel P, Kliche W, Blume A. Hydrophobic interactions are the driving force for the binding of peptide mimotopes and Staphylococcal protein A to recombinant human IgG1. Eur. Biophys. J. 2007;36:647–660. doi: 10.1007/s00249-007-0140-8. [DOI] [PubMed] [Google Scholar]
- 34.Saha K, Bender F, Gizeli E. Comparative study of IgG binding to proteins G and A: nonequilibrium kinetic and binding constant determination with the acoustic waveguide device. Anal. Chem. 2003;75:835–842. doi: 10.1021/ac0204911. [DOI] [PubMed] [Google Scholar]
- 35.Braisted AC, Wells JA. Minimizing a binding domain from protein A. Proc. Natl. Acad. Sci. USA. 1996;93:5688–5692. doi: 10.1073/pnas.93.12.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linhult M, Gülich S, Gräslund T, Simon A, Karlsson M, Sjöberg A, Nord K, Hober S. Improving the tolerance of a protein A analogue to repeated alkaline exposures using a bypass mutagenesis approach. Proteins. 2004;55:407–416. doi: 10.1002/prot.10616. [DOI] [PubMed] [Google Scholar]
- 37.Starovasnik MA, Braisted AC, Wells JA. Structural mimicry of a native protein by a minimized binding domain. Proc. Natl. Acad. Sci. USA. 1997;94:10080–10085. doi: 10.1073/pnas.94.19.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunda MJ, Minden P, Sharpton TR, McClatchy JK, Farr RS. Precipitation of radiolabeled antigen-antibody complexes with protein A-containing Staphylococcus aureus . J. Immunol. 1977;119:193–198. [PubMed] [Google Scholar]