Abstract
Thioredoxin glutathione reductase from Schistosoma mansoni (SmTGR) catalyzes the reduction of both thioredoxin and glutathione disulfides (GSSG), thus playing a crucial role in maintaining redox homeostasis in the parasite. In line with this role, previous studies have demonstrated that SmTGR is a promising drug target for schistosomiasis. To aid in the development of efficacious drugs that target SmTGR, it is essential to understand the catalytic mechanism of SmTGR. SmTGR is a dimeric flavoprotein in the glutathione reductase family and it has a head-to-tail arrangement of its monomers; each subunit has the components of both a thioredoxin reductase (TrxR) domain and a glutaredoxin (Grx) domain. However, the active site of the TrxR domain is composed of residues from both subunits: FAD and a redox-active Cys-154/Cys-159 pair from one subunit and a redox-active Cys-596′/Sec-597′ pair from the other; the active site of the Grx domain contains a redox-active Cys-28/Cys-31 pair. Via its Cys-28/Cys-31 dithiol and/or its Cys-596′/Sec-597′ thiol-selenolate, SmTGR can catalyze the reduction of a variety of substrates by NADPH. It is presumed that SmTGR catalyzes deglutathionylation reactions via the Cys-28/Cys-31 dithiol. Our anaerobic titration data suggest that reducing equivalents from NADPH can indeed reach the Cys-28/Cys-31 disulfide in the Grx domain to facilitate reductions effected by this cysteine pair. To clarify the specific chemical roles of each redox-active residue with respect to its various reactivities, we generated variants of SmTGR. Cys-28 variants had no Grx glutathionylation activity whereas Cys-31 variants retained partial Grx glutathionylation activity, indicating that the Cys-28 thiolate is the nucleophile initiating deglutathionylation. Lags in the steady-state kinetics, found when wild-type (WT) SmTGR was incubated at high concentrations of GSSG, were not present in Grx variants, indicating that this cysteine pair is in some way responsible for the lags. A Sec-597 variant was still able to reduce a variety of substrates, albeit slowly, showing that selenocysteine is important but is not the sole determinant for the broad substrate tolerance of the enzyme. Our data show that Cys-520 and Cys-574 are not likely to be involved in the catalytic mechanism.
Schistosomiasis (bilharzia) caused by parasites in the genus Schistosoma is a serious tropical disease; each year more than 200 million people are infected, resulting in more than 200,000 deaths in tropical and subtropical areas. Praziquantel is the only drug currently administered to treat schistosomiasis. However, praziquantel-resistant strains have been identified both in the laboratory and endemic areas (1). Thus, should praziquantel resistance become widespread, it will be critical to develop new drugs to treat this disease. Unfortunately, development of new drugs for schistosomiasis is hampered by the low financial return on products specifically designed for diseases found in low income populations. The drug, artemether, has been investigated for schistosomiasis therapy, although its use for this therapy may be restricted in areas of malaria transmission to avoid putting its use as an antimalarial at risk (2). Oxamniquine was previously used widely in Brazil, but is no longer manufactured. Moreover, it is effective only against S. mansoni, and resistance to this drug has already been reported (3). Other potential therapies under development employ protease inhibitors and 2-(alkylamino)-1-phenyl-1-ethanethiosulfuric acids (4, 5) but these have yet to reach clinical trials. Recently, proteins involved in the maintenance of the redox status of schistosome worms have been validated as drug targets (6).
Because adult schistosome worms live in the host blood stream, they must express effective defenses in order to neutralize endogenous reactive oxygen species that are generated by mitochondrial respiration and the digestion of red blood cells, as well as exogenous reactive oxygen species that are generated by the host immune system. In most organisms, two major intracellular antioxidant pathways scavenge reactive oxygen species: the glutathione (GSH) and the thioredoxin (Trx) systems. After donating their reducing equivalents, the resulting GSSG (glutathione disulfide) and oxidized Trx molecules can be reduced by glutathione reductase (GR) or thioredoxin reductase (TrxR), respectively. Compared to mammalian cells, schistosome parasites have only limited capacity to reduce reactive oxygen species. For example, although the parasites have high levels of superoxide dismutase to convert the superoxide radical to H2O2, they have no catalase and only low levels of glutathione peroxidase activity to neutralize the resulting H2O2 (7, 8). Instead, the worms rely on peroxiredoxins to reduce H2O2 (7). Oxidized peroxiredoxin-1 can be reduced by Trx, and oxidized peroxiredoxin-2 and peroxiredoxin-3 can be reduced by either Trx or GSH (7, 8). However, in schistosome parasites neither authentic GR nor TrxR proteins are present. Instead, a multifunctional enzyme, thioredoxin glutathione reductase (SmTGR) catalyzes the reduction of both GSSG and oxidized Trx (9, 10). SmTGR is a key enzyme required to maintain redox balance of the parasites. Indeed, silencing of SmTGR by RNA interference leads to worm death, and SmTGR inhibitors have been shown to decrease worm burdens in mice (10–12), validating that SmTGR is a viable drug target. A high-throughput screening of small molecule libraries for SmTGR inhibitors has identified several chemical classes with good activity against cultured ex vivo worms and in experimental laboratory infections (6, 13, 14). A better understanding of the catalytic mechanism of SmTGR would be useful in designing more effective and selective inhibitors targeting SmTGR.
Thioredoxin glutathione reductase (TGR) (E.C. 1.8.1.B1) belongs to the pyridine nucleotide disulfide oxidoreductase family that includes TrxR, GR, peroxiredoxin reductase (AhpF), and lipoamide dehydrogenase (9, 15, 16). TGRs share high sequence similarity with high molecular weight TrxRs (high Mr TrxRs), GRs, and lipoamide dehydrogenases, but are less similar to low molecular weight TrxRs (17, 18). Although TGR in S. mansoni and other parasitic flatworms plays a major role in redox balance, TGR in mammals plays a very restricted role. In mammals, its highest level of expression is in the testes (19). The active sites of known TGRs are virtually identical; therefore, the catalytic mechanisms of TGRs are also thought to be similar. It should be noted, however, that in mammalian TGRs, only one cysteine residue is present in the Grx domain (i.e., a CysXaaXaaSer motif), whereas a CysXaaXaaCys motif exists in the non-mammalian TGR proteins as shown in Fig. S1 (19–22). TGRs are selenoproteins and are similar to selenoprotein TrxRs that have broad substrate tolerance. They are capable of reducing not only GSSG and oxidized Trx, but also low molecular weight compounds such as H2O2 and sodium selenite (10, 23). Unlike other members in the GR family, due to the additional glutaredoxin (Grx) domain at their N-termini, TGRs also can catalyze deglutathionylation reactions. Although the structure of SmTGR has been explored (12, 24), its catalytic mechanism is still unclear.
The catalytic mechanism of GR proteins has been studied extensively. The flow of electrons during GR catalysis is from NADPH to FAD to the active center disulfide (CDistalVNVGCProximal (to the flavin)) to GSSG. Although the enzyme can accept two electron-pairs per subunit (one at the flavin and one at the disulfide), the principal catalytically active forms are the two-electron reduced EH2 and the oxidized Eox forms. In the EH2 state the active-site disulfide is reduced and the FAD is oxidized. This yields a charge-transfer complex (CTC) between the proximal thiolate donor (CP) and the FAD acceptor, giving rise to a characteristic absorption band, best detected at ~540 nm; the flavin absorbance at 460 nm due to the oxidized enzyme decreases and is blue-shifted to peak at ~440 nm. These spectral properties easily distinguish EH2 from Eox or EH4, the fully reduced form, which is nearly colorless (15). The other thiol in EH2 (CD) initiates the dithiol-disulfide interchange with GSSG that involves a mixed disulfide between GSH and CD (25–29).
The catalytic mechanism of the high Mr TrxR is similar to that of GR (18, 30–35). High Mr TrxR is a dimer with each monomer containing an FAD and two pairs of redox active cysteines (or Cys-Sec). Thus, it is like GR, but with an extra C-terminal domain containing a redox active cysteine pair or a redox active Cys-selenocysteine pair that communicates with substrates. Reductive titrations and presteady-state kinetic analyses have shown that the C-terminal pair of cysteines (or Cys-Sec pair) is redox active. The active site dithiol near the flavin of one TrxR subunit reduces the C-terminal center (GlyCysSecGly, CysXaaXaaXaaXaaCys, or SerCysCysSer) of the other subunit (30–32, 35, 36). It is proposed that in high Mr TrxR, catalysis involves electron-pairs being transferred from NADPH to flavin to initially form FADH−, which very rapidly transfers an electron pair to the N-terminal disulfide center, thereby producing EH2, which exhibits a thiolate-FAD charge-transfer spectrum. Redox equilibration between the N-terminal center and the C-terminal redox center of the other subunit constitutes the next step. The 2-electron-reduced enzyme, EH2, can accept a second hydride ion from NADPH to form EH4, and similar electron transfer events lead to a species that also displays thiolate-FAD charge-transfer interactions. EH4 reacts with oxidized Trx to form a mixed disulfide; then reduced Trx is released to return the enzyme to the EH2 state. Therefore, in high Mr TrxR the catalytically active forms are EH4 and EH2. Key to the proposed catalytic mechanism of TrxR is the relatively flexible C-terminal tail that allows the transport of electron pairs from the buried N-terminal redox center to substrates at the surface (30–32, 36).
In contrast, the catalytic mechanism of TGR proteins is not yet adequately understood. Several TGRs from mammals and from microorganisms including S. mansoni have been described (9, 10, 19, 22, 24, 37, 38). Alignments of the amino acid sequences of TGRs display high similarity (greater than 50 % identity) (Fig. S1). Like other members of the GR family, SmTGR is active in a dimeric form with a head-tail arrangement of identical monomers (24). In contrast to TrxR, each subunit has two domains: a TrxR domain and a Grx domain. The active site of SmTGR is composed of residues from each subunit: an FAD, several redox-active cysteine pairs (Cys-154/Cys-159 and Cys-28/Cys-31) from one subunit, and a redox-active cysteine-selenocysteine pair (Cys-596′/Sec-597′) from the other subunit. A catalytic mechanism for TGR has been proposed that is based on the X-ray crystal structure and the mechanisms of TrxR and Grx; reducing equivalents from NADPH are passed to the FAD, subsequently to a cysteine pair adjacent to FAD (Cys-154/Cys-159), then to a cysteine-selenocysteine pair (Cys-596′/Sec-597′), and finally (when appropriate) to a cysteine pair (Cys-28/Cys-31) in the Grx domain (Scheme 1) (12, 24). The C-terminal tail, where the Cys-596′/Sec-597′ redox pair is located, should be highly mobile in order to accept electrons from Cys-154/Cys-159 and to donate electron pairs to Trx or to the N-terminal redox active Cys-28/Cys-31 pair (12, 24, 39, 40). Because TGR has an additional dithiol/disulfide redox center, EH6 forms are possible. However, it is not known which of the species, EH2, EH4, or EH6, reacts with different substrates and inhibitors. Detailed mechanistic studies on TGR have been hampered by the limited availability of recombinant TGR and the low abundance of TGR in mammalian tissues, as well as by the difficulty in obtaining native TGR from other organisms.
Scheme 1.
The proposed mechanism of SmTGR. Two types of charge-transfer complexes (FADH−–NADP+ CTC and thiolate–FAD CTC) are shown by dashed arrows. Residues numbered without a prime are from one subunit and residues numbered with a prime are from the other subunit. The proposed mechanism is based on data in this paper and previous studies (24, 34, 39, 40).
It has been shown that SmTGR reduces substrates via its redox-active dithiols and thiol-selenolate (10), but the specific function of each of the redox-active Cys and selenocysteine residues in the catalytic mechanism of SmTGR have not been established. For instance, the cysteine pair of Cys-28 and Cys-31 in the Grx domain has been proposed to be responsible for the GR and Grx activities of TGRs (24), but it still remains to be determined how this cysteine pair reduces both a mixed disulfide (a glutathionylated substrate) and GSSG. In addition, selenocysteine is thought to give SmTGR broad substrate tolerance. The effects of certain inhibitors against selenoproteins are thought to require the selenocysteine residue, i.e., the inhibitors are pro drugs that are reduced by selenoproteins to active forms that inhibit the enzyme (41–44). However, recently it has been found that auranofin (AF), a gold-containing compound and a potent TGR inhibitor, can inhibit a truncated form of SmTGR that lacks the two C-terminal amino acids, Sec-597 and Gly-598, and that inhibition of GR (an enzyme containing no Sec) by AF could be accelerated by supplementing the reaction with exogenous selenium from benzeneselenol (12). Those results suggested that selenocysteine may not be essential for the action of certain SmTGR inhibitors. Therefore, it is of interest to determine if Sec is crucial for the broad substrate range of SmTGR and for the activity of inhibitors. Two additional Cys residues also may have a role in the catalytic cycle of SmTGR. Angelucci and colleagues suggested that Cys-520 and Cys-574 may be involved in catalysis because this pair has the potential to form a disulfide by bond rotation and is conserved in TrxRs and most TGRs but not GRs (24). In addition, gold atoms from AF were found bound to three sites of SmTGR: at the putative NADPH binding site, near the Cys-154/Cys-159 couple, and to the cysteine pair of Cys-520 and Cys-574 (12, 24).
In order to investigate the functions of each of these cysteine and selenocysteine residues in the catalytic cycle, we have generated variants (Cys to Ala, Cys to Ser, or Sec to Cys), recombinantly expressed them in bacteria, and characterized their biochemical properties. Our results indicate that, as expected, the Grx domain is responsible for deglutathionylation reactions and for the reduction of GSSG; a thiolate anion on Cys-28 nucleophilically attacks the disulfide bonds of GSSG and of glutathionylated proteins, and these reactions constitute the GR and Grx activities, respectively. Sec is not essential for broad substrate tolerance because the Sec-to-Cys variant still is able to reduce low molecular weight compounds. Furthermore, the data suggest that the cysteine pair of Cys-520 and Cys-574 is likely involved in maintaining structural integrity of SmTGR rather than participating in catalysis. The results of reductive spectral titrations suggest that reducing equivalents from NADPH may be transferred to the disulfide in the Grx domain via the C-terminal tail, as suggested by the recent work of Angelucci et al (40).
MATERIALS AND METHODS
Chemicals
5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), GSSG, GSH, L-cysteine, sodium selenite, riboflavin, flavin adenine dinucleotide (FAD), β-hydroxyethyl disulfide (HED), and NADPH were purchased from Sigma-Aldrich. Isopropyl-beta-D-thiogalactopyranoside (IPTG) was from Gold Biotechnology. All other chemicals and reagents were purchased from Fisher Scientific unless stated.
Generation of SmTGR variants by site-directed mutagenesis
The construction of variants was performed using the QuikChange site-directed mutagenesis kit according to manufacturer’s instructions (Stratagene). A pET-24a plasmid encoding cDNA of SmTGR fused with a bacterial-type SECIS element was used as a template (10, 45). The primers used to clone different cysteine variants are listed in Table S1. The sequence of each clone was verified by the DNA services facility at University of Illinois at Chicago and Illinois State University.
Overexpression of SmTGR and its variants and SmTrx-1
Expression and isolation of SmTGR were carried out as described previously (10). For overexpression of SmTGR and its variants, SmTGR in pET-24a and pSUBAC were co-transformed into BL21 (DE3) purchased from Novagen (45). Two antibiotics, kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml), were applied to select cells having pET-24a-SmTGR and pSUBAC, respectively. Transformed cells were cultured in LB media at 37 °C. When OD600 reached 0.8, sodium selenite (5 μM) and L-cysteine (100 μg/ml) were added to the cell culture. Once OD600 reached 2.0, the expression of SmTGR and its variants was induced by adding IPTG (50 μM) with a supplement of riboflavin (20 μg/ml). Cells were cultured at 25 °C for 24 hours. Harvested cells were broken by alternate freeze-thaw cycles followed by sonication. FAD (100μM) was added to the lysates, which were then centrifuged at 14,000 rpm at 4 °C for 1 hour. Sodium chloride was added to the resulting supernatant to prevent E. coli GR from binding, and the mixture was loaded onto a column of 2′,5′ ADP Sepharose 4B purchased from (GE Healthcare). TE buffer was used to remove proteins that were non-specifically; TE buffer with 1 mM NADPH was used to elute SmTGR. The purity of each fraction of enzyme was analyzed by 12% SDS-PAGE. NADP+ (NADPH) was removed by a HiTrap desalting column (matrix: Sephadex G-25 superfine, cross-linked dextran) (GE Healthcare). The concentrations of enzymes were determined spectrally for bound flavin using a value of ε462nm = 11,300 M−1 cm−1 (10). It is known that the efficiency of selenocysteine insertion into selenoproteins from eukaryotic cells does not reach 100% when they are overexpressed in prokaryotic cells, even though cDNAs of selenoproteins were fused with bacterial-type SECIS element (22, 38, 45). Thus, at least two preparations were made for WT SmTGR and each of its variants in order to minimize variations in efficiency of selenocysteine incorporation. Selenium content of selected protein preparations was quantified by inductively coupled plasma atomic emission spectroscopy at the Olson Agricultural Analytical Services Laboratory at South Dakota State University.
Expression of Trx from S. mansoni (SmTrx-1) was carried out as previously described (46). Briefly, SmTrx-1 in pRSETA was expressed in BL21 (DE3). Transformed cells were cultured in LB medium with ampicillin (50 μg/ml) at 37 °C. Once OD600 reached 1, expression of SmTrx-1 was induced by addition of IPTG (1 mM) and the cells were grown for 3 additional hours. Harvested cells were broken by alternating freeze-thaw cycles followed by sonication. Cell lysates were centrifuged and the resulting supernatants were loaded into a Nickel-charged column from GE Health. A step-gradient of imidazole was used to purify SmTrx-1. The purity of SmTrx-1 was analyzed by 15% SDS-PAGE. Desalting columns were used to remove imidazole from proteins. The concentrations of SmTrx-1 were determined using an extinction coefficient at 280 nm (= 9,872 M−1 cm−1) (46).
Steady-state kinetics of WT SmTGR and its variants
All assays were performed in 0.1 M potassium phosphate buffer, 10 mM ETDA, pH 7.4 at 25 °C. In DTNB assays, the reaction rates of enzymes as a function of DTNB (Ellman’s reagent or 5,5′-dithiobis-2-nitrobenzoic acid) concentration were determined by measuring the rates of formation of 2-nitro-5-thiobenzoic acid using various DTNB concentrations and a fixed concentration of NADPH (100 μM). The formation of 2-nitro-5-thiobenzoic acid was determined spectrally using an ε412nm of 13,600 M−1 cm−1 (23). SmTrx-1 and glutathione disulfide (GSSG) were used as substrates to determine TrxR and GR activities of enzymes, respectively. Insulin (1 mg/ml) was added in the assays of TrxR activity. The activities were determined by the rates of NADPH consumption in the presence of 100 μM NADPH and various concentrations of SmTrx or GSSG. The depletion rates of NADPH were determined using an ε340nm of 6,220 M−1 cm−1. The absorbance of samples was followed with a Thermo Scientific Multiskan Spectrum Microplate Spectrophotometer using an experimentally determined pathlength correction factor. Vmax and Km values were derived from graphs of rates versus different concentrations of substrates by using the Michaelis-Menten equation (Sigmaplot).
The Grx glutathionylation activity of enzymes was determined using the method developed by Holmgren et al (47). β-Hydroxyethyl disulfide (HED) (1 mM) and 1 mM GSH were preincubated in the presence of 200 μM NADPH at 25 °C for 3 min. Then, SmTGR and yeast GR were added to the assays. The turnover numbers were determined from the consumption rates of NADPH, indicated by the decrease in absorbance at 340 nm.
Static anaerobic titration of enzymes with NADPH
Enzymes were added to an anaerobic cuvette and degassed by 10 cycles of vacuum followed by flushing with argon. A Hamilton titrating syringe containing degassed NADPH was then attached to the cuvette and small aliquots of NADPH were added. Spectra of enzymes were recorded using a Cary 3 spectrophotometer (Varian) at 25 °C. After each addition of NADPH, the enzyme was allowed to reach equilibrium before the spectrum was recorded.
Determination of substrate specificity of Sec597Cys
In order to investigate substrate specificity of Sec597Cys, different substrates that have been tested against WT enzyme were employed (10). All assays were performed in 0.1 M potassium phosphate buffer, 10 mM ETDA, pH 7.4 at 25 °C. The turnover numbers for different substrates were determined by monitoring consumption rates of NADPH in the presence of different concentrations of substrates and a fixed concentration of NADPH (100 μM). Stock solutions of sodium selenite, dehydroascorbic acid, and hydrogen peroxide were made in water; stock solutions of tert-butyl hydroperoxide, alloxan, lipoic acid, and lipoamide were made in DMSO. A solution with methanol, n-hexane and isopropanol (2:1:1) was used to dissolve ubiquinone (48).
Determination of IC50 values of different inhibitors against WT enzyme and Sec597Cys
The IC50 values of different inhibitors against WT SmTGR and the Sec597Cys variant were determined using DTNB, GR and TrxR activities of the enzymes. In assays, enzymes were preincubated with NADPH (100 μM) and various concentrations of inhibitors at room temperature for 10 min. Then, different substrates were added to the assays to initiate reactions; the concentrations of DTNB, GSSG, and SmTrx-1 were 3 mM, 100 μM, and 20 μM, respectively. The DTNB activity was determined by measuring the formation of 2-nitro-5-thiobenzoic acid which resulted in increased absorbance at 412 nm. The GR and TrxR activities were determined by consumption rates of NADPH based on decreased absorbance at 340nm. The graphs of enzymatic activities versus concentrations of inhibitors were used to determine IC50 values.
RESULTS AND DISCUSSION
The catalytic functions of Cys-28 and Cys-31 in the Grx domain of SmTGR
In addition to the reduction of GSSG, the redox-active cysteine pair of Cys-28 and Cys-31 in the Grx domain is thought to catalyze the deglutathionylation of proteins, which is important to regulatory processes in cells (49). Deglutathionylation catalyzed by Grx proteins occurs using only one of its thiols (Scheme 2A) via a mechanism of deglutathionylation that first involves nucleophilic attack of the N-terminal thiolate anion of Grx on the mixed disulfide between GSH and the substrate, thus forming a Grx-S-SG intermediate (50). At high concentrations, GSH resolves the disulfide bond between GSH and Grx, thereby releasing GSSG and reduced Grx; deprotonation of the resolving GSH thiol (to make it a good nucleophile - not shown) is the rate-limiting step (50). In contrast, at low concentrations of GSH, the Grx-S-SG intermediate can be resolved by the C-terminal cysteine of Grx, resulting in an intramolecular disulfide in oxidized Grx and the concomitant release of GSH. Because the reactions are reversible, oxidized Grx may react with excess GSH to reform the Grx-SSG again. These latter reactions are futile because no net glutathionylated mixed disulfides are consumed (50). In Grx the function of the C-terminal active site cysteine residue in this process is not fully understood (51). It has been observed that in variants involving the C-terminal active site cysteine in Grx-1 from Saccharomyces cerevisiae, and in Grx-1 and Grx-2 from humans, the Grx activity actually increases (52–54). In contrast, Grx activity was lost in analogous variants of Grx-2 from S. cerevisiae and of Grx-1 from Escherichia coli (53, 54).
Scheme 2.
(A) The monothiol mechanism of deglutathionylation catalyzed by authentic Grx. The N-terminal cysteine residue attacks the mixed disulfide bond of a glutathionylated protein; the disulfide bond of the intermediate can be resolved by GSH at high GSH concentrations (path to right), or internally by the thiolate of the C-terminal Cys residue (downward path). (B) The proposed mechanism of deglutathionylation catalyzed by the Grx domain of SmTGR. Cys-28 attacks the mixed disulfide bond of a glutathionylated protein; the disulfide bond of the SmTGR Grx domain intermediate can be resolved by GSH (right path) or internally by Cys-31 (downward path), depending on the availability of GSH. When it is resolved internally, the resulting disulfide in the SmTGR Grx domain subsequently becomes reduced by NADPH via the TrxR domain as shown. Under normal conditions, this reaction is effectively irreversible.
In order to understand the mechanism of deglutathionylation catalyzed by SmTGR, we generated several variants in the Grx domain. As shown in Table 1, Cys28Ala and Cys28Ala/Cys31Ala of SmTGR had no Grx deglutathionylation activity, indicating that Cys-28 is the cysteine residue that initiates nucleophilic attack on mixed disulfide bonds of glutathionylated proteins (or peptides). The deglutathionylation activity of Cys31Ala was only ~17 % of WT activity, indicating that this cysteine residue is also important to the Grx activity of SmTGR. In addition to our findings, Angelucci et al. showed that the Grx activity of truncated SmTGR (without Sec-597 and Gly-598) retained ~ 40 % of WT activity (24). In this truncated form of SmTGR reducing equivalents from NADPH cannot be readily transferred to the redox active disulfide in the Grx domain. It was suggested that the Grx domain of truncated SmTGR can receive reducing equivalents from GSH (without direct donation of reducing equivalents from NADPH via catalysis by the FAD) and thereby catalyze deglutathionylation reactions in a manner similar to authentic Grxs. Thus, based on our data and the findings from Angelucci et al., we propose that there are two mechanisms by which SmTGR catalyzes deglutathionylation reactions. One is a monothiol mechanism in which only Cys-28 is involved; this mechanism functions when the GSH concentration is high (Scheme 2B, top line). In this mechanism, the glutathionylated Cys-28 is resolved by GSH. In the other mechanism, once Cys-28 is glutathionylated, Cys-31 acts as a resolving cysteine residue to break the mixed disulfide bond between GSH and Cys-28, forming a disulfide bond between Cys-28 and Cys-31 of the Grx domain and releasing GSH. The resultant oxidized Grx domain can be reduced by the redox-active cysteine-selenocysteine pair of the TrxR domain (Scheme 2B, bottom reactions). This means that the cell has a functional deglutathionylation system even at low GSH concentrations because the reducing power of NADPH can be brought into play via the cysteine-selenocysteine pair.
Table 1.
The Grx glutathionylation activities of wild-type SmTGR and its variants
| Grx activity (s−1)a | % of WT | |
|---|---|---|
| WT | 27 ± 0.5 | - |
| Cys28Ala | NAb | 0 |
| Cys31Ala | 4 ± 0.2 | 15 |
| Cys28AlaCys31Ala | NAb | 0 |
| Cys31Ser | 6 ± 0.4 | 22 |
| Cys520Ala | 17 ± 1 | 63 |
| Cys574Ala | 19 ± 0.3 | 70 |
| Cys520AlaCys574Ala | 18 ± 0.6 | 67 |
| Sec597Cys | 11 ± 1.2 | 41 |
activities were determined by spectrally the oxidation of NADPH in the presence of 200 μM NADPH, 0.4 units yeast GR and SmTGR were added following 3 min preincubation with 1 mM GSH and 1 mM HED
undetectable activity
The active site of the Grx domain in mouse TGR is CysXaaXaaSer instead of Cys28XaaXaaCys31, which is found in SmTGR. Thus, it is clear that mouse TGR must act via a monothiol mechanism. As mentioned above, however, we found that the Cys31Ala variant of SmTGR had ~ 15 % of the Grx deglutathionylation activity of WT. To clarify whether the Ser in mouse TGR was responsible for making that enzyme active compared with our Cys31Ala variant, we tested whether the Grx deglutathionylation activity in the Cys31Ala variant could be rescued by introduction of serine to position 31 (analogous to mouse TGR). As shown in Table 1, the Grx activity of Cys31Ser was 22 % of WT activity, similar to that of the Cys31Ala variant, showing that Ser does not significantly rescue the Grx glutathionylation activity of Cys31Ala. The results are also consistent with our proposed mechanisms where Cys-31 is involved in the Grx deglutathionylation activity of SmTGR. Even though Cys-31 (or Ser-31) may be able to stabilize the thiolate anion on Cys-28 via a hydrogen bond, we did not observe increased Grx deglutathionylation activity in Cys31Ser. It is possible that because the rate limiting step is deprotonation of GSH, the stabilization of the thiolate anion on Cys-28 may not be as important in the deglutathionylation reactions catalyzed by SmTGR.
Recently, a mechanism for the Grx glutathionylation activity of TGR from Echinococcus granulosus (EgTGR) has been suggested by Bonilla et al. (55). They showed that the Grx glutathionylation activity of EgTGR requires the Grx domain as well as the selenocysteine residue. After the Grx domain of EgTGR interacts with glutathionylated proteins, the resultant mixed disulfide between GSH and Cys-31 (equivalent to Cys-28 of SmTGR) is resolved by the redox-active cysteine-selenocysteine pair from the TrxR domain instead of Cys-34 (equivalent to Cys-31 of SmTGR) (55). However, our data demonstrate that two mechanisms are involved in the Grx glutathionylation activity of SmTGR. The inability of EgTGR to utilize one of the mechanisms may be due to the distance between Cys-31 and Cys-34 in EgTGR being longer than that between Cys-28 and Cys-31 in SmTGR, resulting in limited accessibility of Cys-34 to Cys-31 in EgTGR. Therefore, the mixed disulfide between Cys-34 and GSH can only be resolved by the redox-active cysteine-selenocysteine in EgTGR.
As mentioned previously, unlike TrxR, TGR proteins can reduce GSSG, and the GR activity of TGR is thought to be due primarily to its Grx domain (24, 38). To test this notion, we determined the GR activity of variants of SmTGR that have the Cys-28/Cys-31 pair modified in the Grx domain, but which retain the Cys-154/Cys-159 and Cys-596/Sec-597 pairs. As shown in Table 2, both Cys28Ala and Cys28Ala/Cys31Ala have 3-4 % of the GR activity of WT TGR. Thus, our data corroborate the notion that the Grx domain of SmTGR is responsible for most of the GSSG reduction. Also, Cys-28 is the most likely residue initiating nucleophilic attack on GSSG. Studies from Angelucci et. al. showed that truncated SmTGR (missing Sec-597 and Gly-598) retained some residual GR activity (~ 0.8 %) (24). In addition, it has been suggested that the electrostatic environment surrounding Cys-154 and Cys-159 could facilitate the binding of GSSG (39). These results suggest that the Cys-154/Cys-159 pair could also reduce GSSG even though its catalytic efficiency is relatively low. Thus, our data are consistent with the findings of Angelucci et al. that the redox-active Cys-154/Cys-159 could reduce GSSG, but with low efficiency (24). Cys31Ala retained about 18 % of the GR activity of WT TGR whereas the apparent Vmax of Cys31Ser was comparable to WT TGR (Table 2). These results suggest that in GR activity, the role of Cys-31 (or Ser-31) is to stabilize the thiolate anion on Cys-28 via a hydrogen bond, thus facilitating nucleophilic attack of Cys-28 on GSSG. In Cys31Ala, Cys-28 still is able to attack the disulfide bond of GSSG; the resultant Cys28-GS mixed disulfide can be resolved by electron pairs from the Cys-596/Sec-597 couple (via NADPH and Cys-154/Cys-159). Without stabilization of the thiolate of Cys-28 by Cys-31 (or Ser-31) the activity of Cys31Ala is less than WT TGR, but greater than Cys28Ala. In fact, the Vmax for GR activity of Cys31Ser was significantly greater than that of WT SmTGR, possibly because no substrate inhibition was seen with the Cys31Ser variant, in contrast to WT SmTGR, which does exhibit substrate inhibition (see below).
Table 2.
The kinetic parameters of wild-type SmTGR, Cys28Ala, Cys31Ala, Cys28Ala/Cys31Ala, and Cys31Ser reacting with the substrates, DTNB (DTNB activity), GSSG (GR activity), and Trx (TrxR activity) in the presence of saturating NADPH (100 μM).
| DTNB activity (kcat, s−1) | % of WT | Km for DTNB (μM) | |
|---|---|---|---|
| WT | 16 ± 0.6 | - | 319 ± 47 |
| Cys28Ala | 6 ± 0.2 | 38 | 230 ± 22 |
| Cys31Ala | 6 ± 0.1 | 38 | 43 ± 5 |
| Cys28Ala/Cys31Ala | 8 ± 0.2 | 50 | 406 ± 25 |
| Cys31Ser | 16 ± 0.8 | 100 | 169 ± 24 |
| GR activity (kcat, s−1) | % of WT | Km for GSSG (μM) | |
| WT | 19.4 ± 1.2 | - | 42 ± 6 |
| Cys28Ala | 0.7 ± 0.1 | 4 | 16 ± 0.2 |
| Cys31Ala | 3.5 ± 0.1 | 18 | 13 ± 2 |
| Cys28Ala/Cys31Ala | 0.6 ± 0.1 | 3 | 23 ± 4 |
| Cys31Ser | 25± 1.5 | 129 | 189 ± 27 |
| TrxR activity (kcat, s−1) | % of WT | Km for Trx (μM) | |
| WT | 19 ± 0.2 | - | 8 ± 0.3 |
| Cys28Ala | 14 ± 1.4 | 74 | 22 ± 4.5 |
| Cys31Ala | 19 ± 0.4 | 100 | 8 ± 0.6 |
| Cys28Ala/Cys31Ala | 15± 1.5 | 79 | 19 ± 4 |
| Cys31Ser | 20 ± 0.4 | 105 | 8 ± 0.5 |
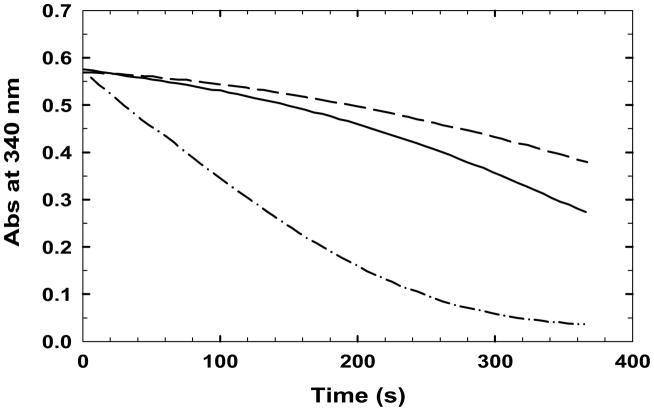
GR activity of TGRs from E. granulosus and Taenia crassiceps is affected by high GSSG concentrations; thus, the steady state of GR activity in TGR occurs only after an initial lag, previously described as hysteresis (37, 38). Preincubation with low concentrations of GSH eliminated the lag that is caused by high concentrations of GSSG. In the study of Bonilla et al., this effect was proposed to be due to glutathionylation of cysteine residues in EgTGR in the presence of high concentrations of GSSG. Two cysteine residues, Cys-88 and Cys-354, were indeed found to be glutathionylated using mass spectrometry analysis (38). However, these two cysteine residues are only relevant to structural integrity rather than to catalysis (38). In addition, as shown in Fig S1, these two cysteine residues are not conserved in all TGRs. Furthermore, because not all redox active Cys and Sec residues were identified in the MS analysis, no clear conclusions can be made on the role of their glutathionylation status during this lag. As shown in Fig. 1, similar to other TGRs, WT SmTGR also displayed this lag; the effect was observed when the concentrations of GSSG were greater than 100 μM (data not shown). Although both Cys31Ala (data not shown) and Cys31Ser (Fig. 1) are active in GSSG reduction, no lag was observed. These observations all point to glutathionylation of Cys-31 being the likely cause of the observed lag of the WT enzyme.
Fig. 1.
The kinetic traces of wild-type SmTGR (—), Cys520Ala (---), and Cys31Ser (— ● —) in the presence of 100 μM NADPH and 500 μM GSSG; the concentrations of wild-type enzyme, Cys520Ala, and Cys31Ser used in the assays were 20, 40, and 20 μM, respectively. Note that the traces for WT and Cys520Ala have lag phases that are referred to as hysteresis.
Most mammalian TGRs have a CysProHisSer motif in their Grx domains, except for that in the TGR of kangaroo rat (20). Glutathionylation is an important mechanism to prevent the over-oxidation of cysteinyl thiols to sulfinic and sulfonic acids (49). Glutathionylation of the CysXaaXaaCys motif in the Grx domain of SmTGR may be a mechanism to prevent its irreversible over-oxidation. To the best of our knowledge, no lags in the steady-state kinetics have been reported in mammalian TGRs, suggesting that glutathionylation might not occur in the Grx domain of mammalian TGRs. Thus, the Grx domain of mammalian TGRs could be more susceptible to oxidative stress than those of TGRs from lower eukaryotes. However, this property is probably not crucial in mammalian cells, because the presence of TrxR and GR enzymes that respond to oxidative stress make TGR less important in maintaining the overall cellular redox balance. However, because in flatworm parasites TGR is the only enzyme capable of reducing Trx and GSSG, and parasites are under constant oxidative stress, mechanisms such as reversible glutathionylation may be important in protecting the activity of parasite TGR. This differential sensitivity could be important in drug development.
The Vmax values, even when measured after steady-state rates are attained, indicate that substrate inhibition still occurs with WT SmTGR at concentrations of GSSG greater than 200 μM (data not shown). However, no substrate inhibition was observed in Cys28Ala, Cys28AlaCys31Ala, Cys31Ala, or Cys31Ser variants, even at concentrations of GSSG as high as 500 μM.
The TrxR activities of Cys28Ala, Cys31Ala, Cys28Ala/Cys31Ala, and Cys31Ser variants of SmTGR were comparable to those of WT enzyme (Table 2), consistent with the theory that the Grx domain does not have a crucial role in Trx reduction. However, the activities of the Grx domain variants with DTNB, which is usually thought to be reduced at the same site as is Trx, were significantly decreased (by ~60%), except in the case of Cys31Ser. These results suggest that reduction of DTNB by SmTGR can occur both at the cysteine Cys-28/Cys-31 pair and the Cys-596/Sec-597 pair. Therefore, reduction of SmTGR by NADPH must also involve reduction of the cysteine couple of Cys-28/Cys-31, which should potentially be able to interact with DTNB.
The catalytic functions of Sec-597 in SmTGR
It has been shown that Sec is very important to the TrxR and GR activities of EgTGR (38); Sec is also thought to be essential to the GR and TrxR activities of SmTGR (9). We have investigated the functions of the analogous Sec-597 residue in the catalytic mechanism of SmTGR. As shown in Table 3, the DTNB, GR, and TrxR activities of the Sec597Cys mutant were approximately 20 % of the wild-type activities. However, unlike Sec597Cys, preparations that are 100 % full length, WT SmTGR preparations are less than 100% full length. This conclusion is based on the selenium content of WT SmTGR (and mutants, excepting Sec597Cys) determined by inductively coupled plasma atomic emission spectroscopy (data not shown). The efficiency of selenocysteine incorporation from seven representative protein samples averaged 27 % (range 20–33 %) and is similar to that previously found for other selenoproteins (22, 38, 45). Therefore, the activities of Sec597Cys are more likely only 5.4 % of WT SmTGR activities, indicating that selenocysteine is very important to the catalytic activity of SmTGR. Our data show that the Grx domain is responsible for the most of GR activity and some of the DTNB activity (see above). To permit GR and DTNB activities at the Grx site, the redox-active Cys-596/Sec-597 pair is presumed to be crucial for transferring reducing equivalents from NADPH through the TrxR domain to the Grx domain; further evidence for this transfer comes from recent structural data (40). It is expected that Sec597Cys should have significantly lower GR and DTNB activities compared to those of WT enzyme. The X-ray crystal structure indicates that the cysteine-selenocysteine pair is responsible for the reduction of Trx (40); therefore, as expected, the TrxR activity of Sec597Cys is significantly less than that of WT (Table 3). However, the Grx deglutathionylation activity of Sec597Cys is ~ 40 % of wild-type activity (Table 1). As described earlier, the truncated enzyme, where both Sec-597 and Gly-598 are missing, still retained 40 % of wild-type Grx deglutathionylation activity. This activity can be attributed to the Grx domain of the truncated enzyme being reduced by GSH rather than by the TrxR domain. As a result, the Grx domain of Sec597Cys is able to function independently of the TrxR domain by virtue of it being be reduced by GSH.
Table 3.
The kinetic parameters of wild-type SmTGR and Sec597Cys reacting with the substrates, DTNB (DTNB activity), GSSG (GR activity), and Trx (TrxR activity)
| DTNB activity (kcat, s−1) | % of WT | Km for DTNB (μM) | |
|---|---|---|---|
| WT | 16 ± 0.6 | - | 319 ± 47 |
| Sec597Cys | 4 ± 0.1 | 25a, 6.8b | 107 ± 10 |
| GR activity (kcat, s−1) | % of WT | Km for GSSG (μM) | |
| WT | 19 ± 1.2 | - | 42 ± 6 |
| Sec597Cys | 4 ± 0.6 | 21a, 5.7b | 71 ± 24 |
| TrxR activity (kcat, s−1) | % of WT | Km for Trx (μM) | |
| WT | 19 ± 1 | - | 8 ± 0.3 |
| Sec597Cys | 3.5 ± 0.1 | 18a, 4.9b | 15 ± 2.3 |
inhibition is determined by the activities of Sec597Cys compared to those of wild-type enzyme
inhibition is determined by the activities of Sec597Cys compared to those of wild-type enzyme when it is considered that only 27 % of wild-type enzyme is full length whereas 100 % of Sec597Cys is full length
The broad substrate range of cytosolic TrxR1 has been attributed to the low pKa value (~5.2) (44, 56, 57) and high nucleophilicity of a selenolate (56, 57). However, the presence of a selenolate is not required for the reduction of many small molecule substrates in the case of mitochondrial TrxR2 (58). While the presence of Sec is also believed to confer broad substrate tolerance on SmTGR (10) for the same reasons as TrxR1 (23, 59–61), substrate electrophilicity has been cited as an important factor for substrate utilization in the case of TrxR2 (62). We tested whether the Sec597Cys variant can also reduce a wide range of substrates. As shown in Table 4, quite surprisingly, although having lower activities in all cases, Sec597Cys nevertheless was able to reduce all of the known TGR substrates. These results are similar to those of the mammalian TrxR2 variant where Sec is replaced by Cys; this variant still was able to reduce selenite, lipoic acid, and lipoamide (58). Thus, selenocysteine is not the sole determinant for the broad substrate tolerance of SmTGR, although it is important to achieve high enzymatic activity.
Table 4.
Substrate specificity of wild-type TGR and Sec597Cys
| Enzymes | Vmax (s−1) | Km (mM) | |
|---|---|---|---|
| NaSeO3 | WT (100 nM)a | 1.3 ± 0.2 | 8.7 ± 2.5 (μM) |
| Sec597Cys (100 nM)a | 0.8 ± 0.1 | 17.3 ± 4.4 (μM) | |
| Alloxan | WT (20 nM)a | 27.7 ± 0.8 | 0.94 ± 0.09 |
| Sec597Cys (100 nM)a | 7.2 ± 0.2 | 0.88 ± 0.07 | |
| H2O2 | WT (20 nM)a | 4.6 ±0.7 | 17.3 ± 7.6 |
| Sec597Cys (100 nM)a | 1.1 ± 0.1 | 52.4 ± 9.8 | |
| Lipoamide | WT (100 nM)a | 4.3 ± 0.4 | 5 ± 1 |
| Sec597Cys (100 nM)a | 2.6 ± 0.6 | 10.7 ± 3.8 | |
| Lipoic acid | WT (100 nM)a | 1.6 ± 0.2 | 2.9 ± 0.7 |
| Sec597Cys (150 nM)a | 1 ± 0.2 | 9.3 ±2.9 | |
| t-Butyl-OOH | WT (20 nM)a | 10.9 ± 0.4 | 75 ±10 |
| Sec597Cys (100 nM)a | 2.9 ± 0.1 | 53 ± 5 | |
| Dehydroascorbic acid | WT (20 nM) | NDb | NDb |
| Sec597Cys (100 nM) | NDb | NDb | |
| Ubiquinone | WT (20 nM) | NDb | NDb |
| Sec597Cys (100 nM) | NDb | NDb |
concentrations in parentheses represent the concentrations of enzymes used in the assays
undetectable activity
Selenocysteine was believed to be required for some inhibitors to be effective. For example, some gold compounds need selenocysteine to exert their inhibitory effects on selenoproteins (42, 44, 63). However, recent data indicate that the selenocysteine residue is not essential for inhibition of SmTGR by AF. AF still is able to inhibit the GR activity of truncated SmTGR that does not contain Sec-597 or Gly-598, although the degree of and rate of inhibition can be increased when exogenous selenium (benzeneselenol) is supplied (12). Thus, we tested three SmTGR inhibitors: Furoxan (4-phenyl-1,2,5-oxadiazole-3-carbonitrile-2-oxide, Fx), which has been shown to release nitric oxide that inhibits SmTGR (11, 14), potassium antimony tartrate (PAT), an anti-schistosomal drug shown previously to inhibit SmTGR (10), and AF, a gold (I) compound. All of the compounds inhibited both Sec597Cys and WT enzyme, and the IC50 values were in the 8–300 nM range for PAT and AF (Table 5). These results indicate that selenocysteine is not critical for the inhibitory actions of these inhibitors. These results are similar to the observations from Lothrop et al. (58). We propose that the increased rates of inactivating the truncated SmTGR by AF with the supplements of exogenous selenium (12) are due to accessibility of the selenium to AF to release the gold atoms. Thus, in Sec597Cys, the cysteine pair of Cys-596 and Cys-597 could interact with AF to facilitate the transfer of gold atoms from AF to redox-active cysteines in the protein. Since a thiolate anion on Cys-597 is required for disulfide-dithiol interchange reactions, it is reasonable to speculate that Cys-597 has a low pKa value allowing it to release gold from AF resulting in the inhibition of Sec597Cys.
Table 5.
IC50 values of furoxan (Fx), potassium antimony tartrate (PAT), and auranofin (AF) against DTNB, GR, and TrxR activities of wild-type and Sec597Cys SmTGRs
| WT | Sec597Cys | ||
|---|---|---|---|
| DTNBa | Fx | 3.0 μM | 6 μM |
| PAT | 9 nM | 20 nM | |
| AF | 3.2 nM | 20 nM | |
| GRb | Fx | 3.2 μM | 2.5 μM |
| PAT | 8 nM | 8 nM | |
| AF | 5.4 nM | 9 nM | |
| TrxRc | Fx | 5.4 μM | 2.1 μM |
| PAT | 45 nM | 29 nM | |
| AF | 8.3 nM | 16 nM |
The concentrations of wild-type enzyme and Sec597Cys used were 5 and 20 nM, respectively
The concentrations of wild-type enzyme and Sec597Cys used were 20 and 50 nM, respectively
The concentrations of wild-type enzyme and Sec597Cys used were 10 and 20 nM, respectively
In order to test the hypothesis that reducing equivalents can pass readily from the reduced TrxR domain to the Grx domain, we have investigated the reductive half-reaction in SmTGR Sec597Cys using static, anaerobic titrations with NADPH to ascertain whether all of the cysteine pairs can be reduced by NADPH. The limitation of this type of experiment is that one does not observed the transient states, but only the spectral characteristics of the products of the reductive half-reaction, including products that may have been formed slowly. The spectra of Sec597Cys recorded after the addition to the oxidized enzyme of 1, 2, and 6 moles NADPH/mole FAD are shown in Fig. 2. The Inset shows the changes in absorbance at three critical wavelengths, 462 nm (flavin reduction), 540 nm (thiolate-FAD CTC, i.e., a thiolate anion on Cys-159 interacts with FAD, Scheme 1), and 360 nm (NADPH oxidation). The oxidation of NADPH can be followed most clearly at 360 nm where the spectra of all of the enzyme forms are virtually isosbestic. The data show that all of the added NADPH reacted with the enzyme until at least 2.6 moles NADPH/mole FAD had been added. This means that 2.6 eq react before free NADPH starts to accumulate. The data at the three wavelengths shown suggest three phases in the titration. During the addition of the first equivalent of NADPH, the spectrum remained the same as the spectrum of oxidized enzyme; that is, it did not lead to decreased absorbance at 463 nm or increased absorbance at 540 nm, as would be expected for the formation of the thiolate-FAD CTC. The lack of the CTC indicates that the added reducing equivalents were neither in FAD nor in the proximal cysteine pair, Cys-154/Cys-159. Thus, the electron pair must have passed on to the Cys-596′/Cys-597′ pair in the flexible tail, and possibly to the Cys-28/Cys-31 pair in the Grx domain. Precedence for this interpretation comes from studies of mercuric reductase, another member of this enzyme superfamily that has three redox-active centers (65, 66). Further addition of NADPH (1.1–2.6 eq) resulted in decreased absorbance at 463 nm, increased absorbance at 540 nm, but still almost no change at 360 nm; this indicates that in this phase the N-terminal redox-active disulfide that is proximal to the FAD was partially reduced. The final phase (2.6 – 6 equiv) involved further reduction of the N-terminal disulfide and enhancement of the charge-transfer complex by bound NADPH. Given the flexibility of the C-terminal tail, as indicated in the latest crystal structure (40) and our interpretation (above) of the NADPH titration, the cysteine pair of Cys-28 and Cys-31 in the Grx domain appears to be able to react with DTNB regardless of the equilibria existing between the three redox-active cysteine pairs.
Fig. 2.

Anaerobic titration of Sec597Cys. The concentrations of enzymes were 10 μM. Spectra are as follows: oxidized enzyme (—), after addition of 1 equiv of NADPH (---), after addition of 2 equiv of NADPH (— ● —), after addition of 6 equiv of NADPH (— ● ● —) (Inset) absorbance at 462 nm (●), left y-axis; 540 nm (□), right y-axis; and 360 nm (▲), right offset y-axis as function of equiv of NADPH added (x-axis).
It has been proposed that the transfer of reducing equivalents from the Cys-597/Sec-598 pair in the C-terminal “tail” to the Cys pair in the Grx domain involves substantial structural changes in the protein (24, 39, 40). Reduction of the C-terminal Cys-Sec pair may be the signal for rotation of the flexible C-terminus to a position close to the Grx domain of the other subunit. In this configuration, reducing equivalents could be transferred to the Cys-28/Cys-31 pair, resulting in the oxidation of the Cys-Sec pair; this would be followed by a return to a conformation in which the Cys-Sec pair is away from the Grx domain but close to the cysteine pair proximal to the flavin. In this latter conformation, reducing equivalents are effectively trapped in the Grx domain.
Because only about 27 % of the expressed wild-type SmTGR is full-length protein whereas Sec597Cys preparations are 100 % full-length protein, we were surprised to find that titrations of both enzymes with NADPH resulted in similar titration patterns (data not shown). We do not fully understand why the largely truncated enzyme took up more than 2 eq of NADPH. One possibility is that the redox-active Cys-596-Sec-597 in the full-length SmTGR present in the enzyme population can pass electrons (slowly) to Grx domains of truncated SmTGRs intermolecularly as well as to its own Grx domain intramolecularly. The similarity of titrations of wild-type enzyme and the Sec597Cys variant suggests that reduction of the Grx domain disulfide is by a C-terminal tail redox pair for both Sec597Cys and the 27 % full-length wild-type enzyme preparations.
The possible functions of Cys-520 and Cys-574 in SmTGR
It has been suggested that the Cys-520/Cys-574 pair are also redox-active in catalysis because after incubation of wild-type SmTGR with AF and NADPH, gold atoms were found bound to this pair with an occupancy of 50 % (12). To investigate this issue, the Cys520Ala, Cys574Ala, and Cys520Ala/Cys574Ala variants were generated and studied. As shown in Table 6, the DTNB, GR, and TrxR activities of the three variants ranged from 31–47 % of WT, whereas Grx deglutathionylation activities of three variants were 63–70 % of WT activity (Table 1). The lower DTNB, GR and TrxR activities of the three variants can be explained by the TrxR domain in these three variants not being fully functional. As described above, only a small portion of the Grx deglutathionylation activity is dependent on the TrxR domain; because the Grx domain in these three variants, in principle, still could be reduced by either GSH or the TrxR domain. In our proposed electron flow within SmTGR, the DTNB, GR, and TrxR activities of these variants require the TrxR domain. We still observed lags in the GR steady-state activity of all three of these variants, showing that glutathionylation by GSSG in these two cysteine residues (rather than the Cys-28/Cys-31 pair) is unlikely (Fig. 1). The results above (Table 6) suggest that the Cys-520/Cys-574 couple can affect the electron flow in the TrxR domain to some degree but indirectly.
Table 6.
The kinetic parameters of wild-type SmTGR, Cys520Ala, Cys574Ala, and Cys520Ala/Cys574Ala reacting with DTNB (DTNB activity), GSSG (GR activity), and Trx (TrxR activity)
| DTNB activity (kcat, s−1) | % of WT | Km of DTNB (μM) | |
|---|---|---|---|
| WT | 16 ± 0.6 | - | 319 ± 47 |
| Cys520Ala | 5 ± 0.2 | 31 | 375 ± 56 |
| Cys574Ala | 6 ± 0.2 | 38 | 230 ± 27 |
| Cys520Ala/Cys574la | 6 ± 0.2 | 38 | 187 ± 28 |
| GR activity (kcat, s−1) | % of WT | Km of GSSG (μM) | |
| WT | 19 ± 1.2 | - | 42 ± 6 |
| Cys520Ala | 5 ± 0.5 | 26 | 31 ± 6 |
| Cys574Ala | 7 ± 0.7 | 37 | 36 ± 8 |
| Cys520Ala/Cys574la | 5 ± 0.4 | 26 | 24 ± 5 |
| TrxR activity (kcat, s−1) | % of WT | Km of Trx (μM) | |
| WT | 19 ± 0.2 | - | 8 ± 0.3 |
| Cys520Ala | 9 ± 0.1 | 47 | 7 ± 0.4 |
| Cys574Ala | 8 ± 0.1 | 42 | 7 ± 0.2 |
| Cys520Ala/Cys574la | 8 ± 0.2 | 42 | 15 ± 1.1 |
We reduced the three Cys-520 and Cys-574 variants anaerobically with 6 eq of NADPH. The reduced spectra for these three variants (Fig. 3B shows spectra for Cys520Ala/Cys574Ala as an example) were similar to those of wild-type enzyme (cf. Fig. 3B and Fig 3A). However, more flavin reduction (462 nm) was observed, and less FAD-thiolate CTC was evident (540 nm) in these three variants than in wild-type SmTGR. These observations indicate that the relative redox potentials of the FAD and the proximal cysteine pair in the three variants had changed. These results suggest that Cys-520 and Cys-574 are close enough to the flavin, or at least they affect the structure of the enzyme sufficiently, to influence its redox potential relative to that of the proximal redox-active cysteine pair. Such a change could influence the Vmax of the enzyme. Structures of SmTGR (pdb files, 3H4K and 2V6O) show that the Cys-520/Cys-574 pair is on the strand that contains His-571 and Glu-576, a catalytic dyad conserved in all members of this enzyme superfamily. This dyad has been shown to facilitate deprotonation of one cysteine residue of the N-terminal redox-active cysteine pair adjacent to FAD. Thus, in GRs, TrxRs, and lipoamide dehydrogenases, mutation of this dyad resulted in significant loss of enzymatic activity (64, 67–71). Similarly, in SmTGR, His-571, and Glu-576 are thought to facilitate deprotonation of Cys-159 (24). The results with the Cys-520/Cys-574 TGR variant are similar to those with a variant of TrxR from Drosophila melanogaster. In this variant, mutation of His-464, equivalent to His-571 in SmTGR, changed the redox potential of FAD (70). It is possible that in the Cys-520 and/or Cys-574 variants of SmTGR, the redox potential and the alignment of His-571, Glu-576, and Cys-159 is distorted, thereby affecting the catalytic efficiency of the TrxR domain of SmTGR.
Fig. 3.
Spectra of oxidized and reduced wild-type enzyme (A) and Cys520Ala/Cys574Ala (B). The concentrations of enzymes were 10 μM. The spectra of reduced enzymes were recorded after addition of 6 equiv of NADPH. Spectra are: oxidized enzyme (—) and after addition of 6 equiv of NADPH (– •• –).
Concluding remarks
Schistosomiasis is the second most important parasitic disease after malaria. Because praziquantel-resistant parasites are an imminent threat, it is urgent that new drugs against schistosomiasis are developed. SmTGR is essential for the survival of S. mansoni, has a different functional role in the worm than in its host, and can be selectively inhibited relative to the host flavoenzyme reductases, indicating that SmTGR could be an excellent drug target. To facilitate the development of inhibitors of SmTGR, it is vital to understand its catalytic mechanism.
Although a catalytic mechanism for TGR proteins has been proposed (21, 22), experimental details characterizing this process have been lacking. The most intriguing questions are the particular chemical functions of cysteine residues that are involved in catalysis. Such questions led us to investigate these catalytic functions of the cysteine residues as well as the selenocysteine residue. It should be noted that the kinetic parameters of WT enzyme and its variants reported here were based on protein concentrations determined by using the extinction coefficient of FAD; we did not correct the parameters with the factor of selenocysteine incorporation. Such comparisons among WT enzyme, Cys-28, Cys-31, Cys-520 and Cys-574 variants should not affect our interpretations; selenium content in a protein (excepting Sec597Cys) is independent of the mutation and averages 27%. In contrast, the comparisons between WT enzyme and Sec597Cys were adjusted to account for the relative incorporation of selenocysteine (shown in Table 3). Our results show that the redox active Cys-28/Cys-31 pair in the Grx domain of TGR is responsible for glutathione reductase and deglutathionylation activities, as expected. On the basis of our results, we conclude that the deglutathionylation activity of the Grx domain can occur via two parallel mechanisms that are partitioned according to the particular conditions extant. The Cys-28 thiolate is the nucleophile that attacks the disulfide bond between GSH and a cysteine residue of proteins (or peptides), thus forming a glutathionylated mixed disulfide linked enzyme intermediate in both mechanisms. In the mechanism occurring under normal metabolic conditions of high GSH concentration, this is followed by attack of a free GSH on the mixed disulfide bond between GSH and Cys-28 to release GSSG. At low GSH concentrations, in a second mechanism, the disulfide bond of the mixed disulfide can be resolved by Cys-31 to form a disulfide bond between Cys-28 and Cys-31; this disulfide can then be reduced by the redox-active selenocysteine-cysteine of the TrxR domain. In addition to the deglutathionylation reactions, the Cys-28/Cys-31 pair in the Grx domain is responsible for reduction of GSSG. The GR activity of the Cys-31 variants displayed no lags in the steady-state kinetics that are thought to be due to glutathionylation on cysteine residues caused by high concentrations of GSSG. Given that some Grx enzymes catalyze the deglutathionylation of proteins using a single active site thiol, we suggest that the role of Cys-31 is mainly to stabilize the thiolate anion on Cys-28 via a hydrogen bond, thus facilitating nucleophilic attack of Cys-28 on GSSG. Our results showing that the Sec597Cys variant could reduce a variety of known SmTGR substrates, albeit at much reduced rates, suggests that the presence of selenocysteine is not essential for the broad substrate tolerance of SmTGR. We also find that the action of inhibitors did not require selenocysteine, because the Sec597Cys variant was susceptible to several known inhibitors. We also investigated whether another cysteine pair, Cys-520/Cys-574, is involved in catalysis by SmTGR. Our data indicate that this particular cysteine pair may affect acid-base catalysis in the TrxR domain of SmTGR but does not directly participate in catalysis. The results from spectrally monitored reductive titrations suggest that reducing equivalents from NADPH can be transferred to the disulfide in the Grx domain via the C-terminal tail, as suggested by the recent structure of SmTGR (40).
This study further elucidates the catalytic mechanism of SmTGR and has made a beginning toward assigning specific roles to Cys-28 and Cys-31 in the Grx domain, in confirming the assumed role of Sec-597 as the reductant of Trx and the Grx domain active site cysteines, and in eliminating Cys-520 and Cys-574 as participants in catalysis. These results will help the planning to design differential inhibitors against SmTGR and human TrxR.
Supplementary Material
Acknowledgments
This work was supported by NIH/National Institute of Allergy and Infectious Diseases (NIAID) grant AI065622 (D.L.W.).
The authors appreciate critical comments from Prof. Andrea Bellelli and Dr. Francesco Angelucci (the Sapienza University of Rome), whose work has made important contributions to our knowledge of the structure and mechanism of SmTGR; we are also grateful to the reviewers for important suggestions.
Abbreviations
- AF
auranofin
- CTC
charge transfer complex
- DTNB
5, 5′-dithiobis (2-nitrobenzoic acid)
- EgTGR
thioredoxin glutathione reductase from Echinococcus granulosus
- FAD
flavin adenine dinucleotide
- Furoxan (Fx)
4-phenyl-1,2,5-oxadiazole-3-carbonitrile-2-oxide
- GSSG
glutathione disulfide
- GR
glutathione reductase
- Grx
glutaredoxin
- high Mr TrxR
high molecular weight thioredoxin reductase
- TGR
thioredoxin glutathione reductase
- TrxR
thioredoxin reductase
- Sec
selenocysteine
- SmTGR
thioredoxin glutathione reductase from Schistosoma mansoni
- SmTrx-1
thioredoxin-1 from S. mansoni
- Trx
thioredoxin
- WT
wild-type
Footnotes
SUPPORTING INFORMATION AVAILABLE
The alignment of sequences of TrxR from different species (Figure S1) and the primers used to clone Cys28Ala, Cys31Ala, Cys31Ser, Cys520Ala, Cys574Ala, and Sec597Cys SmTrxR (Table S1). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Doenhoff MJ, Cioli D, Utzinger J. Praziquantel: mechanisms of action, resistance and new derivatives for schistosomiasis. Curr Opin Infect Dis. 2008;21:659–667. doi: 10.1097/QCO.0b013e328318978f. [DOI] [PubMed] [Google Scholar]
- 2.Utzinger J, Xiao SH, Tanner M, Keiser J. Artemisinins for schistosomiasis and beyond. Curr Opin Investig Drugs. 2007;8:105–116. [PubMed] [Google Scholar]
- 3.Cioli D, Pica-Mattoccia L, Archer S. Drug resistance in schistosomes. Parasitology today (Personal ed) 1993;9:162–166. doi: 10.1016/0169-4758(93)90138-6. [DOI] [PubMed] [Google Scholar]
- 4.Abdulla MH, Lim KC, Sajid M, McKerrow JH, Caffrey CR. Schistosomiasis mansoni: novel chemotherapy using a cysteine protease inhibitor. PLoS Med. 2007;4:e14. doi: 10.1371/journal.pmed.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreira LS, Pilo-Veloso D, de Mello RT, Coelho PM, Nelson DL. A study of the activity of 2-(alkylamino)-1-phenyl-1-ethanethiosulfuric acids against infection by Schistosoma mansoni in a murine model. Trans R Soc Trop Med Hyg. 2007;101:385–390. doi: 10.1016/j.trstmh.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Simeonov A, Jadhav A, Sayed AA, Wang Y, Nelson ME, Thomas CJ, Inglese J, Williams DL, Austin CP. Quantitative high-throughput screen identifies inhibitors of the Schistosoma mansoni redox cascade. PLoS Negl Trop Dis. 2008;2:e127. doi: 10.1371/journal.pntd.0000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sayed AA, Williams DL. Biochemical characterization of 2-Cys peroxiredoxins from Schistosoma mansoni. J Biol Chem. 2004;279:26159–26166. doi: 10.1074/jbc.M401748200. [DOI] [PubMed] [Google Scholar]
- 8.Sayed AA, Cook SK, Williams DL. Redox balance mechanisms in Schistosoma mansoni rely on peroxiredoxins and albumin and implicate peroxiredoxins as novel drug targets. J Biol Chem. 2006;281:17001–17010. doi: 10.1074/jbc.M512601200. [DOI] [PubMed] [Google Scholar]
- 9.Alger HM, Williams DL. The disulfide redox system of Schistosoma mansoni and the importance of a multifunctional enzyme, thioredoxin glutathione reductase. Mol Biochem Parasitol. 2002;121:129–139. doi: 10.1016/s0166-6851(02)00031-2. [DOI] [PubMed] [Google Scholar]
- 10.Kuntz AN, Davioud-Charvet E, Sayed AA, Califf LL, Dessolin J, Arner ES, Williams DL. Thioredoxin glutathione reductase from Schistosoma mansoni: an essential parasite enzyme and a key drug target. PLoS Med. 2007;4:e206. doi: 10.1371/journal.pmed.0040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sayed AA, Simeonov A, Thomas CJ, Inglese J, Austin CP, Williams DL. Identification of oxadiazoles as new drug leads for the control of schistosomiasis. Nat Med. 2008;14:407–412. doi: 10.1038/nm1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angelucci F, Sayed AA, Williams DL, Boumis G, Brunori M, Dimastrogiovanni D, Miele AE, Pauly F, Bellelli A. Inhibition of Schistosoma mansoni thioredoxin-glutathione reductase by auranofin: structural and kinetic aspects. J Biol Chem. 2009;284:28977–28985. doi: 10.1074/jbc.M109.020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lea WA, Jadhav A, Rai G, Sayed AA, Cass CL, Inglese J, Williams DL, Austin CP, Simeonov A. A 1,536-well-based kinetic HTS assay for inhibitors of Schistosoma mansoni thioredoxin glutathione reductase. Assay Drug Dev Technol. 2008;6:551–555. doi: 10.1089/adt.2008.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rai G, Sayed AA, Lea WA, Luecke HF, Chakrapani H, Prast-Nielsen S, Jadhav A, Leister W, Shen M, Inglese J, Austin CP, Keefer L, Arner ES, Simeonov A, Maloney DJ, Williams DL, Thomas CJ. Structure mechanism insights and the role of nitric oxide donation guide the development of oxadiazole-2-oxides as therapeutic agents against schistosomiasis. J Med Chem. 2009;52:6474–6483. doi: 10.1021/jm901021k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams CH. Lipoamide Dehydrogenase, Glutathione Reductase, Thioredoxin reductase, and Mercuric ion reductase-A family of Flavoenzyme Transhydrogenase. Vol. 3. CRC press, Inc; Boca Raton. FL: 1992. [Google Scholar]
- 16.Poole LB, Reynolds CM, Wood ZA, Karplus PA, Ellis HR, Li Calzi M. AhpF and other NADH:peroxiredoxin oxidoreductases, homologues of low Mr thioredoxin reductase. Eur J Biochem. 2000;267:6126–6133. doi: 10.1046/j.1432-1327.2000.01704.x. [DOI] [PubMed] [Google Scholar]
- 17.Williams CH., Jr Thioredoxin-thioredoxin reductase--a system that has come of age. Eur J Biochem. 2000;267:6101. doi: 10.1046/j.1432-1327.2000.01700.x. [DOI] [PubMed] [Google Scholar]
- 18.Williams CH, Arscott LD, Muller S, Lennon BW, Ludwig ML, Wang PF, Veine DM, Becker K, Schirmer RH. Thioredoxin reductase two modes of catalysis have evolved. Eur J Biochem. 2000;267:6110–6117. doi: 10.1046/j.1432-1327.2000.01702.x. [DOI] [PubMed] [Google Scholar]
- 19.Su D, Novoselov SV, Sun QA, Moustafa ME, Zhou Y, Oko R, Hatfield DL, Gladyshev VN. Mammalian selenoprotein thioredoxin-glutathione reductase. Roles in disulfide bond formation and sperm maturation. J Biol Chem. 2005;280:26491–26498. doi: 10.1074/jbc.M503638200. [DOI] [PubMed] [Google Scholar]
- 20.Gerashchenko MV, Su D, Gladyshev VN. CUG start codon generates thioredoxin/glutathione reductase isoforms in mouse testes. J Biol Chem. 285:4595–4602. doi: 10.1074/jbc.M109.070532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun QA, Kirnarsky L, Sherman S, Gladyshev VN. Selenoprotein oxidoreductase with specificity for thioredoxin and glutathione systems. Proc Natl Acad Sci USA. 2001;98:3673–3678. doi: 10.1073/pnas.051454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun QA, Su D, Novoselov SV, Carlson BA, Hatfield DL, Gladyshev VN. Reaction mechanism and regulation of mammalian thioredoxin/glutathione reductase. Biochemistry. 2005;44:14528–14537. doi: 10.1021/bi051321w. [DOI] [PubMed] [Google Scholar]
- 23.Arner ES, Zhong L, Holmgren A. Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Methods Enzymol. 1999;300:226–239. doi: 10.1016/s0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- 24.Angelucci F, Miele AE, Boumis G, Dimastrogiovanni D, Brunori M, Bellelli A. Glutathione reductase and thioredoxin reductase at the crossroad: the structure of Schistosoma mansoni thioredoxin glutathione reductase. Proteins. 2008;72:936–945. doi: 10.1002/prot.21986. [DOI] [PubMed] [Google Scholar]
- 25.Arscott LD, Veine DM, Williams CH., Jr Mixed disulfide with glutathione as an intermediate in the reaction catalyzed by glutathione reductase from yeast and as a major form of the enzyme in the cell. Biochemistry. 2000;39:4711–4721. doi: 10.1021/bi9926431. [DOI] [PubMed] [Google Scholar]
- 26.Rietveld P, Arscott LD, Berry A, Scrutton NS, Deonarain MP, Perham RN, Williams CH., Jr Reductive and oxidative half-reactions of glutathione reductase from Escherichia coli. Biochemistry. 1994;33:13888–13895. doi: 10.1021/bi00250a043. [DOI] [PubMed] [Google Scholar]
- 27.Karplus PA, Pai EF, Schulz GE. A crystallographic study of the glutathione binding site of glutathione reductase at 0.3-nm resolution. Eur J Biochem. 1989;178:693–703. doi: 10.1111/j.1432-1033.1989.tb14500.x. [DOI] [PubMed] [Google Scholar]
- 28.Pai EF, Schulz GE. The catalytic mechanism of glutathione reductase as derived from x-ray diffraction analyses of reaction intermediates. J Biol Chem. 1983;258:1752–1757. [PubMed] [Google Scholar]
- 29.Arscott LD, Thorpe C, Williams CH., Jr Glutathione reductase from yeast. Differential reactivity of the nascent thiols in two-electron reduced enzyme and properties of a monoalkylated derivative. Biochemistry. 1981;20:1513–1520. doi: 10.1021/bi00509a016. [DOI] [PubMed] [Google Scholar]
- 30.Arscott LD, Gromer S, Schirmer RH, Becker K, Williams CH., Jr The mechanism of thioredoxin reductase from human placenta is similar to the mechanisms of lipoamide dehydrogenase and glutathione reductase and is distinct from the mechanism of thioredoxin reductase from Escherichia coli. Proc Natl Acad Sci USA. 1997;94:3621–3626. doi: 10.1073/pnas.94.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gromer S, Arscott LD, Williams CH, Jr, Schirmer RH, Becker K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J Biol Chem. 1998;273:20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 32.Wang PF, Arscott LD, Gilberger TW, Muller S, Williams CH., Jr Thioredoxin reductase from Plasmodium falciparum: evidence for interaction between the C-terminal cysteine residues and the active site disulfide-dithiol. Biochemistry. 1999;38:3187–3196. doi: 10.1021/bi982674g. [DOI] [PubMed] [Google Scholar]
- 33.Zhong L, Arner ES, Holmgren A. Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc Natl Acad Sci USA. 2000;97:5854–5859. doi: 10.1073/pnas.100114897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauer H, Massey V, Arscott LD, Schirmer RH, Ballou DP, Williams CH., Jr The mechanism of high Mr thioredoxin reductase from Drosophila melanogaster. J Biol Chem. 2003;278:33020–33028. doi: 10.1074/jbc.M303762200. [DOI] [PubMed] [Google Scholar]
- 35.Gromer S, Johansson L, Bauer H, Arscott LD, Rauch S, Ballou DP, Williams CH, Jr, Schirmer RH, Arner ES. Active sites of thioredoxin reductases: why selenoproteins? Proc Natl Acad Sci USA. 2003;100:12618–12623. doi: 10.1073/pnas.2134510100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong L, Holmgren A. Essential role of selenium in the catalytic activities of mammalian thioredoxin reductase revealed by characterization of recombinant enzymes with selenocysteine mutations. J Biol Chem. 2000;275:18121–18128. doi: 10.1074/jbc.M000690200. [DOI] [PubMed] [Google Scholar]
- 37.Rendon JL, del Arenal IP, Guevara-Flores A, Uribe A, Plancarte A, Mendoza-Hernandez G. Purification, characterization and kinetic properties of the multifunctional thioredoxin-glutathione reductase from Taenia crassiceps metacestode (cysticerci) Mol Biochem Parasitol. 2004;133:61–69. doi: 10.1016/j.molbiopara.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Bonilla M, Denicola A, Novoselov SV, Turanov AA, Protasio A, Izmendi D, Gladyshev VN, Salinas G. Platyhelminth mitochondrial and cytosolic redox homeostasis is controlled by a single thioredoxin glutathione reductase and dependent on selenium and glutathione. J Biol Chem. 2008;283:17898–17907. doi: 10.1074/jbc.M710609200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma M, Khanna S, Bulusu G, Mitra A. Comparative modeling of thioredoxin glutathione reductase from Schistosoma mansoni: a multifunctional target for antischistosomal therapy. J Mol Graph Model. 2009;27:665–675. doi: 10.1016/j.jmgm.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 40.Angelucci F, Dimastrogiovanni D, Boumis G, Brunori M, Miele AE, Saccoccia F, Bellelli A. Mapping the catalytic cycle of Schistosoma mansoni thioredoxin glutathione reductase by x-ray crystallography. J Biol Chem. 2010;285:32557–32567. doi: 10.1074/jbc.M110.141960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gromer S, Urig S, Becker K. The thioredoxin system--from science to clinic. Med Res Rev. 2004;24:40–89. doi: 10.1002/med.10051. [DOI] [PubMed] [Google Scholar]
- 42.Rigobello MP, Scutari G, Folda A, Bindoli A. Mitochondrial thioredoxin reductase inhibition by gold(I) compounds and concurrent stimulation of permeability transition and release of cytochrome c. Biochem Pharmacol. 2004;67:689–696. doi: 10.1016/j.bcp.2003.09.038. [DOI] [PubMed] [Google Scholar]
- 43.Witte AB, Anestal K, Jerremalm E, Ehrsson H, Arner ES. Inhibition of thioredoxin reductase but not of glutathione reductase by the major classes of alkylating and platinum-containing anticancer compounds. Free Radic Biol Med. 2005;39:696–703. doi: 10.1016/j.freeradbiomed.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 44.Lu J, Chew EH, Holmgren A. Targeting thioredoxin reductase is a basis for cancer therapy by arsenic trioxide. Proc Natl Acad Sci USA. 2007;104:12288–12293. doi: 10.1073/pnas.0701549104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arner ES, Sarioglu H, Lottspeich F, Holmgren A, Bock A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J Mol Biol. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 46.Alger HM, Sayed AA, Stadecker MJ, Williams DL. Molecular and enzymatic characterisation of Schistosoma mansoni thioredoxin. Int J Parasitol. 2002;32:1285–1292. doi: 10.1016/s0020-7519(02)00108-x. [DOI] [PubMed] [Google Scholar]
- 47.Holmgren A, Aslund F. Glutaredoxin. Methods Enzymol. 1995;252:283–292. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- 48.Xia L, Nordman T, Olsson JM, Damdimopoulos A, Bjorkhem-Bergman L, Nalvarte I, Eriksson LC, Arner ES, Spyrou G, Bjornstedt M. The mammalian cytosolic selenoenzyme thioredoxin reductase reduces ubiquinone. A novel mechanism for defense against oxidative stress. J Biol Chem. 2003;278:2141–2146. doi: 10.1074/jbc.M210456200. [DOI] [PubMed] [Google Scholar]
- 49.Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Gallogly MM, Starke DW, Leonberg AK, Ospina SM, Mieyal JJ. Kinetic and mechanistic characterization and versatile catalytic properties of mammalian glutaredoxin 2: implications for intracellular roles. Biochemistry. 2008;47:11144–11157. doi: 10.1021/bi800966v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iversen R, Andersen PA, Jensen KS, Winther JR, Sigurskjold BW. Thiol-disulfide exchange between glutaredoxin and glutathione. Biochemistry. 2010;49:810–820. doi: 10.1021/bi9015956. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y, Jao S, Nanduri S, Starke DW, Mieyal JJ, Qin J. Reactivity of the human thioltransferase (glutaredoxin) C7S, C25S, C78S, C82S mutant and NMR solution structure of its glutathionyl mixed disulfide intermediate reflect catalytic specificity. Biochemistry. 1998;37:17145–17156. doi: 10.1021/bi9806504. [DOI] [PubMed] [Google Scholar]
- 53.Discola KF, de Oliveira MA, Rosa Cussiol JR, Monteiro G, Barcena JA, Porras P, Padilla CA, Guimaraes BG, Netto LE. Structural aspects of the distinct biochemical properties of glutaredoxin 1 and glutaredoxin 2 from Saccharomyces cerevisiae. J Mol Biol. 2009;385:889–901. doi: 10.1016/j.jmb.2008.10.055. [DOI] [PubMed] [Google Scholar]
- 54.Gallogly MM, Starke DW, Mieyal JJ. Mechanistic and kinetic details of catalysis of thiol-disulfide exchange by glutaredoxins and potential mechanisms of regulation. Antiox Redox Signal. 2009;11:1059–1081. doi: 10.1089/ars.2008.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonilla M, Denicola A, Marino SM, Gladyshev VN, Salinas G. Linked Thioredoxin-Glutathione Systems in Platyhelminth Parasites: Alternative Pathways for Glutathione Reduction and Deglutathionylation. J Biol Chem. 2011;286:4959–4967. doi: 10.1074/jbc.M110.170761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Copeland PR. Making sense of nonsense: the evolution of selenocysteine usage in proteins. Genome Biol. 2005;6:221. doi: 10.1186/gb-2005-6-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wessjohann LA, Schneider A, Abbas M, Brandt W. Selenium in chemistry and biochemistry in comparison to sulfur. Biol Chem. 2007;388:997–1006. doi: 10.1515/BC.2007.138. [DOI] [PubMed] [Google Scholar]
- 58.Lothrop AP, Ruggles EL, Hondal RJ. No selenium required: reactions catalyzed by mammalian thioredoxin reductase that are independent of a selenocysteine residue. Biochemistry. 2009;48:6213–6223. doi: 10.1021/bi802146w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rundlof AK, Carlsten M, Giacobini MM, Arner ES. Prominent expression of the selenoprotein thioredoxin reductase in the medullary rays of the rat kidney and thioredoxin reductase mRNA variants differing at the 5′ untranslated region. Biochem J. 2000;347(Pt 3):661–668. [PMC free article] [PubMed] [Google Scholar]
- 60.Arner ES. Focus on mammalian thioredoxin reductases--important selenoproteins with versatile functions. Biochim et Biophy Acta. 2009;1790:495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 61.Lu J, Holmgren A. Selenoproteins. J Biol Chem. 2009;284:723–727. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- 62.Hondal RJ, Ruggles EL. Differing views of the role of selenium in thioredoxin reductase. Amino acids. 2010 doi: 10.1007/s00726-010-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith AD, Guidry CA, Morris VC, Levander OA. Aurothioglucose inhibits murine thioredoxin reductase activity in vivo. J Nutr. 1999;129:194–198. doi: 10.1093/jn/129.1.194. [DOI] [PubMed] [Google Scholar]
- 64.McMillan PJ, Arscott LD, Ballou DP, Becker K, Williams CH, Jr, Muller S. Identification of acid-base catalytic residues of high-Mr thioredoxin reductase from Plasmodium falciparum. J Biol Chem. 2006;281:32967–32977. doi: 10.1074/jbc.M601141200. [DOI] [PubMed] [Google Scholar]
- 65.Miller SM, Moore MJ, Massey V, Williams CH, Jr, Distefano MD, Ballou DP, Walsh CT. Evidence for the participation of Cys558 and Cys559 at the active site of mercuric reductase. Biochemistry. 1989;28:1194–1205. doi: 10.1021/bi00429a037. [DOI] [PubMed] [Google Scholar]
- 66.Miller SM, Massey V, Williams CH, Jr, Ballou DP, Walsh CT. Communication between the active sites in dimeric mercuric ion reductase: an alternating sites hypothesis for catalysis. Biochemistry. 1991;30:2600–2612. doi: 10.1021/bi00224a006. [DOI] [PubMed] [Google Scholar]
- 67.Karplus PA, Schulz GE. Substrate binding and catalysis by glutathione reductase as derived from refined enzyme: substrate crystal structures at 2 A resolution. J Mol Biol. 1989;210:163–180. doi: 10.1016/0022-2836(89)90298-2. [DOI] [PubMed] [Google Scholar]
- 68.Benen J, van Berkel W, Zak Z, Visser T, Veeger C, de Kok A. Lipoamide dehydrogenase from Azotobacter vinelandii: site-directed mutagenesis of the His450-Glu455 diad. Spectral properties of wild type and mutated enzymes. Eur J Biochem. 1991;202:863–872. doi: 10.1111/j.1432-1033.1991.tb16444.x. [DOI] [PubMed] [Google Scholar]
- 69.Benen J, van Berkel W, Dieteren N, Arscott D, Williams C, Jr, Veeger C, de Kok A. Lipoamide dehydrogenase from Azotobacter vinelandii: site-directed mutagenesis of the His450-Glu455 diad. Kinetics of wild-type and mutated enzymes. Eur J Biochem. 1992;207:487–497. doi: 10.1111/j.1432-1033.1992.tb17075.x. [DOI] [PubMed] [Google Scholar]
- 70.Huang HH, Arscott LD, Ballou DP, Williams CH., Jr Acid-Base Catalysis in the Mechanism of Thioredoxin Reductase from Drosophila melanogaster. Biochemistry. 2008;47:1721–1731. doi: 10.1021/bi702040u. [DOI] [PubMed] [Google Scholar]
- 71.Huang HH, Arscott LD, Ballou DP, Williams CH. Function of Glu-469′ in the acid-base catalysis of thioredoxin reductase from Drosophila melanogaster. Biochemistry. 2008;47:12769–12776. doi: 10.1021/bi801449h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.