Abstract
NR2E3 (PNR), a nuclear receptor specifically expressed in photoreceptors, represses cone-specific genes and activates several rod-specific genes. In humans, mutations in NR2E3 have been associated with the recessively inherited enhanced short wavelength sensitive (S-) cone syndrome (ESCS) and, recently, with autosomal dominant retinitis pigmentosa (adRP). In the present work, we describe two additional families affected by adRP that carry a heterozygous c.166G>A (p.G56R) mutation in the NR2E3 gene. Functional analysis determined dominant negative activity of the p.G56R mutant protein as the molecular mechanism of adRP. Interestingly, in one pedigree, the most common causal variant for ESCS (p.R311Q) co-segregated with the adRP-linked p.G56R mutation, and, the compound heterozygotes exhibited an ESCS-like phenotype, which in one of the 2 cases was strikingly “milder” than the patients carrying the p.G56R mutation alone. Impaired repression of cone-specific genes by the corepressors atrophin-1 (dentatorubral-pallidoluysian atrophy DRPLA gene product) and atrophin-2 (RERE repeat protein) appeared to be a molecular mechanism mediating the beneficial effect of the p.R311Q mutation. Finally, the functional dominance of the p.R311Q to the p.G56R mutation is discussed.
Keywords: retinal degeneration, transcriptional regulation, cofactor assembly, corepressor binding, NR2E3, photoreceptor-specific nuclear receptor, PNR
Introduction
NR2E3 (MIM# 604485), also called PNR (photoreceptor-specific nuclear receptor), is a photoreceptor-specific transcription factor of the nuclear hormone receptor superfamily [Kobayashi et al., 1999]. NR2E3 exhibits the evolutionarily conserved modular structure of nuclear receptors, namely a highly conserved DNA-binding domain (DBD) that specifically binds to consensus binding sites located in promoters of target genes, and a ligand-binding domain (LBD) [Chen et al., 2005; Kobayashi et al., 1999]. Although NR2E3 acts as a transcriptional activator of several rod-specific genes, in synergy with the cone-rod homeobox (CRX; MIM# 602225) and the neural retina leucine zipper (NRL; MIM# 162080) transcription factors [Cheng et al., 2006; Cheng et al., 2004; Peng et al., 2005], a physiological function of NR2E3 is thought to be repression of cone-specific genes in rods [Chen et al., 2005; Peng et al., 2005]. For instance, transgenic mice ectopically expressing NR2E3 under the control of the CRX promoter in photoreceptor precursor cells showed a suppression of cone-specific gene expression; consequently, differentiation of cone photoreceptor cells was suppressed, but non-functional rod-like photoreceptor cells were generated [Cheng et al., 2006]. The critical role of NR2E3 for the regulation of photoreceptor generation became evident in the rd7/rd7 mice, a spontaneous loss-of-function model of NR2E3 [Akhmedov et al., 2000; Haider et al., 2001]. In rd7/rd7 retinas, blue (S-) opsin expressing cone cells are generated throughout postnatal life. This abnormal number of cones leads to laminar disorganization and fragmentation of the retina [Akhmedov et al., 2000; Haider et al., 2006; Haider et al., 2001]. NR2E3 might therefore be needed to suppress the cone generation program in late mitotic retinal progenitor cells [Haider et al., 2006].
In humans, mutations in NR2E3 have been associated with the recessively inherited enhanced short wavelength sensitive (S-) cone syndrome (ESCS; MIM# 268100) [Haider et al., 2000]. ESCS is characterized by unique full-field and spectral electroretinographic findings with hyperfunction of S-cones (‘blue’ cones) and impaired M- and L-cone and rod functions [Gouras, 1982; Gouras et al., 1985; Jacobson et al., 1990; Marmor et al., 1990]. To date, twenty different mutations located in the evolutionary conserved DBD and LBD of NR2E3 have been linked to ESCS with c.932G>A (p.R311Q) being the most prevalent [Haider et al., 2000; Hayashi et al., 2005; Milam et al., 2002; Sharon et al., 2003; Wright et al., 2004].
Recently, a c.166G>A (p.G56R) mutation in the NR2E3 gene was reported as one of the more common single causal mutations for autosomal dominant retinitis pigmentosa (adRP) and termed RP37 (MIM# 611131) [Coppieters et al., 2007; Gire et al., 2007]. The phenotype corresponded to that seen in classic adRP, with progressive degeneration of rods and subsequent involvement of cones.
In the present work, we functionally characterized this p.G56R mutation that we had previously identified [Bouayed-Tiab et al., 2006], and showed that dominant negative activity of the mutant protein was the molecular mechanism underlying adRP. We also analyzed a pedigree in which adRP and ESCS segregate due to the presence of a p.G56R/wild-type or a p.G56R/p.R311Q genotype. We addressed the mechanisms by which the recessive trans-acting p.R311Q (ESCS) mutation has a protective effect when uncovered by the dominant cis-acting p.G56R (adRP) mutation. Additionally, we identified atrophin-1, the dentatorubral-pallidoluysian atrophy (DRPLA) gene product [Yazawa et al., 1995], and the related atrophin-2 (RERE repeat protein) [Yanagisawa et al., 2000] as corepressors of NR2E3 and showed that impaired corepressor activity may be the molecular basis of ESCS.
Materials and Methods
Patient Information and Examination
Studies were conducted under protocols approved by the Institutional Review Boards of Hôpital Ophtalmique Jules–Gonin and Columbia University. Written informed consent for clinical examination and DNA analysis was obtained from all individuals. Each patient and several unaffected family members were examined by standard ophthalmological methods including fundus photography and electroretinography (ERG), the older subjects over a 25-year-period. To determine dark-adapted thresholds, a Goldmann-Weekers adaptometer (Haag-Streit, Bern) was used. Since only the final dark-adapted threshold, and not dark adaptation curves, was being determined, a pre-adapting light was unnecessary. Final dark-adapted thresholds were obtained in Foot Lamberts and were compared to values of normal controls on the same adaptometer. Color vision was normal in all subjects on the Farnsworth Panel D-15.
For ERG measurements, the light source was a Grass stimulator recessed in a ganzfeld dome. The subjects were first studied in the dark-adapted state using spectral stimuli obtained by the use of four Kodak Wratten filters (29 (red), 21 (yellow), 61 (green) and 98 (blue)). Then the subjects were light-adapted with a steady ganzfeld background of 53 foot-lamberts and the responses to the four spectral stimuli obtained again (Supplementary Figure S1A). A complete description of this methodology is published [Gouras et al., 1985]. In addition ERG responses were obtained to white light stimulation. Responses were detected at the cornea by Burian-Allen bipolar contact lens electrodes and amplified by a Nicolet computer system with low noise amplifiers, averaging usually 50-100 responses to the same flash presented every second. In one case (proband II-1) a spectral sensitivity function for the ERG was obtained in the dark- and light-adapted state by defining the relationship between ERG amplitude and the energy of stimulation using four additional Wratten filters and then determining the energy required for each filter to produce a constant slightly supra-threshold response in the dark- and light-adapted state (Supplementary Figure S1B).
Reagents
If not stated otherwise, chemical reagents were from Fluka-Sigma (Buchs, Switzerland), oligonucleotides from Eurogentec (Liège, Belgium) and Microsynth (Balgach, Switzerland), DNA purification kits from Macherey-Nagel (Düren, Germany), plasticware from Techno Plastic Products (Trasadingen, Switzerland) and cell culture reagents from Invitrogen.
Genotyping, linkage analysis and mutation detection
Blood samples were obtained for DNA analysis from 11 out of 13 affected and 16 out of 18 non affected members of the Swiss family and all members of the American family. DNA was isolated from leucocytes (Nucleon Bacc2, Amersham Biosciences). As inheritance was clearly dominant in the Swiss family, linkage analysis was performed in a 2-step approach. First, two markers flanking each of the fifteen most frequent adRP loci (CA4, CRX, FSCN2, GUCA1B, IMPDH1, NRL, PRPF3, PRPF8, PRPF31, RDS, RHO, ROM1, RP1, RP9, SEMA4) were chosen and haplotypes were established. Second, because all loci were excluded, a genome-wide linkage analysis was performed with 380 microsatellites from the ABI linkage mapping set 2.5 (average distance between markers: 10 cM). Haplotypes were determined with Autoscan V1.01B and lod (logarithm of odds) scores were calculated using Mlink V5.1 under the assumption of autosomal dominant inheritance with complete penetrance, neo mutation rate of 0.001 and equal allele frequency. Heteroduplex analysis of NR2E3 amplicons was performed using an automated denaturing high performance liquid chromatography (DHPLC) instrument (WAVE, Transgenomic). Amplicons with abnormal retention time were directly sequenced on an ABI 3100XL DNA sequencer (Big Dye Terminator Labeling Kit). Finally, after identification of the p.G56R mutation, the potential promoter region, exonic and intronic sequences of the NR2E3 gene were directly sequenced for all patients after PCR amplification.
NR2E3 cDNA mutagenesis
The cDNA bases were numbered according to the human NR2E3 reference sequence in GenBank NM_014249.2, where +1 corresponds to the A of the ATG translation initiation codon. Mutagenesis was performed according to the QuikChange®II Site-Directed Mutagenesis Kit using PfuUltra polymerase (Stratagene). Briefly, pcDNA3.1/HisC-hNR2E3 [Peng et al., 2005] was amplified with forward 5′-GTGCGGAGACAGCAGCAGCAGGAAGCACTATGGC-3′ and reverse 5′-GCCATAGTGCTTCCTGCTGCTGCTGTCTCCGCAC-3′ oligonucleotides to obtain the pcDNA3.1/HisC-hNR2E3-G56R plasmid. To obtain the pcDNA3.1/HisC-hNR2E3R311Q plasmid, oligonucleotides 5′-CCTGCAGGAAACTATCTCTCGGTTCCAGGCATTGGCGG-3′ and 5′-CCGCCAATGCCTGGAACCGAGAGATAGTTTCCT GCAGG -3′ were used.
Transactivation assays
A mouse S-opsin promoter fragment (opn1sw) spanning nucleotides −736/+17 relative to the ATG was amplified with oligos 5′-GGGGTACCGGTGTGATGATGCTGACAAG-3′ and 5′-CGGGATCCTCATCCTCTC CTGACATCTC-3′, verified by sequencing and then subcloned into KpnI/BglII sites of the pGL2-promoter luciferase reporter vector (Promega). Human embryonic kidney (HEK) 293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10 % FCS and penicillin/streptomycin (Invitrogen). Cells were plated in 12-well plates and transfected at a confluence of 30% with the Calcium Phosphate method (ProFection®, Promega). Per well, 30 ng each of the expression vectors pcDNA3.1/HisC-hNR2E3 (wild-type, p.R311Q and p.G56R), pcDNA3.1/HisC-hCRX and pMT-NRL were used [Peng et al., 2005], together with 500 ng of the luciferase reporter constructs for rhodopsin promoter BR225-Luc [Peng et al., 2005], for M-opsin promoter Mop250-Luc [Peng et al., 2005] or for S-opsin promoter (see above). As internal standard, 50 ng of plasmid CMVβ (Clontech) encoding β-galactosidase was used. For corepressor studies, 33, 66 or 100 ng of the pCMX expression vectors encoding FLAG-tagged atrophin-1, atrophin-2 and NCoR were transfected [Hörlein et al., 1995; Wang et al., 2006]. To keep the total transfected DNA quantity constant, appropriate quantities of pcDNA3.1/HisC and pCMX-FLAG empty vectors were added in all experiments. Enzymatic activities were assessed with Luciferase Assay System (Promega) and standard β-Gal assay. For corepressor studies, we considered only experiments where the β-galactosidase activities were comparable between all wells.
Electrophoretic Mobility Shift Assay (EMSA)
The pcDNA3.1/HisC-hNR2E3 expression vectors were used for in vitro transcription/translation (TNT, Promega). In vitro DNA-binding was tested on the annealed oligonucleotides NR2E3REfor (5′-CCTTTAAAAGTCAAAAGTCAACTTCCAA-3′) and NR2E3RErev (5′-TTCCGTTGGAAGTTGACTTTTGACTTTT-3′). Radiolabeling by Klenow fill-in with 30 μCi of [α-32P]dATP (3000 Ci/mmol) (Hartmann Analytik, Braunschweig) and subsequent probe purification on Sephadex G-50 columns was according to manufacturers instructions (Roche). DNA-binding reactions were carried out in 20 μl of 10 mM Tris (pH 7.5), 160 mM KCl, 1mM DTT, 10% glycerol, 10 μg of sonicated salmon sperm DNA (Roche) and 2 μg of poly(dI-dC) with indicated amounts of programmed reticulocyte lysate. For competition experiments, unlabeled probes were added in 2- to 100-fold molar excess. After an 15-min incubation on ice, 1 ng of 32P-labeled probe was added, and incubations were continued for an additional 15 min at room temperature. For ‘supershift’ experiments, 2 μg of rabbit polyclonal anti-human NR2E3 (1 μg/μl; Chemicon, Millipore) was added to the reaction and incubation proceeded for another 10 min. DNA-protein complexes were separated from free probe on native 4% polyacrylamide gel in 0.5xTBE buffer, gels were dried and revealed by phosphorimaging (Molecular Dynamics).
Animal handling
All experiments performed in this study were in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by the Veterinary Service of the State of Valais (Switzerland). C57BL/6 mice (RCC, Basel, Switzerland) were kept in a 12-h light-dark cycle with unlimited access to food and water.
Chromatin-immunoprecipitations
Experiments were carried out on 6 retinas exactly as described [Peng and Chen, 2005], except protein-protein crosslinking of cells in 1.5 mM ethylene glycolbis[succinimidyl succinate] (EGS) preceding formaldehyde fixation [Zeng et al., 2006]. For immunoprecipitation, a sheep polyclonal anti-atrophin-1 (A-19) antibody was used (Santa Cruz).
In situ hybridization
Eyes were enucleated, rinsed in 1xPBS-DEPC, fixed for 2 h with 4% paraformaldehyde-1xPBS-DEPC and included for overnight in 30% sucrose-1xPBS-DEPC. Eyes were sectioned at −21° C on a Leica CM1900 cryostat and 12-μm sections recovered on SuperFrost®Plus microscope slides (Menzel Gläser, Braunschweig, Germany) pretreated with Vectabond (Vector Laboratories). A 250 bp-probe for atrophin-1 was amplified from reverse transcribed mouse brain mRNA with primers 5′-CTTCGTCACCAGCTTTTTGC-3′ and 5′-GCTTGTCACTCTCCTTCTTC-3′, and then subcloned into the pGEM®-T Easy vector (Promega). DIG-labeled sense and anti-sense probes were tested by immuno-dot blotting. In situ hybridizations were carried out at 48°C as previously described [Braissant and Wahli, 1998].
Results
Mutations in NR2E3 cause dominant or recessive retinal diseases
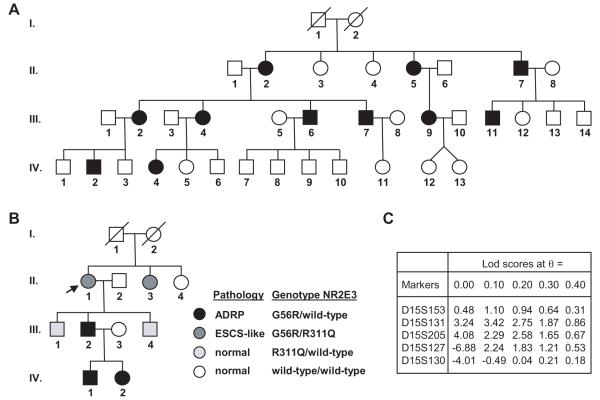
During our systematic screening of families with various forms of RP in Switzerland, we identified a large family where 11 members were affected with classical autosomal dominant RP (Fig. 1A). In this family, a genome-wide linkage analysis had resulted in a strong positive lod score of 4.08 for marker D15S205 (Fig. 1C). Haplotype analysis restricted the shared interval to markers D15S153 and D15S127 (data not shown). As this interval contained the NR2E3 gene, the putative promoter, exons and introns of NR2E3 were directly sequenced and the causal mutation for adRP37 (MIM# 611131) c.166G>A (p.G56R) was identified in all affected individuals. None of the non-affected family members possessed this variant nor was it found in more than 200 ethnic matched control individuals. The patients of the Swiss family did not possess any other NR2E3 variants.
Figure 1.
Pedigrees of affected families of Swiss (A) and Jewish-American (B) origin. All patients with black symbols harbor the G56R heterozygous mutation and are affected with adRP, while the two patients with dark gray symbols are compound heterozygous G56R/R311Q and are affected with ESCS syndrome. Individuals with the heterozygous R311Q mutation (light gray symbols) have no clinical phenotype. Arrow denotes the proband of the American family. C) Lod scores for microsatellite markers obtained in the Swiss family.
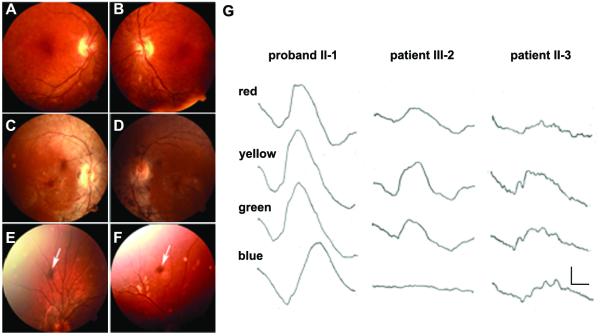
We have followed an American family since 1980 with evidence of retinal degeneration in two generations (Fig. 1B)[Gouras, 1982]. Two affected grandchildren in the third generation were included with time. The proband II-1 had a most unusual ERG that showed supernormal responses to short wavelength stimuli [Gouras et al., 1985], which subsequently became known as the enhanced S-cone syndrome (ESCS) [Jacobson et al., 1990; Marmor et al., 1990] (Fig. 2G; Supplementary Figure S1). The proband had night blindness since childhood but, remarkably, had no evidence of RP at 65 years of age with full visual fields (Fig. 2A and 2B; Table 1). Indeed, a dominant form of RP characterized by retinal pigmentary changes, visual field defects, night blindness and a profound reduction in the electroretinogram (ERG), was present in her son (III-2) and her two grand-children (IV-1, IV-2) (Fig. 2E, 2F, and 2G; Table 1). The parents I.1 and I.2 were long deceased, but there was anecdotal evidence that the mother (I.2) and her sister had vision problems, the latter being blind at elder age. The proband’s sister (II-3) had attenuated retinal vessels and massive clumped intraretinal pigmentation in her mid-peripheral retina (Fig. 2C and 2D). However, an electrophysiological finding suggested a more ESCS-than adRP-like phenotype. Her response to blue light had greater amplitude than that of her nephew’s (III-2), whereas the converse was the case for red light (Fig. 2G). Her dark-adapted threshold to peripheral testing was lower than that of her younger nephew’s (Table 1) and her visual fields in the 1980s were larger than her nephew (III-1). The field of II-3 extended slightly beyond 20 degrees of visual angle while that of her much younger nephew extended to only 10 degrees in the 1980s. Now their visual fields are less than 5 degrees. Therefore both II-3 and III-1 progressed with time but they had different phenotypes. II-3 had more peripheral field function and a more ESCS-like ERG than her younger nephew. Both II-1 and II-3 had evidence of foveal dysfunction as demonstrated by their slightly reduced acuities (Table 1) which has declined very slowly since 1980. The other affected members of the pedigree have retained normal visual acuity.
Figure 2.
Clinical data of the American family. A-F) Fundus photographs. The proband II-1 shows a few small spots of clumped pigmentation in her peripheral retina, mild atrophic changes of the retinal epithelium in the macula, but no attenuation of retinal vessels (A, B). Her sister II-3 shows attenuated retinal vessels and clumped intraretinal pigmentation in her mid-peripheral retina (C, D). Patients IV-1 (E) and IV-2 (F) show a retinal pigmentary clump (arrow) in the peripheral retina. G) ERGs of patients in the American family. The subjects were studied in the dark-adapted state using Wratten filters providing red (Wratten 29), yellow (Wratten 21), green (Wratten 61) and blue (Wratten 98) light (see Methods). The proband, her son and her sister were analyzed in 1980, when the two most severely affected individuals had detectable responses. The proband, who is the oldest of all affected, has much better responses than her sister II-3 and her son III-2. Patient II-3 response to blue light has greater amplitude than that of III-2 (arrows). The converse is true for red light (arrows). The calibration, lower right, indicates 7 microvolts vertically and 20 milliseconds horizontally.
Table 1.
Clinical features of patients in the American family.
| year | 1980 | 2003 | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| dark-adapted thresholds | acuity | visual field | |||||
| subject | age | Central Ft.L |
Peripheral Ft.L |
Central Ft.L |
OD | OS | OU |
| II-1 | 39/62 | 4×10−3 | 4×10−3 | 3×10−3 | 20/40 | 20/50 | full |
| II-3 | 33/56 | 7×10−3 | 7×10−2 | difficult | 20/50 | 20/50 | constricted |
| III-2 | 18/37 | 7×10−3 | 8×10−2 | difficult | 20/20 | 20/20 | constricted |
| IV-1 | 13 | - | - | 4×10−4 | 20/20 | 20/20 | ring scotoma |
| IV-2 | 10 | - | - | 3×10−4 | 20/20 | 20/20 | ring scotoma |
| normal | - | 2-8×10−6 | 1-5×10−6 | 2-8×10−6 | 20/20 | 20/20 | full |
Note: Dark-adapted thresholds (Foot Lamberts =Ft.L) were determined with a Goldmann-Weekers adaptometer (see Methods). All patients had normal color vision. The proband II-1 and her sister II-3 presented macular scotomas. The probands grandchildren IV-1 and IV-2 presented ring scotomas.
Importantly, the proband II-1 and her sister II-3 were determined compound heterozygotes for two NR2E3 mutations: the most common causal mutation for ESCS p.R311Q [Haider et al., 2000] and the causal mutation for adRP p.G56R (Fig. 1B). Her son (III-2) and both grandchildren (IV-1, IV-2) affected by adRP had solely inherited the p.G56R mutation from their mother/grandmother and did not possess any other NR2E3 variants.
On the basis of these different clinical and genetic data, we therefore aimed at elucidating (i) the molecular mechanisms leading to p.G56R-linked adRP, and (ii) the molecular mechanisms underlying the ESCS-like phenotype in the two p.G56R/p.R311Q compound heterozygous patients.
The NR2E3-G56R mutant acts as a dominant negative protein
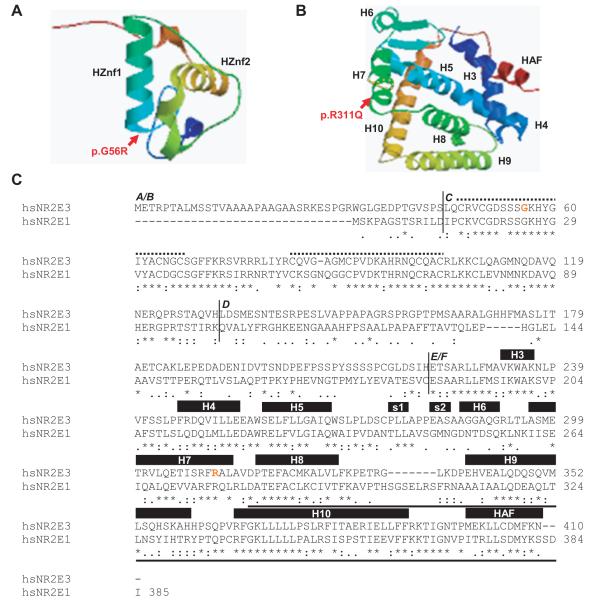
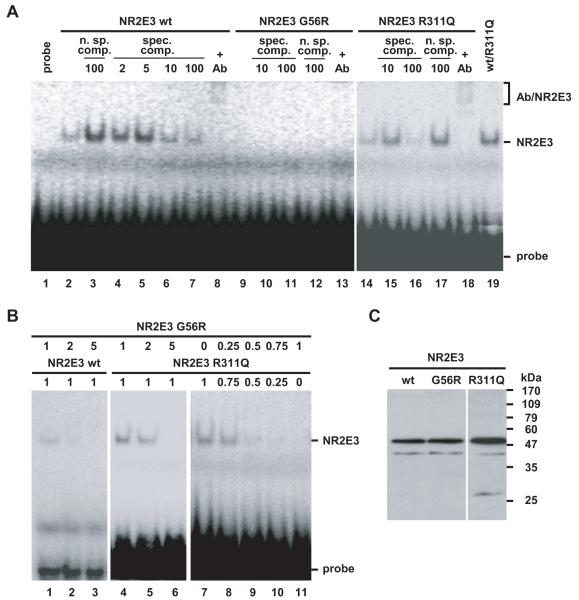
Glycine 56 is located in the highly conserved DBD of NR2E3 at the basis of the α-helical DNA-binding motif of the first so-called ‘zinc-finger’-like structure (Fig. 3). To test whether the p.G56R mutation abolished DNA binding by disrupting the α-helical DNA-binding motif, we performed electrophoretic mobility shift assays with in vitro translated NR2E3 wild-type and mutant proteins on a synthetic NR2E3 response element, consisting of a direct repeat of two NR2E3 consensus binding sites spaced by one nucleotide (Fig. 4). We observed in presence of the NR2E3 wild-type protein a specific complex that had been suggested to be a NR2E3 homodimer (Fig. 4A, lane 2) [Kobayashi et al., 1999]. The specificity of binding was demonstrated by specific competition with increasing amounts of unlabeled oligonucleotides (Fig. 4A, lanes 4-7) and by non-specific competition with an unrelated response element for Ets transcription factors (Fig. 4A, lane 3). Further evidence of specificity was provided by the binding of anti-NR2E3 antisera to the NR2E3 complex, resulting in a slower migrating complex, appearing as a smear in the uppermost part of the lane (Fig. 4A, lane 8). On the contrary to the NR2E3 wild-type protein, no complex was observed in presence of an equivalent amount of the p.G56R protein, indicating that the mutant protein was unable to bind DNA (Fig. 5A, lanes 9-13). Similar to NR2E3 wild-type protein, the p.R311Q protein bound the NR2E3 response element as a specific complex (Fig. 5A, lanes 14-18). We observed a strong signal corresponding to the NR2E3 dimer in presence of both NR2E3 wild-type and p.R311Q proteins (Fig. 4A, lane 19), suggesting that the p.R311Q mutation located in the LBD did not impair dimerization with wild-type NR2E3. Increasing amounts of the p.G56R protein abolished binding of both NR2E3 wild-type and p.R311Q proteins (Fig. 4B, lanes 1-6). By decreasing the amounts of p.R311Q proteins and increasing those of p.G56R, dimer binding to the radiolabeled probe became gradually weaker. Notably, in presence of equivalent amounts of the two NR2E3 mutant proteins, that is potentially mimicking the situation in the heterozygous patients, binding of dimers to DNA was barely detected (Fig. 4B, lanes 7-11). Taken together, this suggested that the p.G56R mutant protein was competent in dimerization, but had a dominant negative effect towards both NR2E3 wild-type and p.R311Q proteins through its inability to bind DNA.
Figure 3.
NR2E3 protein structure. A) The NR2E3 DNA-binding domain (DBD) folds into two ‘zinc finger’-like structures. The α-helical DNA binding motif (HZnf1 and HZnf2) of each zinc finger lies perpendicular to the other. The p.G56R mutation is located at the basis of the α-helix of the first zinc finger. B) The NR2E3 ligand-binding domain (LBD) presents a canonical α-helical structure forming a hydrophobic ligand-binding pocket. R311 is located at the end of helix 7 (H7). Modelling of the mutant protein suggests a slight destabilization of the α-helical turn immediately C-terminal of R311 (orange dots). Ligand-binding involves residues of H7, but is mainly mediated by H3, H6 and HAF (AF-2: Activator Function-2). The dimerization interface is mainly located in H9 and H10. Modeling was performed with SWISSMODEL [Schwede et al., 2003]. C) Human (hs) NR2E1 and NR2E3 amino acid sequences were aligned with ClustalW 1.83 algorithm [Chenna et al., 2003]. A/B: N-terminal A/B domain; C: DBD; D: hinge region; E/F: LBD. Identical amino acids are denoted by arrows, conserved amino acids by dots. In the DBD, the two zinc fingers are indicated by dotted lines. In the LBD, α-helices (H) and β-sheets (s) are indicated by black boxes. A bold line underlines the C-terminal part of the NR2E1 ligand-binding domain sufficient for NR2E1/atrophin-1 interaction [Wang et al., 2006; Zhang et al., 2006]. The sites of p.G56R and p.R311Q mutations are indicated by red letters in the NR2E3 sequence.
Figure 4.
The adRP-linked mutation p.G56R results in a dominant negative NR2E3 mutant protein. A) The p.G56R mutant protein is defective in DNA-binding. Binding of NR2E3 wild-type and mutant proteins to a radiolabeled response element was tested in EMSA by nonspecific (n. sp.) and specific (spec.) competition (comp.). Fold excess of cold probe is indicated. A specific anti-NR2E3 antibody (Ab) recognizes the NR2E3 dimer, inducing the formation of a slower migrating complex (‘supershift’ Ab/NR2E3). Contrast for lanes 1 to 13 was increased to show absence of binding of the p.G56R mutant protein and presence of the ‘supershift’ in lane 8. B) Dominant negative activity of p.G56R towards NR2E3 wild-type and p.R311Q proteins. Amounts of reticulocyte lysates are indicated in μl. C) Western Blot with anti-NR2E3 antisera on 10 μl of in vitro translated proteins. A major band at the expected size of 48 kDa for His-tagged NR2E3 is detected. The higher translation efficiency of pcDNA3.1-NR2E3R311Q contributes to a stronger signal detected by EMSA.
Figure 5.
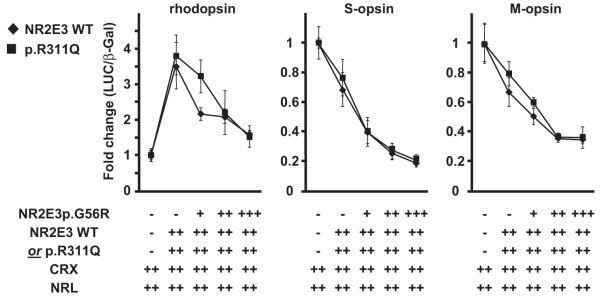
Impaired regulation of opsin genes by the p.G56R mutant protein. 293T cells were transiently transfected with rhodopsin, S-opsin and M-opsin luciferase reporter constructs. CRX and NRL expression vectors (30 ng each) were present in all wells. Activity of NR2E3 wild-type (WT) or p.R311Q mutant protein (30 ng each) was tested in presence of increasing amounts of the p.G56R mutant protein (+:15 ng; ++:30 ng; +++:45 ng). Three independent experiments were performed in duplicates in 12-well plates using the luciferase assay system (Promega). Luciferase activity was normalized to β-galactosidase activity. Normalized luciferase activities of cells transfected with CRX and NRL alone were set to 1. Error bars represent S.E.M.
We further tested the p.G56R mutant protein in 293T cells, using luciferase reporter constructs for opsin promoter fragments (Fig. 5). NR2E3 enhanced CRX/NRL-mediated transactivation of the rhodopsin promoter in these heterologous transactivation assays [Peng et al., 2005]. The p.G56R mutant protein did show dominant negative activity, by abolishing NR2E3-driven potentiation of rhodopsin promoter transactivation, in presence of either NR2E3 wild-type or p.R311Q proteins. In contrast to the transactivation of the rhodopsin promoter, NR2E3 repressed S- and M-opsin promoter activity in HEK293 cell-based assays [Peng et al., 2005]. Surprisingly, the p.G56R mutant further enhanced the transrepression of S- and M-opsin promoters, in presence of either wild-type or p.R311Q proteins. Importantly, there was no difference in transactivation levels of all tested opsin promoters, whether the p.G56R protein was expressed at equal amounts than the wild-type, or the p.R311Q protein, i.e. potentially mimicking NR2E3 expression of patients with genotype p.G56R/wild-type or p.G56R/p.R311Q.
Taken together, these experiments showed that dominant negative activity, i.e. competition for dimerization by a DNA binding-defective mutant, was a molecular mechanism intrinsic to the adRP-linked p.G56R mutant protein, but that this mechanism could not account for the clinical differences observed in patients with genotype p.G56R/p.R311Q affected by ESCS-like degeneration versus patients with genotype p.G56R/wild-type affected by adRP.
Atrophins are corepressors of NR2E3
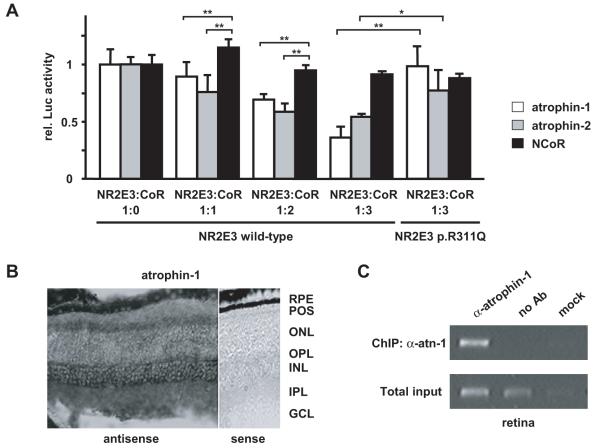
With the aim to elucidate the effect of the p.R311Q mutation in the proband II-1 of the American family and her sister II-3 (Fig. 1B), we analyzed in more details the effect of this second mutation. Arginine 311 is located in helix 7 of the NR2E3 LBD (Fig. 3B). This region participates in the binding of specific ligands to nuclear receptors [Li et al., 2005]. Ligand binding, in turn, regulates cofactor recruitment on nuclear receptors [Nettles and Greene, 2005]. As NR2E3 represses cone-specific genes in rod photoreceptors [Chen et al., 2005; Cheng et al., 2006; Peng et al., 2005], we hypothesized that the p.R311Q mutation might alter the binding to corepressors. The recent description of atrophins as corepressors of NR2E1 [Wang et al., 2006; Zhang et al., 2006] and the high amino acid sequence homology between NR2E1 and NR2E3 (Fig. 3C), prompted us to test atrophins in 293T cell-based transactivation assays for repressor activity on NR2E3 (Fig. 6A). Atrophins enhanced the NR2E3-mediated repression on the M-opsin promoter by over 30% when expressed in 2-fold excess and by over 50% when present in 3-fold excess. In contrast, NCoR, a reported corepressor for class I nuclear hormone receptors [Hörlein et al., 1995], did not enhance M-opsin repression. By in situ hybridization experiments, atrophin-1 was detected throughout the retina, with highest levels in ganglion cells, at the inner limit of the inner nuclear layer where the amacrine cells are preferentially located, and, in the inner segments of the photoreceptors (Fig. 6B). In contrast, atrophin-2 mRNA was not detected in photoreceptors (data not shown). Consistent with a role in the repression of cone-specific genes, atrophin-1 was associated with the proximal M-opsin promoter, as tested by chromatin-immunoprecipitations in adult mouse retinas (Fig. 6C). Expression of atrophin-1 in photoreceptors where NR2E3 is expressed, and association of atrophin-1 to a NR2E3-regulated promoter in vivo, suggested that atrophin-1 was a physiologically relevant corepressor of NR2E3.
Figure 6.
Atrophins are corepressors of NR2E3. a) Atrophins enhanced NR2E3-mediated repression of the M-opsin promoter in presence of NR2E3 wild-type, but not NR2E3 p.R311Q protein. 293T cells were transiently transfected with the M-opsin reporter construct, in presence of NRL, CRX, and, respectively, NR2E3 wild-type or p.R311Q (30 ng each), along with increasing amounts of atrophin-1, atrophin-2 and NCoR (33 ng, 66 ng or 100 ng). At least three independent experiments were performed in duplicates in 12-well plates using the luciferase assay system (Promega). Luciferase activity was normalized to β-galactosidase activity. Normalized luciferase activities of cells transfected without corepressors were set to 1. Error bars represent S.E.M; *: p<0.05; **: p<0.01. B) In situ hybridization for atrophin-1 mRNAs on retinal sections of 2-months-old Bl6/C57. RPE: retinal pigment epithelium; POS: photoreceptor outer segments; ONL: outer nuclear layer; OPL: outer plexiform layer; INL: inner nuclear layer; IPL: inner plexiform layer; GCL: ganglion cell layer. Note the absence of signal in presence of the control sense probe. Scale bar: 50 μm. C) Atrophin-1 associates with the M-opsin promoter in vivo. Chromatin-immunoprecipitation for the M-opsin promoter was performed with anti-atrophin-1 antisera (α-atn-1) on retinas of 2-months-old C57/BL6 mice. As controls, immunoprecipitation without antisera (no Ab) or with the buffer alone (mock) were used. PCR amplifications on total input chromatin (Total input) are shown in the lower panel.
The NR2E3-R311Q mutant is defective in atrophin-dependent M-opsin repression
To test our hypothesis, that NR2E3 wild-type and p.R311Q proteins differentially interacted with corepressors, we performed transactivation assays in 293T cells (Fig. 6A). When present in a 3-fold excess, atrophin-1 repressed the M-opsin promoter by over 50% in presence of wild-type NR2E3 protein, but failed to repress the M-opsin promoter in presence of the NR2E3-R311Q protein (p=0.009). The same was observed to a lesser extent with atrophin-2 (p=0.047). Consistent with a specific binding of atrophins to NR2E3, no difference in M-opsin repression between NR2E3 wild-type and p.R311Q proteins was observed in presence of NCoR. These results suggested that impaired corepressor binding is the molecular basis of ESCS.
Discussion
The aims of the present work were (i) to characterize the p.G56R mutation causing adRP, and (ii) to elucidate why the presence of two mutations in the compound heterozygotes (p.G56R/p.R311Q) resulted in an ESCS-like phenotype and in one of the two cases a much milder phenotype, whereas the presence of the p.G56R mutation alone resulted in rapidly progressing adRP.
Dominant negative activity of the adRP-causing NR2E3-p.G56R mutant protein
We have reported two new families affected by adRP caused by the p.G56R mutation in the NR2E3 gene. This mutation is unique among the reported causal NR2E3 mutations, because it acts in a dominant manner and is located in the first Zn-finger of the DBD. Indeed, the majority of ESCS patients carry mutations in the LBD. The few ESCS patients with mutations in the DBD are either homozygous (G88V, R97H) or compound heterozygous for the common intron 1 splice acceptor mutation (IVS1-2A>C) and R76Q, R76W, R104W or deletion p.65-67 [Haider et al., 2000; Wright et al., 2004]. This latter mutation deletes the P-box located at the C-terminus of the first zinc finger, whereas the other mutations are located in the second zinc finger or in the C-terminal extension of the DBD. Whether these ESCS-linked mutations affect DNA-binding and/or dimerization needs to be determined.
Our functional data showed that absence of DNA-binding, but competition for dimer formation, explained the dominant negative activity exhibited by the p.G56R mutant protein. This provided a molecular basis of adRP observed in the patients. Indeed, if the mutant protein had failed to dimerize, the NR2E3 wild-type protein transcribed from the healthy allele in heterozygous individuals might have been sufficient to maintain normal NR2E3 activity.
The cell-based transactivation studies were useful to functionally characterize the dominant negative activity of the p.G56R mutant protein, but physiological interpretation of these data should be done with caution. On the one hand, the modest decrease by less than 50% in rhodopsin promoter transactivation by transfecting equivalent amounts of NR2E3 wild-type and p.G56R proteins, i.e. the ratio one might expect to occur in heterozygous adRP patients, could be significant in vivo. Indeed, mice expressing about 50% of the rhodopsin levels observed in wild-type mice, showed a clear reduction in rod function, decreased length of photoreceptor outer segments at P15 and thinning of the outer nuclear layer by 1-2 rows at P90 [Lem et al., 1999]. On the other hand, the surprising observation that the p.G56R mutant protein increased the repression of both S- and M-opsin genes in the heterologous transactivation assays, was different from in vivo data, where S-opsin mRNA expression was derepressed in rd7/rd7 mice, but not that of M-opsin [Chen et al., 2005; Corbo and Cepko, 2005]. Alternatively, the increase of S- and M-opsin repression by the p.G56R mutant protein could be attributed to inhibition of an activator protein. The competition between DNA-binding and activator-binding existing for the NR2E3 wild-type protein would then be shifted solely towards activator-binding for the p.G56R mutant protein, resulting in a drop of promoter activity. With respect to this latter potential mechanism, it will be interesting to determine if the p.G56R mutation alters physical interactions with the transcriptional activator CRX [Peng et al., 2005].
Impaired corepressor-binding by the ESCS-linked NR2E3-p.R311Q mutant protein
Individuals carrying the ESCS-causing p.R311Q mutation at a heterozygous state, did not show any clinical phenotype [Haider et al., 2000]. But a beneficial effect of this mutation was uncovered in the compound heterozygous patients of the American family, where it cosegregated with the adRP-causing p.G56R mutation. Strikingly, the ERG of the proband II-1 remained stationary for 25 years with a classical ESCS phenotype. This also indicated that the p.R311Q mutation was functionally dominant to the p.G56R mutation.
The heterologous transactivation assays clearly showed that the p.R311Q mutant protein failed to repress the M-opsin promoter in presence of atrophin corepressors, suggesting that impaired corepressor-binding was the molecular basis of ESCS. A beneficial effect of the recessive p.R311Q mutation in presence of the dominant p.G56R mutation, might therefore be due to increased photoreceptor-specific gene expression caused by impaired repression. As ligands for NR2E3 are not available so far [Wolkenberg et al., 2006], we could not test (i) if the p.R311Q mutation altered the specificity and the affinity towards endogenous ligands and the altered ligand binding caused corepressor release, (ii) if the p.R311Q mutation induced a subtle conformational change in the LBD, permissive for a corepressor/coactivator exchange and, (iii) if corepressors were bound to unliganded and/or ligand-bound NR2E3. It has to be mentioned that the p.R311Q mutation is located outside the atrophin-binding region identified for NR2E1 [Wang et al., 2006; Zhang et al., 2006]. Consistently, in transactivation assays in Y79 human retinoblastoma cells, the corepressor Ret-CoR, structurally unrelated to atrophins, also failed to repress the cyclin D1 promoter in presence of the p.R311Q mutant protein [Takezawa et al., 2007]. These results also showed that NR2E3 repressive activity might be mediated by distinct corepressor complexes, namely during development and in the adult retina.
Whatever the exact molecular mechanisms mediated by the p.R311Q mutation are, they cannot explain alone the clinical differences observed in the proband II-1 and her sister II-3. Modifier effects of unknown (or not tested) genes and/or environmental influences are likely to be involved. Interestingly, an extensive variability in ESCS disease expression has been described in patients with homozygous p.R311Q mutations, v.g. abundance of clumped retinal pigments and rates of retinal degeneration [Audo et al., 2008; Chavala et al., 2005; Gerber et al., 2000; Haider et al., 2000; Hayashi et al., 2005; Milam et al., 2002; Sharon et al., 2003; Wright et al., 2004]. Our observation that atrophins had to be overexpressed in cell-based transactivation assays in order to see an effect of the p.R311Q mutation on transactivation of target genes, suggests that corepressor levels could contribute to these clinical differences in p.R311Q/p.G56R compound heterozygotes and p.R311Q/p.R311Q homozygotes.
In conclusion and based on our experimental data, loss of interaction with a corepressor is a plausible model for how the p.R311Q mutant protein causes ESCS. Understanding the exact mode of action of the p.R311Q mutant protein may open the provoking perspective of introducing by gene therapy the mutant p.R311Q protein in adRP patients carrying the p.G56R mutation, to slow down the progression of adRP.
Supplementary Material
Acknowledgments
We thank affected individuals and their family for participation to this study, Nathalie Voirol, Tatiana Favez, Céline Agosti, Loriane Moret and Dr. Tanja Egener-Kuhn for technical help. The Flag-tagged NCoR vector was a gift of Drs. Manfred Kögl and Andreas Hörlein, DKFZ, Heidelberg, Germany. PG and RA are supported by an unrestricted grant to the Department of Ophthalmology, Columbia University from Research to Prevent Blindness, Inc, New York, USA, and in part by NIH Grants EY015293 (to PG) and EY13435 (to RA). SC is supported by NIH grant EY12543. DFS and FLM are supported by the Swiss National Science Foundation (#31-111948).
References
- Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc. Natl. Acad. Sci. USA. 2000;97(10):5551–5556. doi: 10.1073/pnas.97.10.5551. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audo I, Michaelides M, Robson AG, Hawlina M, Vaclavik V, Sandbach JM, Neveu MM, Hogg CR, Hunt DM, Moore AT. Phenotypic Variation in Enhanced S-cone Syndrome. Invest. Ophthalmol. Vis. Sci. 2008;49(5):2082–2093. doi: 10.1167/iovs.05-1629. others. [DOI] [PubMed] [Google Scholar]
- Bouayed-Tiab L, Delarive T, Agosti C, Borruat F-X, Munier FL, Schorderet DF. A Heterozygous Mutation in the NR2E3 Gene Is Associated With an Autosomal Dominant Retinitis Pigmentosa. Invest. Ophthalmol. Vis. Sci. 2006;47 E-Abstract 1033. [Google Scholar]
- Braissant O, Wahli W. A simplified in situ hybridization protocol using non-radioactive labeled probes to detect abundant and rare mRNAs on tissue sections. Biochemica. 1998;(1):10–16. [Google Scholar]
- Chavala SH, Sari A, Lewis H, Pauer GJ, Simpson E, Hagstrom SA, Traboulsi EI. An Arg311Gln NR2E3 mutation in a family with classic Goldmann-Favre syndrome. Br. J. Ophthalmol. 2005;89(8):1065–1066. doi: 10.1136/bjo.2005.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J. Neurosci. 2005;25(1):118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Aleman TS, Cideciyan AV, Khanna R, Jacobson SG, Swaroop A. In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum. Mol. Genet. 2006;15(17):2588–2602. doi: 10.1093/hmg/ddl185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum. Mol. Genet. 2004;13(15):1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters F, Leroy BP, Beysen D, Hellemans J, De Bosscher K, Haegeman G, Robberecht K, Wuyts W, Coucke PJ, De Baere E. Recurrent Mutation in the First Zinc Finger of the Orphan Nuclear Receptor NR2E3 Causes Autosomal Dominant Retinitis Pigmentosa. Am. J. Hum. Genet. 2007;81:147–157. doi: 10.1086/518426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo JC, Cepko CL. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005;1(2):e11. doi: 10.1371/journal.pgen.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber S, Rozet JM, Takezawa SI, dos Santos LC, Lopes L, Gribouval O, Penet C, Perrault I, Ducroq D, Souied E. The photoreceptor cell-specific nuclear receptor gene (PNR) accounts for retinitis pigmentosa in the Crypto-Jews from Portugal (Marranos), survivors from the Spanish Inquisition. Hum. Genet. 2000;107(3):276–284. doi: 10.1007/s004390000350. others. [DOI] [PubMed] [Google Scholar]
- Gire AI, Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Daiger SP. The Gly56Arg mutation in NR2E3 accounts for 1-2% of autosomal dominant retinitis pigmentosa. Mol. Vis. 2007;13:1970–1975. [PubMed] [Google Scholar]
- Gouras P. Invest. Ophthalmol. Vis. Sci. 1982;20:21. Abstract. [Google Scholar]
- Gouras P, MacKay C, Evers H, Eggers H. In: Retinal Degenerations: Experimental & Clinical Studies. LaVail MM, Hollyfield JG, Anderson RE, editors. 1985. pp. 115–130. [Google Scholar]
- Haider NB, Demarco P, Nystuen AM, Huang X, Smith RS, McCall MA, Naggert JK, Nishina PM. The transcription factor Nr2e3 functions in retinal progenitors to suppress cone cell generation. Vis. Neurosci. 2006;23(6):917–929. doi: 10.1017/S095252380623027X. [DOI] [PubMed] [Google Scholar]
- Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat. Genet. 2000;24(2):127–131. doi: 10.1038/72777. others. [DOI] [PubMed] [Google Scholar]
- Haider NB, Naggert JK, Nishina PM. Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum. Mol. Genet. 2001;10(16):1619–1626. doi: 10.1093/hmg/10.16.1619. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Gekka T, Goto-Omoto S, Takeuchi T, Kubo A, Kitahara K. Novel NR2E3 mutations (R104Q, R334G) associated with a mild form of enhanced S-cone syndrome demonstrate compound heterozygosity. Ophthalmology. 2005;112(12):2115. doi: 10.1016/j.ophtha.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Hörlein AJ, Näär AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Söderström M, Glass CK. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor corepressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. others. [DOI] [PubMed] [Google Scholar]
- Jacobson SG, Marmor MF, Kemp CM, Knighton RW. SWS (blue) cone hypersensitivity in a newly identified retinal degeneration. Invest. Ophthalmol. Vis. Sci. 1990;31:827–838. [PubMed] [Google Scholar]
- Kobayashi M, Takezawa S, Hara K, Yu RT, Umesono Y, Agata K, Taniwaki M, Yasuda K, Umesono K. Identification of a photoreceptor cell-specific nuclear receptor. Proc. Natl. Acad. Sci. USA. 1999;96:4814–4819. doi: 10.1073/pnas.96.9.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolò M, Makino CL, Sidman RL. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc. Natl. Acad. Sci. USA. 1999;96(2):736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Suino K, Daugherty J, Xu HE. Structural and biochemical mechanisms for the specificity of hormone binding and coactivator assembly by mineralocorticoid receptor. Mol. Cell. 2005;19(3):367–80. doi: 10.1016/j.molcel.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Marmor MF, Jacobson SG, Foerster MH, Kellner U, Weleber RG. Diagnostic clinical findings of a new syndrome with night blindness, maculopathy, and enhanced S cone sensitivity. Am. J. Ophthalmol. 1990;110(2):124–134. doi: 10.1016/s0002-9394(14)76980-6. [DOI] [PubMed] [Google Scholar]
- Milam AH, Rose L, Cideciyan AV, Barakat MR, Tang WX, Gupta N, Aleman TS, Wright AF, Stone EM, Sheffield VC. The nuclear receptor NR2E3 plays a role in human retinal photoreceptor differentiation and degeneration. Proc. Natl. Acad. Sci. USA. 2002;99(1):473–478. doi: 10.1073/pnas.022533099. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettles KW, Greene GL. Ligand control of coregulator recruitment to nuclear receptors. Annu. Rev. Physiol. 2005;67:309–333. doi: 10.1146/annurev.physiol.66.032802.154710. [DOI] [PubMed] [Google Scholar]
- Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum. Mol. Genet. 2005;14(6):747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis. Neurosci. 2005;22(5):576–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D, Sandberg MA, Caruso RC, Berson EL, Dryja T. Shared mutations in NR2E3 in enhanced S-cone syndrome, Goldmann-Favre syndrome, and many cases of clumped pigmentary retinal degeneration. Arch. Ophthalmol. 2003;121:1316–1323. doi: 10.1001/archopht.121.9.1316. [DOI] [PubMed] [Google Scholar]
- Takezawa S, Yokoyama A, Okada M, Fujiki R, Iriyama A, Yanagi Y, Ito H, Takada I, Kishimoto M, Miyajima A. A cell cycle-dependent co-repressor mediates photoreceptor cell-specific nuclear receptor function. EMBO J. 2007;26(3):764–774. doi: 10.1038/sj.emboj.7601548. others. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang L, Rajan H, Pitman JL, McKeown M, Tsai CC. Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev. 2006;20(5):525–530. doi: 10.1101/gad.1393506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkenberg SE, Zhao Z, Kapitskaya M, Webber AL, Petrukhin K, Tang YS, Dean DC, Hartman GD, Lindsley CW. Identification of potent agonists of photoreceptor-specific nuclear receptor (NR2E3) and preparation of a radioligand. Bioorg. Med. Chem. Lett. 2006;16(19):5001–5004. doi: 10.1016/j.bmcl.2006.07.056. [DOI] [PubMed] [Google Scholar]
- Wright AF, Reddick AC, Schwartz SB, Ferguson JS, Aleman TS, Kellner U, Jurklies B, Schuster A, Zrenner E, Wissinger B. Mutation analysis of NR2E3 and NRL genes in enhanced S-cone syndrome. Hum. Mutat. 2004;24(5):439–450. doi: 10.1002/humu.9285. others. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Bundo M, Miyashita T, Okamura-Oho Y, Tadokoro K, Tokunaga K, Yamada M. Protein binding of a DRPLA family through arginine-glutamic acid dipeptide repeats is enhanced by extended polyglutamine. Hum. Mol. Genet. 2000;9(9):1433–1442. doi: 10.1093/hmg/9.9.1433. [DOI] [PubMed] [Google Scholar]
- Yazawa I, Nukina N, Hashida H, Goto J, Yamada M, Kanazawa I. Abnormal gene product identified in hereditary dentatorubral-pallidoluysian atrophy (DRPLA) Nat. Genet. 1995;10:99–103. doi: 10.1038/ng0595-99. [DOI] [PubMed] [Google Scholar]
- Zeng P-Y, Vakoc CR, Chen Z-C, Blobel GA, Berger SL. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. BioTechniques. 2006;41(6):694–698. doi: 10.2144/000112297. [DOI] [PubMed] [Google Scholar]
- Zhang CL, Zou Y, Yu RT, Gage FH, Evans RM. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 2006;20(10):1308–1320. doi: 10.1101/gad.1413606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.