Abstract
Lamins are intermediate filament proteins that assemble into a meshwork underneath the inner nuclear membrane, the nuclear lamina. Mutations in the LMNA gene, encoding lamins A and C, cause a variety of diseases collectively called laminopathies. The disease mechanism for these diverse conditions is not well understood. Since lamins A and C are fundamental determinants of nuclear structure and stability, we tested whether defects in nuclear mechanics could contribute to the disease development, especially in laminopathies affecting mechanically stressed tissue such as muscle. Using skin fibroblasts from laminopathy patients and lamin A/C-deficient mouse embryonic fibroblasts stably expressing a broad panel of laminopathic lamin A mutations, we found that several mutations associated with muscular dystrophy and dilated cardiomyopathy resulted in more deformable nuclei; in contrast, lamin mutants responsible for diseases without muscular phenotypes did not alter nuclear deformability. We confirmed our results in intact muscle tissue, demonstrating that nuclei of transgenic Drosophila melanogaster muscle expressing myopathic lamin mutations deformed more under applied strain than controls. In vivo and in vitro studies indicated that the loss of nuclear stiffness resulted from impaired assembly of mutant lamins into the nuclear lamina. Although only a subset of lamin mutations associated with muscular diseases caused increased nuclear deformability, almost all mutations tested had defects in force transmission between the nucleus and cytoskeleton. In conclusion, our results indicate that although defective nuclear stability may play a role in the development of muscle diseases, other factors, such as impaired nucleo-cytoskeletal coupling, likely contribute to the muscle phenotype.
INTRODUCTION
The mammalian nucleus is the largest organelle within the cell. It is separated from the cytoplasm by the nuclear envelope. The nuclear envelope consists of the outer nuclear membrane, which is continuous with the rough endoplasmic reticulum, the inner nuclear membrane and the nuclear lamina (1). The lamina is a proteinaceous network located underneath the inner nuclear membrane and is tightly connected to nuclear pore complexes and nuclear envelope transmembrane proteins. The lamina is primarily formed by two distinct types of proteins, referred to as A- and B-type lamins (2). Lamins are type-V intermediate filaments (3,4), i.e. fibrous proteins with a characteristic tripartite structural organization: an extended, central α-helical ‘rod’ domain flanked by non-α-helical N- and C-terminal domains. Lamins form coiled-coil dimers through interactions of the central rod heptad sequence repeats and further assemble into higher order structures (5,6). In mammalian somatic cells, the most abundant isoforms are lamins A and C, which arise from a single gene, LMNA, by alternative splicing (7), and lamin B1 and B2, encoded by the genes LMNB1 and LMNB2, respectively (8,9). Lamins A/C form interactions with a multitude of nuclear components and are involved in many major nuclear processes, including DNA replication and repair, chromatin organization, transcriptional regulation and stem-cell maintenance and differentiation (reviewed in 10). The recent discovery that mutations in the LMNA gene cause a large variety of human diseases, collectively termed laminopathies, resulted in a rapidly growing interest in the biological functions of lamins A and C. Laminopathies include the autosomal dominant form of Emery–Dreifuss muscular dystrophy (EDMD), limb–girdle muscular dystrophy, dilated cardiomyopathy (DCM), familial partial lipodystrophy (FPLD) and the segmental aging disease Hutchison–Gilford progeria syndrome (11–13). Despite much progress, it remains unclear how mutations in a single, nearly ubiquitously expressed gene can cause such a variety of disorders and why the majority of the more than 400 mutations identified to date primarily affect muscle tissue, whereas other laminopathies mostly lack muscular phenotypes (11,13). Intriguingly, lamin mutations resulting in the same disease are often scattered across the length of the gene, whereas in other cases, different mutations in the same amino acid can cause distinct disease phenotypes (13).
Different, non-mutually exclusive hypotheses have been proposed to explain the broad range of laminopathies: the ‘structural hypothesis’ postulates that mutated lamins assemble into a structurally impaired lamina and lead to more fragile nuclei that rupture and result in cell death, especially in mechanically stressed tissue such as muscle. A variation of the ‘structural hypothesis’ is that mutations in lamins do not affect nuclear stability directly, but rather affect lamin interactions with components of the linker of nucleoskeleton and cytoskeleton (LINC) complex (14), which provides a physical connection between the nuclear interior and the cytoskeleton (15,16). Decreased LINC complex formation might result in impaired nucleo-cytoskeletal coupling, and thereby cause disease. The ‘gene expression hypothesis’ proposes that mutations in lamins A/C can alter gene regulation, and that misregulated, tissue-specific signaling pathways give rise to the various disease phenotypes (17–20). Other hypotheses include altered stem-cell conservation and differentiation, which could result in impaired tissue maintenance and regeneration in laminopathies (21).
Previous studies have identified lamins A and C as fundamental determinants of nuclear mechanical properties and demonstrated that loss of lamin A/C causes weaker, more deformable nuclei (19,22,23). However, the effect of specific disease-causing lamin mutations on nuclear mechanics has never been systematically tested. Therefore, it remains unclear whether mutations responsible for muscular phenotypes have more severe effects on nuclear mechanics compared with mutations linked to other laminopathies. In this study, we systematically tested the effect of laminopathic mutations on various aspects of nuclear mechanics, including nuclear stiffness (i.e. the extent to which the nucleus resists deformation) and nuclear fragility, both aspects of the stability of a nucleus under mechanical stress, as well as nucleo-cytoskeletal coupling, which describes the ability to transmit intracellular forces between the cytoskeleton and nuclear interior. We evaluated nuclear stiffness in skin fibroblasts from patients with EDMD and FPLD, in a panel of Lmna+/− mouse embryonic fibroblasts (MEFs) genetically modified to stably express physiological levels of mutant or wild-type lamin A, and in intact Drosophila melanogaster larval body wall muscle tissue expressing various EDMD mutations. We complemented these studies with assays to evaluate the effect of specific mutations on lamin assembly in vivo and in vitro and on force transmission between the nucleus and cytoskeleton. Our results suggest that specific myopathic lamin A mutations interfere with normal lamin assembly and result in a loss of nuclear stability that likely contributes to muscle-specific phenotypes, but that also other factors, such as impaired nucleo-cytoskeletal coupling, play a role in the development of muscular laminopathies.
RESULTS
Fibroblasts from EDMD patients show increased nuclear deformability
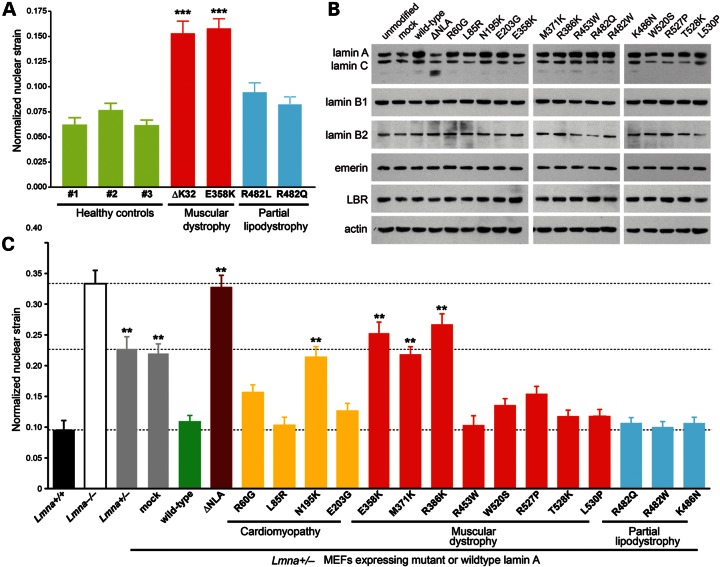
Cells and biopsies from laminopathy patients and mouse models often display misshapen or ruptured nuclei (24–29), providing anecdotal support for the hypothesis that mutated lamins may alter the lamina structure in a way that renders nuclei more susceptible to mechanical stress (30,31). To test whether lamin A/C mutations linked to muscular diseases (e.g. EDMD, DCM) cause particularly strong defects in nuclear mechanics and thereby promote muscular phenotypes, we assessed the mechanical properties of nuclei in primary skin fibroblasts from EDMD patients and compared them with cells from FPLD patients and healthy controls. Nuclear stiffness of adherent cells was inferred from the induced nuclear deformations in response to substrate strain application (32). We found that nuclei of cells from two EDMD patients, carrying the LMNA mutations ΔK32 and E358K, respectively, deformed significantly more than those from passage-matched healthy controls (Fig. 1A), indicating increased nuclear deformability in the EDMD cells. In contrast, cells from two FPLD patients with the LMNA mutations R482Q and R482L, respectively, had normal nuclear stiffness. In addition, we observed that cells from EDMD patients became more frequently damaged during the strain application than cells from healthy controls, indicating increased sensitivity to mechanical stress (Supplementary Material, Fig. S1A and B). In particular, close to 12% of EDMD fibroblasts with the LMNA E358K mutation were damaged during strain application, whereas <1% of cells were damaged in the three control cell lines (Supplementary Material, Fig. S1C).
Figure 1.
Myopathic mutations in LMNA have variable effects on nuclear stiffness. (A) Normalized nuclear strain measurements of skin fibroblasts from healthy controls (green) and human patients with LMNA mutations that cause EDMD (red) or FPLD (blue). Measurements were performed at passage numbers between P8 and P15. In our experiments, we observed no correlation between passage number and nuclear deformability. ***P < 0.001, versus healthy controls. (B) Expression levels of nuclear envelope proteins in Lmna+/− MEFs stably expressing the empty vector (mock), wild-type lamin A, head-truncated ΔNLA or disease-specific lamin A mutations. Actin was used as loading control. (C) Normalized nuclear strain measurements of unmodified Lmna+/+, Lmna−/− and Lmna+/− fibroblasts, as well as of Lmna+/− fibroblasts expressing specific lamin A mutations, wild-type lamin A, head-truncated ΔNLA or the empty vector (mock). Expression of wild-type lamin A rescued nuclear stiffness to levels comparable with Lmna+/+ cells; in contrast, ΔNLA resulted in stiffness comparable with Lmna−/− fibroblasts. **P < 0.01 and ***P < 0.001, compared with Lmna+/− MEFs expressing wild-type lamin A.
Lamin mutations causing EDMD or DCM, but not FPLD, fail to restore nuclear stiffness
Since analysis of patient fibroblasts is limited by the heterogeneous genetic background and the limited availability of samples, we developed a comprehensive approach to test a wider panel of lamin A mutations associated with EDMD, DCM and FPLD in a genetically uniform background. We ectopically expressed mutant or wild-type lamin A in MEFs lacking a single Lmna allele (Lmna+/− MEFs), which express only ∼50% of the normal levels of lamin A/C (33,34). The lamin mutations were introduced with a bicistronic retroviral construct consisting of non-tagged human lamin A and a ZsGreen fluorescent reporter. Since both proteins are translated from a single bicistronic mRNA transcript and expressed at similar levels, we were able to sort for cells with physiological levels of ectopically expressed lamin A by fluorescence-activated cell sorting for ZsGreen. The resulting model system closely resembles the situation in laminopathy patients, who typically carry autosomal dominant LMNA mutations and express approximately equal amounts of mutant and wild-type lamins. Furthermore, expressing the mutations in the Lmna+/− MEF background and comparing them with Lmna+/+ and Lmna−/− MEFs enabled us to distinguish whether any observed effects of specific mutations were dominant-negative or caused by a loss-of-function of the mutant lamin A protein (i.e. haploinsufficiency). We tested a total of 15 mutations, representing EDMD (8 mutations), DCM (4 mutations) and FPLD (3 mutations) (summarized in Table 1). For comparison, we evaluated Lmna+/− MEFs stably expressing either the empty vector (mock control) or wild-type lamin A. As an additional control, we assessed the effect of a lamin A mutant lacking the N-terminal 33 amino acids (ΔNLA), which are critical for the assembly of higher order lamin structures. When expressed in mammalian cells, ΔNLA fails to assemble into the lamina and disrupts the existing lamin network, thus acting in a dominant-negative manner (35).
Table 1.
Human disease-associated LMNA mutations used in this study
| Mutation | Disease | Reference |
|---|---|---|
| ΔK32 | EDMD | (68) |
| R60G | DCM-CD, DCM-CD + FPLD, DCM-CD + FPLD + CMT2 | (62) |
| L85R | DCM-CD | (62) |
| N195K | DCM-CD | (62) |
| E203G | DCM-CD | (62) |
| E203K | DCM-CD | (77) |
| E358K | EDMD | (78) |
| M371K | EDMD | (78) |
| R386K | EDMD | (78) |
| R453W | EDMD, EDMD + FPLD, LGMD1B | (78) |
| R482Q | FPLD | (79) |
| R482W | FPLD, FPLD + LGMD | (80) |
| K486N | FPLD | (81) |
| W520S | EDMD | (78) |
| R527P | EDMD, EDMD + FPLD, LGMD1B | (82) |
| T528K | EDMD, LGMD1B | (78) |
| L530P | EDMD | (82) |
| ΔNLA | Synthetic dominant-negative construct | (35) |
The tested mutations were R60G, L85R, N195K E203K and E203G, causing DCM (62,77); ΔK32, E358K, M371K, R386K, R453W, W520S, R527P, T528K and L530P, found in patients with EDMD (68,78,82); and R482Q, R482W and K486N, identified in patients with FPLD (79–81). In addition, we expressed the engineered dominant-negative construct ΔNLA, which disrupts endogenous lamin organization (35). Note that, although most mutations affect both lamin A and lamin C, we expressed only modified lamin A in MEF cells.
EDMD, Emery–Dreifuss muscular dystrophy; LGMD1B, limb–girdle muscular dystrophy type 1B; DCM-CD, dilated cardiomyopathy with conduction defect; CMT2, autosomal recessive Charcot–Marie–Tooth disease; FPLD, familial partial lipodystrophy type Dunnigan (FPLD2). For additional information on these mutations, see also http://www.umd.be/LMNA/.
We confirmed that the modified cells expressed the lamin constructs at physiological levels by western analysis (Fig. 1B and Supplementary Material, Fig. S2). Moreover, expression levels of lamin B1 and other nuclear envelope proteins such as emerin and lamin B receptor were not notably altered by ectopic expression of the lamin A variants (Fig. 1B). However, we found a decrease in the expression levels of lamin B2 for some of the mutants, most prominently for R453W, R482Q and L530P. Nuclear strain experiments confirmed that the nuclear stiffness of non- and mock-modified Lmna+/− MEFs has an intermediate value compared with that of Lmna+/+ and Lmna−/− cells (19), consistent with the lower levels of lamins A/C in these cells. Ectopic expression of wild-type lamin A in Lmna+/− MEFs completely restored nuclear stiffness to levels of Lmna+/+ MEFs (Fig.1C), whereas expression of the dominant negative ΔNLA mutant caused an increase in nuclear deformability in the modified Lmna+/− MEFs, resulting in values comparable to Lmna−/− cells. In contrast, none of the disease-causing lamin mutations resulted in dominant-negative effects on nuclear deformability. Four of the tested lamin A mutants associated with muscular disease, i.e. N195K (DCM), E358K (EDMD), M371K (EDMD) and R386K (EDMD), failed to restore nuclear stiffness and caused nuclear deformations similar to those observed in mock and non-modified Lmna+/− MEFs. These findings imply that these mutant lamins result in a loss of structural function and fail to form a lamina network that can withstand mechanical forces as efficiently as a lamina formed from wild-type lamin. Although these findings implicate increased nuclear deformability in the pathogenesis of myopathic laminopathies, other DCM and EDMD mutations partially or completely restored nuclear stiffness (Fig. 1C), indicating that additional mechanisms may contribute to the disease mechanism. Importantly, all three tested FPLD mutations completely restored nuclear stiffness as efficiently as wild-type lamin A, suggesting that the FPLD mutations have no effect on the structural function of lamin A.
Myopathic lamin A mutations that cause defects in nuclear stability are more soluble
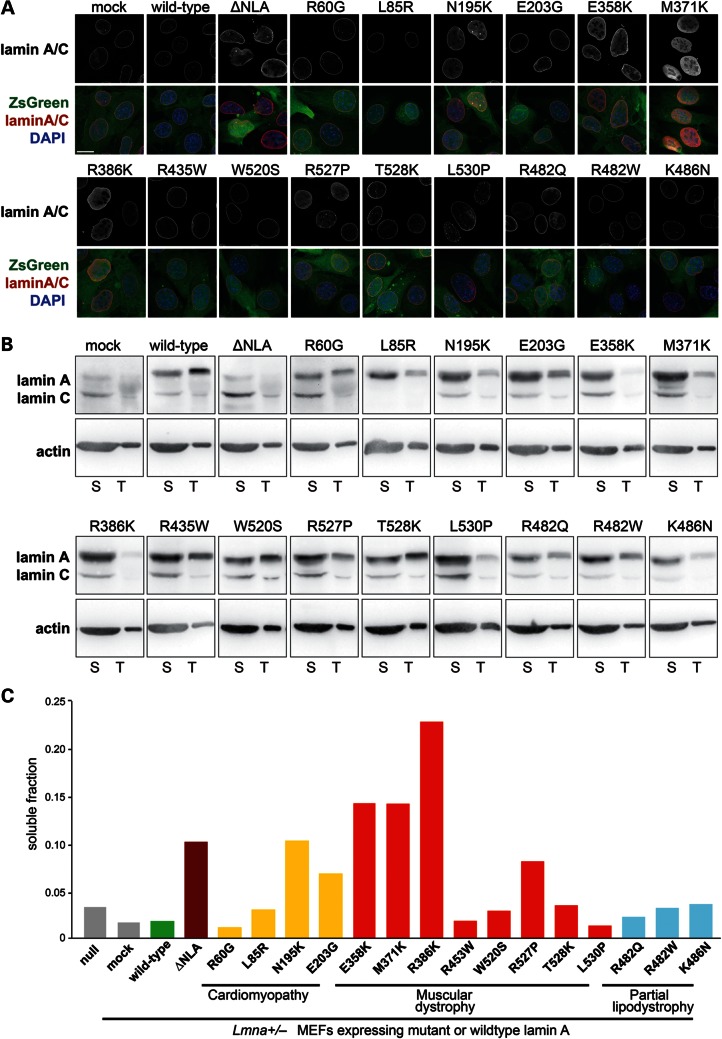
To investigate possible molecular mechanisms for the loss of structural functions in some of the EDMD and DCM mutations, we investigated the intranuclear localization of the various mutants, as increased nucleoplasmic localization could indicate impaired incorporation of mutant proteins into the nuclear lamina. In wild-type cells and in Lmna+/− MEFs expressing wild-type lamin A, lamins A/C were predominantly localized at the nuclear rim (Fig. 2A). In contrast, cells expressing any of the mutations associated with the loss of nuclear stability (i.e. N195K, E358K, M371K and R386K and ΔNLA) had a prominent nucleoplasmic localization of lamins A and C (Fig. 2A). Since lamin A/C-deficient cells have an abnormal distribution of emerin (34), we also assessed expression and localization of emerin in our panel of cells. However, we did not observe any obvious defects in emerin expression (Fig. 1B) or localization (Supplementary Material, Fig. S3). Since the nucleoplasmic pool of lamin A was increased in cells expressing the lamin A variants N195K, E358K, M371K and R386K and the ΔNLA construct, we next quantified the soluble fraction of lamin A in these cells by a sequential protein extraction procedure using mild detergents (36). Cells expressing ΔNLA and the N195K, E358K, M371K and R386K mutants had substantially increased fractions of soluble lamin A compared with wild-type expressing cells (Fig. 2B and C), suggesting that these lamin A mutations fail to correctly incorporate into the nuclear lamina and thereby result in impaired nuclear stability.
Figure 2.
Myopathic lamin A mutations that cause defects in nuclear stability have increased nucleoplasmic distribution and are more soluble. (A) Immunofluorescence staining for lamin A/C in Lmna+/− fibroblasts stably expressing the empty vector (mock), wild-type lamin A, head-truncated ΔNLA or disease-specific lamin A mutations (upper panels) and overlaid with the ZsGreen signal and DAPI chromatin staining (lower panels). Cells expressing the ΔNLA, N195K, E358K, M371K and R386K mutations have increased nucleoplasmic localization of lamin A/C. Scale bar: 20 µm. (B) Soluble lamin A protein fraction (S) versus total lamin A and C levels in Lmna+/− fibroblasts stably expressing the empty vector (mock), wild-type lamin A, head-truncated ΔNLA or disease-specific lamin A mutations, as detected by western analysis. The soluble fraction contains lamin protein that is not incorporated into the nuclear lamina, and was therefore extracted by treatment of cells with mild detergent. Note that only 1/30 of the total lamin A fraction, compared with the soluble fraction, was loaded on the gel. Similar protein levels were loaded, as reflected by actin staining. Since different amount of cells were loaded onto gels for each cell line, a direct comparison of amounts of soluble or total Lamin A fraction between cell lines is not possible, and it is the ratio between soluble to total protein that is used for the interpretation of the results. Extraction was performed three independent times; one representative panel is shown. (C) Quantification of the soluble lamin A fraction of the cells analyzed in (B), indicating that ΔNLA and the disease-specific lamin variants N195K, E358K, M371K and R386K are more soluble than wild-type lamin A and other lamin variants.
Different amino acid substitutions in the same amino acid positions can have distinct effects on lamin structural function
All lamins share a conserved, tripartite organization comprising a central coiled-coil rod domain, as well as non-helical head and tail domains (37,38). The coiled-coil rod is the principal subunit for all structural states, both for extended filaments and for more complex arrays formed from filaments as revealed early on by in vitro assembly studies (39) and recently developed further in detail by cryoelectron tomography (40). It is clear from these studies that these coiled coils interact by multiple longitudinal and lateral interactions mediated by the many ionic side chains that are found on the surface of the coiled coil, to form higher order structures. Strikingly, the four mutations that had the most severe effects on nuclear structure were localized within, or in the case of R386K just three amino acids after the predicted coiled-coil rod domain of lamin A (41) (Supplementary Material, Fig. S4A). Moreover, the mutated amino acids were located in the outward-facing positions f or g of the heptad repeat of the coiled-coil dimer (Supplementary Material, Fig. S4B) and were substitutions to the positively charged lysine. On the other hand, substitution of E203, another outward-facing position in the central rod heptad repeat, to the neutral glycine had no effect on the structural function of the protein. Based on these findings, we hypothesized that mutations to lysine in these outward-facing positions might interfere with the higher order assembly of lamin dimers. We therefore tested whether substitutions to small uncharged amino acids (alanine, glycine) could restore normal nuclear stiffness. In the case of E358, both alanine and glycine substitutions rescued nuclear stiffness, supporting our hypothesis. However, in the case of M371, only the glycine substitution restored nuclear stiffness, whereas substitution to alanine resulted in increased nuclear deformability; conversely, in the case of R386, only alanine but not glycine rescued nuclear stiffness (Supplementary Material, Fig. S4C). In addition, we tested whether mutation of amino acid E203 to lysine resulted in increased deformability, i.e. a loss of structural function. Surprisingly, neither lysine nor alanine at position E203 caused nuclei to become more deformable. These data suggest that each individual amino acid position and each substitution can have different effects on structural functions of lamin A and the corresponding nuclear stiffness, consistent with the finding that different amino acid substitutions in the same position can result in different laminopathies (13).
Lamin mutations that cause impaired nuclear stability have disturbed filament and paracrystal assembly in vitro
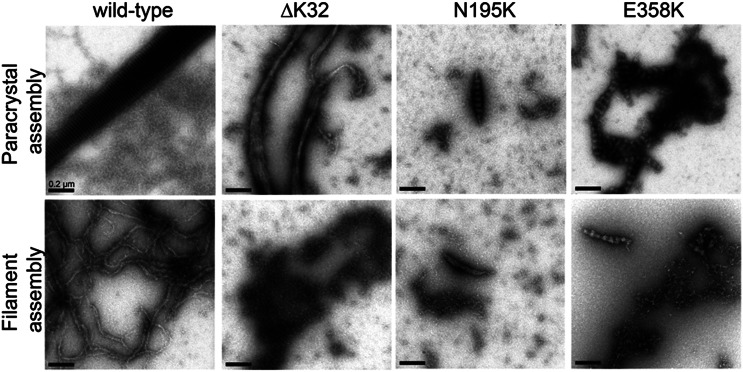
Since our solubility studies suggested that lamin A mutants that have impaired structural function fail to correctly incorporate into the lamina network, we decided to directly assess their assembly into higher order structures. Recombinant human lamin A can be induced to assemble into either paracrystalline structures or filaments in vitro, depending on buffer conditions and dialysis procedure (42). We evaluated wild-type lamin A and three lamin mutations that caused the loss of structural function in our nuclear strain assay, namely ΔK32, N195K and E358K mutants (Fig. 1A and C). Although wild-type lamin A formed the expected large structures of laterally organized fibers (paracrystalline arrays), the three lamin A mutants failed to assemble into well-organized structures and instead formed small aggregates lacking the typical 24.5 nm repeat pattern (Fig. 3, top row and Supplementary Material, Fig. S5). Similarly, only wild-type lamin A assembled into regular extended filaments in vitro (Fig. 3, bottom row and Supplementary Material, Fig. S5). These results suggest that the loss of structural properties of lamins through distinct mutations associated with more deformable nuclei is caused by impaired assembly interactions of the mutant proteins, consistent with recent reports on Caenorhabditis elegans lamin carrying amino acid substitutions that correspond to laminopathic mutations (43,44).
Figure 3.
Impaired in vitro assembly of myopathic lamin A mutations that alter nuclear mechanical properties. Recombinant human wild-type lamin A ΔC18 (i.e. mature lamin A) as well as the single-amino-acid deletion ΔK32 and the point mutations N195K and E358K were induced to assemble into paracrystalline structures (upper panel) or filaments (lower panel) in vitro and imaged by transmission electron microscopy. In contrast to wild-type lamin A, the lamin A variants ΔK32, N195K and E358K failed to assemble into regularly-organized structures and instead formed irregular aggregates. Scale bar: 0.2 µm.
Nuclei of D. melanogaster larvae body wall muscle expressing disease-causing Lamin C variants are more deformable
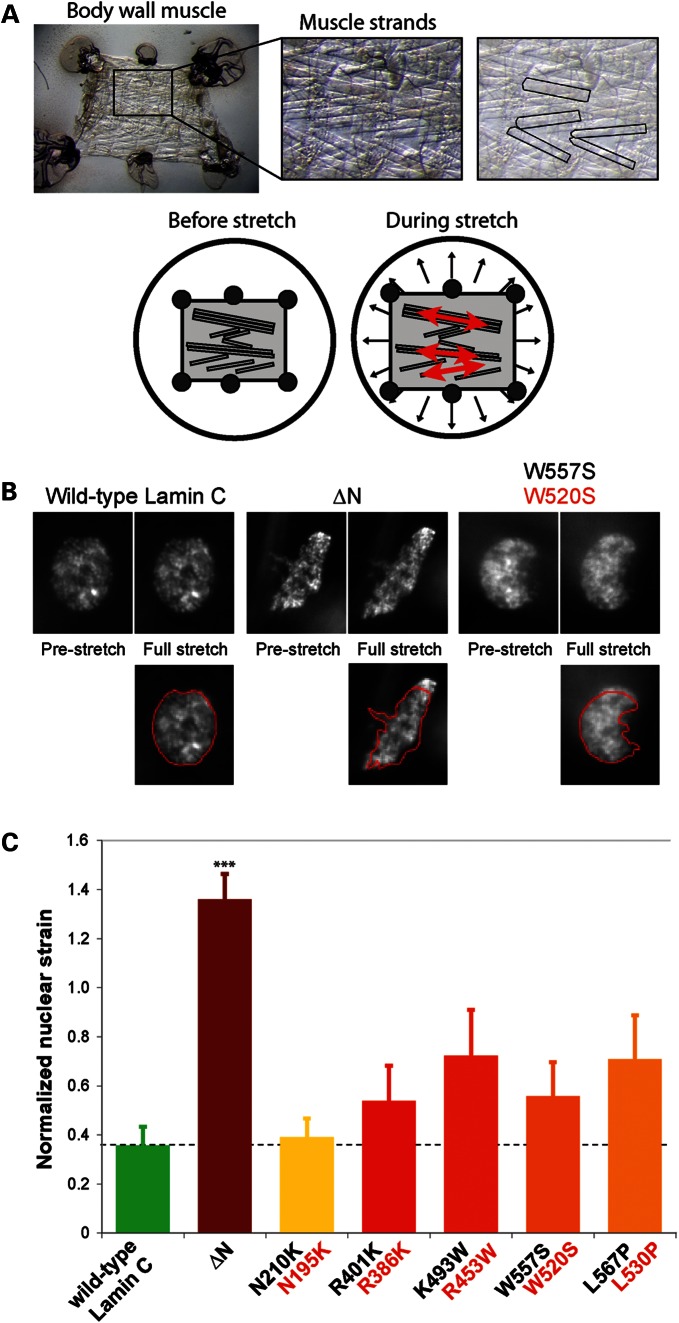
To test whether lamin mutations associated with muscular dystrophy also cause defects in nuclear stability in muscle tissue, we developed a novel experimental assay that enables us to examine nuclear deformability in the intact body wall muscle of D. melanogaster third instar larvae subjected to mechanical strain (Fig. 4A and B). D. melanogaster possesses genes encoding Lamin C and lamin Dm0, considered to share properties with mammalian A- and B-types, respectively. D. melanogaster can serve as a model to study human laminopathies (45–47), and flies and larvae lacking Lamin C show similar defects in nuclear structure as cells from Lmna−/− mice and patients with EDMD (47). Muscle-specific expression of Drosophila Lamin C possessing mutations modeled after those causing human muscle laminopathies leads to larval locomotion defects, nuclear and cytoplasmic abnormalities and, in some cases, lethality at the pupal stage (46). We assessed nuclear deformability in the body wall muscles of six different D. melanogaster strains, expressing wild-type Lamin C, the headless variant of Lamin C lacking the first 42 amino acids (Lamin C ΔN) (47), and the Lamin C variants N210K, R401K, K493W, W557S and L567P, corresponding to the human point mutations N195K, R386K, R453W, W520S and L530P, respectively, which cause DCM (N195K) and EDMD (R386K, R453W, W520S and L530P) in humans. The mutant lamins were expressed using the Gal4/UAS system (48), with the C57 Gal4 driver stock providing the muscle specificity (49). Note that the wild-type and mutant lamins were expressed in an otherwise wild-type genetic background, allowing for tests of dominant-negative function. Body wall muscle expressing the ΔN mutation had misshapen nuclei (Fig. 4B), consistent with previous reports (46). These nuclei also deformed significantly more under applied strain, compared with muscle nuclei expressing wild-type lamin and non-transgenic larvae (Fig. 4C). Similar to our results in MEFs, where only the ΔNLA mutation caused dominant-negative effects on nuclear stiffness, muscle-specific expression of lamin point mutations resulted in substantially milder disturbances in nuclear deformability. Although the differences were not statistically significant compared with the expression of wild-type Drosophila Lamin C, we observed a clear trend toward softer nuclei of larvae muscle expressing EDMD mutations (Fig. 4C). This reduced effect on nuclear stiffness in Drosophila muscle cells, compared with MEF cells, could be attributed to the fact that in the Drosophila system mutations were expressed in a wild-type background of Lamin C expression, whereas in MEF cells mutations were expressed in a Lmna+/− background. Importantly, nuclear stiffness of epidermal cells, which did not express the mutant lamins, was indistinguishable between all strains, including larvae with muscle-specific expression of the ΔN mutant (1.248 ± 0.019 for unmodified larvae, 1.239 ± 0.017 and 1.248 ± 0.023 for wild-type and ΔN-expressing larvae, respectively), confirming that the observed defects in the myonuclei were a direct consequence of the muscle-specific expression of Lamin C mutants.
Figure 4.
Decreased nuclear stiffness in body wall muscle of D. melanogaster larvae expressing Lamin C variants causing EDMD. (A) Overview of the experimental approach to observe nuclear deformations in D. melanogaster larval filet subjected to strain application. Upper panel: the larval filet, consisting of the body wall muscle, is glued to a transparent silicone membrane (left panel); individual muscle fibers are easily detectable (middle and right panel, with distinct muscle fibers highlighted in the right panel). Lower panel: although biaxial strain is applied to the Drosophila larval filet, the tissue strain in the muscle fibers is almost completely uniaxial in the direction of the muscle fiber. (B) Change in nuclear shape in pre-stretched and fully stretched muscle strands of larvae expressing wild-type Lamin C (the A-type lamin of D. melanogaster), head-truncated ΔN Lamin C or the W557S Lamin C mutation, which corresponds to the human LMNA W520S mutation. Muscle cells expressing the ΔN lamin mutation deformed significantly more under applied strain. Images in the lower panel represent nuclei under strain, overlaid with the nuclear contour from the pre-stretch state in red. (C) Normalized nuclear stiffness of D. melanogaster larvae expressing wild-type D. melanogaster Lamin C, head-truncated Lamin C ΔN or Lamin C mutations (labeled in black) that correspond to human EDMD-causing LMNA mutations (labeled in red). ***P < 0.001, compared with muscle nuclei expressing wild-type Lamin C.
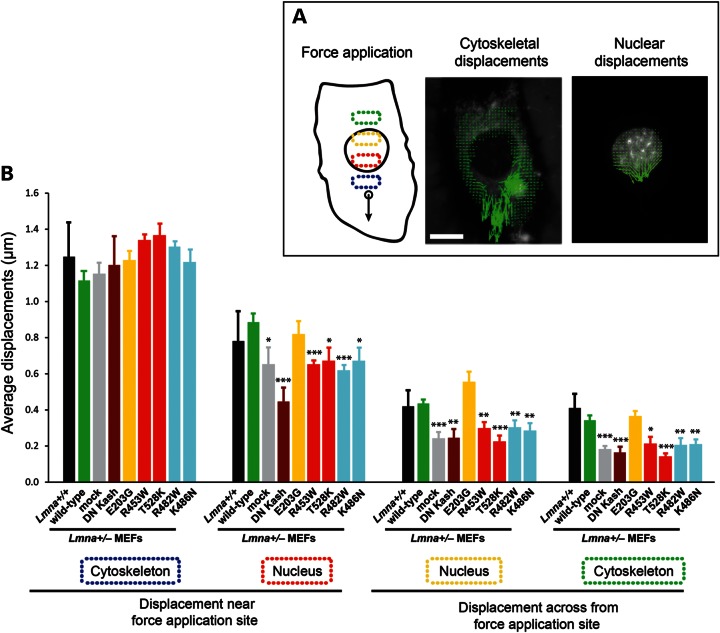
Laminopathic lamin A mutations disrupt nucleo-cytoskeletal coupling
Our findings that only a subset of the DCM- or EDMD-causing mutations resulted in significantly impaired nuclear stability suggests that additional mechanisms may contribute to the muscular phenotype. One such mechanism could be that lamin mutations disrupt the physical coupling between the nucleus and cytoskeleton, which plays important roles in muscle function. Lamins A/C interact with inner nuclear membrane proteins containing a SUN (Sad1p and Unc-84 homology) domain such as Sun1 and Sun2 (50). Through the SUN domain, these proteins associate across the perinuclear space with the KASH (Klarsicht, Anc-1 and Syne homology) domain of a protein family termed nesprins (51–53) located on the outer nuclear membrane; nesprins in turn connect to cytoskeletal components including actin, microtubules (via dynein) and intermediate filaments (via plectin) (14,54). Lamin mutations could interfere with this nucleo-cytoskeletal connection, and defects in nucleo-cytoskeletal coupling and anchoring have been reported in Lmna−/− mice (16) and MEFs (15,55). To directly test the effect of specific lamin mutations on intracellular force transmission between the nucleus and cytoskeleton, we applied a technique recently developed in our laboratory (15), in which we locally apply force to the cytoskeleton while measuring the induced nuclear and cytoskeletal displacements across the cell (Fig. 5A). We performed these measurements on Lmna+/− MEFs stably expressing the empty vector (mock), wild-type lamin A or five different laminopathic lamin A mutations: E203G causing DCM; R453W and T528K causing EDMD; and the FPLD mutations R482W and K486N. Importantly, all of these mutations displayed normal nuclear deformability in our nuclear strain assays (Fig. 1C). For comparison, we included Lmna+/+ MEFs, which serve as control for normal nucleo-cytoskeletal coupling, and Lmna+/− MEFs stably expressing a dominant-negative nesprin mutant consisting of the KASH domain of human nesprin 1α (DN KASH) that is sufficient to disrupt nucleo-cytoskeletal coupling by displacing endogenous nesprins into the endoplasmic reticulum (15). Lmna+/− MEFs expressing either the empty vector (mock) or DN KASH had significantly impaired force transmission between the nucleus and cytoskeleton, as reflected by smaller nuclear and cytoskeletal displacements in areas away from the force application site (Fig. 5B) compared with Lmna+/+ MEFs. This indicates that lamin A expression levels from only one functional Lmna allele is not sufficient to establish the LINC complexes necessary to provide as strong a coupling between the nucleus and the cytoplasm as in Lmna+/+ MEFs. Reintroduction of wild-type lamin A was sufficient to completely restore intracellular force transmission in Lmna+/− cells to levels comparable with Lmna+/+ MEFs (Fig. 5B). In contrast, all tested mutations, with the exception of the E203G mutant, failed to rescue intracellular force transmission (Fig. 5B), indicating that these mutations interfere with nucleo-cytoskeletal coupling despite having apparently normal incorporation into the nuclear lamina and maintaining normal nuclear stiffness. Surprisingly, these defects were also visible in the two FPLD mutations.
Figure 5.
Laminopathic lamin A mutations disrupt force transmission between the nucleus and cytoskeleton. (A) Schematic overview of the microneedle manipulation experiments to assess intracellular force transmission between the nucleus and cytoskeleton. A fine microneedle is inserted into the cytoplasm near the nucleus and moved toward the periphery (left panel). Displacement maps of the cytoskeleton (middle panel) and the nucleus (right panel) are plotted by tracking intracellular fluorescent markers; measurements of the average displacements within four defined areas inside the cytoplasm and nucleus (indicated by dotted boxes in the left panel) are used to evaluate the intracellular force transmission between the nucleus and cytoskeleton. Scale bar: 20 µm. (B) Average displacements at the indicated regions in Lmna+/+ MEFs, and in Lmna+/− fibroblasts stably expressing the empty vector (mock), wild-type lamin A, head-truncated ΔNLA or disease-specific lamin A mutations. The displacements in the first cytoskeletal region were comparable in all cells, illustrating similar strain application with the microneedle. Lmna+/− cells expressing mutant lamins (except for the E203G mutations) and the empty vector had significantly smaller nuclear and cytoskeletal displacements in the three cellular regions observed away from the site of strain application compared with Lmna+/− cells expressing wild-type lamin A or non-modified Lmna+/+ cells, indicating reduced intracellular force transmission between the nucleus and cytoskeleton in the mutant cells. *P < 0.05; **P < 0.01; ***P < 0.001 versus Lmna+/− MEFs expressing wild-type lamin A.
DISCUSSION
The finding that different mutations in the LMNA gene cause a variety of human diseases and that many of these mutations result in tissue-specific defects has continued to puzzle researchers for years. Gross alterations in nuclear morphology are often noted as a hallmark of laminopathies (24–29,31), and biopsies from muscular dystrophy and DCM patients have revealed defects in nuclear envelope continuity, loss of compartmentalization and even the presence of mitochondria within the nuclear interior (25,30,56), suggesting physical damage to mechanically fragile nuclei. Nonetheless, it has remained unclear to what extent mechanical defects, i.e. impaired nuclear structure and stability or disruption of nucleo-cytoskeletal connections, contribute to the development of muscular phenotypes observed in many laminopathies.
We performed a comparative study on the effect of a panel of lamin mutations, representing EDMD, DCM and FPLD, on nuclear mechanical properties. We used human patient fibroblasts as well as Lmna+/− MEFs modified to express different disease-causing lamin A mutations. The latter cell line was previously reported to still express a truncated fragment of lamin A (57). In our studies, we found Lmna+/+, Lmna+/− and Lmna−/− MEFs to be a valid system to test the effect of laminopathic mutations on nuclear stability: we found that nuclear stiffness shows a dose-dependent increase with the amount of wild-type lamin A, and nuclear deformability increases with loss of its expression (Fig. 1C). Moreover, the reintroduction of wild-lamin A into Lmna+/− MEFs completely restored nuclear stiffness to values comparable with Lmna+/+ MEFs. Therefore, the truncated lamin A fragment present in the Lmna+/− and Lmna−/− MEFs had no effect on nuclear deformability.
In our studies on both patient cells and modified Lmna+/− MEFs, we found that all lamin mutants responsible for FPLD had normal nuclear stiffness. In contrast, several mutations associated with EDMD and DCM resulted in a loss of nuclear stability, as evidenced by increased deformation of the nuclei and enhanced sensitivity to mechanical strain. The findings are consistent with previous reports that FPLD mutations, which are often clustered around a small surface region of the lamin Ig-fold, do not affect lamin diffusional mobility (58,59). Of note, we found the most severe defects in EDMD and DCM mutations affecting the N-terminal domain of A-type lamins and specific amino acid substitutions in outward-facing positions of the coiled coil of the central rod, implying that defects in the higher order assembly of lamins are responsible for the loss of structural function. We speculate that distinct myopathic mutations might have effects on the lamina network and nuclear integrity due to impaired assembly into the lamina network. Therefore, a large fraction of these mutant proteins remains nucleoplasmic and more soluble, so that the lamina cannot adequately support nuclear stability. Other mutations might have more subtle effects on assembly or produce minor irregularities in the lamina lattice, which are not easily detectable in our nuclear strain assays. Unlike the nuclear strain experiments, our in vitro assembly studies, which revealed severed defects in filament and paracrystal assembly of lamin mutants causing EDMD, were performed in the absence of wild-type lamin A. Although we cannot exclude the possibility that the presence of wild-type lamin A could assist in the formation of more regular-appearing structures comprised of mutant and wild-type lamins (which would likely still be mechanically weaker), previous co-assembly studies with wild-type and mutant desmin found that the mutant protein segregates in vitro and in vivo (60,61), which would also provide an explanation for the increased solubility of the mutant lamins in our studies (Fig. 2).
We found that different amino acid substitutions at the same amino acid position within the central rod domain can have very distinct effects on nuclear deformability. This observation may at least in part explain why some amino acid substitutions at specific positions are more prevalent among patients with LMNA mutations than others. For example, for position E203, both E203K and E203G substitutions have been identified in patients with DCM, whereas substitution to E203A, which had normal structural function in our nuclear strain assay, has never been reported in patients. Similarly, for position E358, the E358K mutation is the only reported amino acid change reported in severe muscular dystrophy, and position R386 has been reported mutated to lysine (K), threonine (T), or methionine (M), but not to glycine (G) or alanine (A). Although this preference for specific amino acid substitutions may simply represent underreporting of disease mutations or the rarity of LMNA mutations, it is intriguing to consider the possibility that the fact that some amino acid substitutions have not been reported in patients may indicate that these changes either have no profound effects or are so deleterious that they are lethal during development.
Using muscle-specific expression of myopathic lamin mutants in D. melanogaster, we provided the first direct evidence that lamin mutations impair nuclear stability in muscle fibers subjected to mechanical strain. Interestingly, the N210K mutation, which corresponds to the human N195K mutation that causes DCM, had no effect on nuclear stability in the Drosophila body wall muscle, which may indicate that lamin mutations can differentially affect nuclear structure and mechanics in skeletal and cardiac muscle. This idea is further supported by the fact that patients carrying the LMNA N195K mutation show no clinical symptoms typical for muscular dystrophy, and skeletal muscle biopsies exhibit no pathology (62).
Although our findings strongly support a role for impaired nuclear mechanics in the development of muscular phenotypes in laminopathy patients, it is important to note that only four of the 12 EDMD and DCM mutations caused severe defects in nuclear stability, whereas other mutations displayed normal nuclear stiffness in our assays and yet cause EDMD or DCM in humans. For cases in which no obvious defect in nuclear stability was observed, we cannot exclude the possibility that abnormalities are masked by impaired nucleo-cytoskeletal coupling. However, considering the strong correlation between nuclear deformation (Fig. 1C) and solubility of mutant lamins (Fig. 2C), it seems likely that our measurements reflect specific differences in structural function of different lamin mutations. This idea is supported by prior observations using C2C12 myoblasts that demonstrated diffuse, nucleoplasmic localization in transfected cells for the same four mutations we found to be defective in providing nuclear stability (63). For cases where we did not observe negative effects on nuclear stiffness, we favor the explanation that additional factors must contribute to the development of muscular disease, specifically a disruption of nucleo-cytoskeletal connections. In support of this hypothesis, we observed defects in intracellular force transmission (Fig. 5B). Our findings are consistent with a recent report in which laminopathic mutations associated with DCM and EDMD caused impaired nuclear movement in MEFs, which was attributed to defects in the attachment of cytoskeletal actin cables to the nuclear lamina via nesprin-2 giant/Sun2 complexes termed transmembrane actin-associated nuclear lines (55).
One potential limitation of the current studies is that we investigated mutations in lamin A only, whereas most human mutations affect both lamins A and C. However, we found that the expression of lamin A was sufficient to completely restore nuclear stiffness in Lmna+/− MEFs, suggesting some redundancy between these two isoforms, which is consistent with the lack of phenotype in mice lacking either lamin A or C (64–66). In addition, we obtained similar results in Drosophila muscle fibers which express only a single A-type lamin.
Taken together, our findings demonstrate the complexity of lamin mutations and their effect on nuclear structure and stability in muscle diseases. Many, but not all, mutations associated with EDMD and DCM are characterized by defects in structural function, resulting in more deformable and more fragile nuclei, which could render cells more sensitive to mechanical stress. In addition, all tested EDMD mutations disrupted nucleo-cytoskeletal coupling, which could further impair muscle function, for example, by failure of myonuclei to anchor at neuromuscular junctions (16). In addition to these mechanical aspects, it is likely that lamin mutations can interfere with a broad range of other cellular processes requiring lamins, including replication, gene expression, DNA repair, proliferation and stem-cell differentiation (10,21). In this scenario, different lamin mutations may specifically impact one or more of these functions, thus explaining the broad spectrum of human diseases caused by the diverse LMNA mutations.
MATERIALS AND METHODS
Cell lines
Lmna+/+, Lmna+/− and Lmna−/− MEFs were cultured in D10 medium (DMEM supplemented with 10% fetal bovine serum, 4 mm l-glutamine, 50 U/ml penicillin and 50 μg/ml streptomycin). 293GPG cells (67) were cultured in D10 supplemented with 1 µg/ml tetracycline, 2 µg/ml puromycin and 300 µg/ml G418. Human patient fibroblasts (68) were kindly provided by Howard Worman (Columbia University, New York, NY, USA) and maintained in D10 containing 5 μg/ml Plasmocin™ (InvivoGen) for prophylactic treatment against mycoplasma contamination.
Plasmids
cDNAs of wild-type and point-mutated human prelamin A, as well as the ΔNLA lamin variant lacking the first 33 amino acids, were kindly provided by Howard Worman [Columbia University, New York, NY, USA (63)]. The point-mutated cDNAs used in this study cause amino acid substitutions as shown in Table 1. Lamin variants were either amplified by PCR and ligated into pRetroX-IRES-ZsGreen1 using NotI and BamHI restriction sites, or subcloned from a shuttle vector as described previously (69). Mutagenesis of the codons of amino acids E203, E358, M371 and R386 to either glycine or alanine was performed by amplifying human prelamin A in two fragments and inducing codon changes over the respective primers. After PCR, the two fragments were ligated together via an introduced restriction site that does not change the primary sequence of the prelamin A protein (introducing BspDI at amino acid positions 195–196: 583–588acaggc→atcgat results in N195N, G196G; introducing EcoRV at amino acid positions 364 and 365: 1090–1095gacatc→gatatc results in D364D, I365I; or introducing XhoI at amino acid positions 380 and 381: 1138–1142ttggag→ctcgag results in L380L, E381E). As control, primers with the same introduced restriction sites but without amino acid change in positions E203, E358, M371 and R386 were used to amplify the two prelamin A fragments that were ligated together. Ligated full-length prelamin A variants were ligated into pRetroX-IRES-ZsGreen1 using NotI and BamHI restriction sites. The dominant-negative nesprin KASH (DN KASH) plasmid was described previously (15). Constructs encoding the mutant forms of Drosophila Lamin C were generated using the QuikChange II Site-Directed Mutagenesis Kit. The primers contain nucleotide substitutions that result in single-amino-acid substitutions within Lamin C. The mutated Lamin C sequences were cloned into the P-element transformation vector pUAST (70) and used to generate transgenic stocks (45). For all mutagenesis procedures, introduced mutations were confirmed by sequencing.
Retroviral infection and cell sorting
Lmna+/− MEFs were modified by retroviral infection. 293GPG cells were transfected with pRetroX-IRES-ZsGreen1 vectors containing lamin A variants, using Lipofectamine™ 2000 (Invitrogen). Starting 6 h after transfection, cells were maintained in D10 without tetracycline, puromycin and G418. For 6 subsequent days, cell medium was collected and replaced. This medium containing assembled virus was filtered through 0.45 µm pores, supplemented with 8 µg/µl Polybrene and added to Lmna+/− MEFs for 24 h. At 5–10 days after retroviral infection, positive cells were sorted for the bicistronically expressed ZsGreen, which allowed us to finely control expression levels of the ectopically expressed lamins translated from the same bicistronic mRNA. Cell sorting was performed on a BD FACSARIA Special Order 11 color sorter, using a 488 nm laser to excite ZsGreen1 and with a sort pressure of 70 psi.
Protein extraction
Preparation of total cell extracts containing 3 × 105 cells per microliters was performed as previously described by resuspending pelleted cells in Laemmli sample buffer (LSB, 10% glycerol, 3% SDS, 62.5 mm Tris–HCl, 50 mm DTT and 0.05% bromphenol blue) and subsequent boiling for 5 min (71). Extraction of the soluble lamin A/C fraction of modified and unmodified Lmna+/− MEFs was performed following a modified protocol for differential protein extraction (36). Confluent cells were washed with phosphate-buffered saline (PBS), trypsinized and resuspended in 1.2 ml of growth medium. An amount of 33 µl of the cell suspension was immediately boiled with 267 µl of LSB (‘total cell fraction’). Another 1 ml of aliquot (‘soluble lamin fraction’) of the cell suspension was transferred into a new tube; cells were briefly spun down and resuspended in 200 µl of lysis buffer containing 0.5× PBS, 50 mm MOPS (pH 7.0), 10 mm MgCl2, 1 mm EGTA, 1% NP40, protease inhibitor (SIGMAFAST™ Protease Inhibitor, Sigma-Aldrich) and 0.75% saturated PMSF in ethanol. Extraction with 1% NP40 detergent was previously shown to release the soluble lamin A/C fraction but not lamina-associated A-type lamins (36). Extraction was performed at room temperature for 5 min; cell remnants were spun down, and 150 µl of the supernatant were boiled with 75 µl of 3× LSB. Extraction was performed three independent times. Notably, owing to slight changes in experimental conditions for each isolation, e.g. cell density, buffer composition, vortexing or extraction timing, the absolute results of the soluble lamin fractions varied between individual experiments, even though the relative difference between cell lines within each experiment were very similar.
Immunofluorescence analysis
Immunofluorescence analysis was performed as previously described (71). In brief, cells were plated on sterile coverslips and grown overnight, briefly washed with PBS, fixed in 4% formaldehyde (pH 7.4) and permeabilized with 0.5% Triton X-100 in PBS. Fixed cells were incubated either with mouse anti-lamin A/C antibody [LaZ (72)] or with guinea pig anti-emerin antibody [Em-N-term (73)], diluted 1:2 and 1:300, respectively, in PBS containing 10% normal donkey serum (Jackson ImmunoResearch). Cells were washed and incubated with secondary antibodies: Cy3-coupled donkey anti-mouse or donkey anti-guinea pig IgG, diluted 1:500 in PBS containing 10% normal donkey serum. Cells were then incubated in 2 µg/ml Hoechst 33342 and embedded in Eukitt quick-hardening mounting medium (Fluka). Confocal laser scanning microscopy was performed on a TCS-SP II microscope (Leica Microsystems). Images were acquired with a 63.0× oil-immersion objective (NA 1.40) with a 1 airy unit pinhole.
Western analysis
Western analysis of modified and unmodified Lmna+/− MEFs was performed as previously described (71). Polyvinylidene difluoride membranes were incubated with the following antibodies diluted in PBS containing 0.5% Tween-20 and 5% milk powder: mouse anti-lamin A/C antibody (LaZ) at 1:5 dilution, goat anti-lamin A/C antibody (N-18, Santa Cruz #sc-6215) at 1:500 dilution, goat anti-lamin B1 antibody (M-20, Santa Cruz #sc-6217) at 1:500 dilution, mouse anti-lamin B2 antibody (X223, Progen #65147C) at 1:5 dilution, guinea pig anti-emerin antibody (Em-N-term) at 1:2000 dilution and rabbit anti-actin antibody (Sigma-Aldrich, #20–33) at 1:5000 dilution. Membranes were washed in PBS containing 0.5% Tween-20 and incubated with secondary antibodies: peroxidase-coupled donkey anti-guinea pig, donkey anti-rabbit, donkey anti-goat or donkey anti-mouse IgG (Jackson ImmunoReseach) diluted 1:5000 in PBS containing 0.5% Tween-20 and 5% milk powder. Enhanced luminescence reaction was performed and membranes were exposed to X-ray films.
Drosophila stocks
Generation of transgenic stocks was performed as previously described (45). Stocks expressing wild-type and mutant lamins under the control of a UAS element were crossed to the C57 Gal4 driver stock (49) and the resulting larval progeny were analyzed for muscle defects (45). The lamin stocks encode wild-type Lamin C, headless Lamin C (lacking the first 42 amino acids, Lamin C ΔN) or the mutations N210K, R401K, K493W, W557S and L567P (corresponding to human mutations N195K, R386K, R453W, W520S and L530P, respectively) (46,47,74). Drosophila stocks were maintained on standard corn meal media at room temperature; the Gal4/UAS crosses were performed at 25°C.
Nuclear strain analysis and microneedle manipulation assay of cultured cells
Nuclear strain analysis and microneedle manipulation of patient fibroblasts and Lmna+/− MEFs, as well as data analysis, were performed as described previously (32,75).
Nuclear strain analysis in muscle tissue
For nuclear strain analysis of D. melanogaster muscle tissue, third instar larvae were placed in a drop of cold low-potassium buffer containing 108 mm NaCl, 5 mm KCl, 2 mm CaCl2, 8 mm MgCl2, 1 mm NaH2PO4, 4 mm NaHCO3, 10 mm sucrose, 5 mm trehalose, 5 mm HEPES and 100 μg/ml Hoechst 33342 nuclear stain on a glass slide. Subsequently, the head and tail were removed and the body wall muscle was opened with a horizontal incision down the length of the longitudinal axis to remove the organs. The body wall muscle filet was then transferred onto an elastic silicone membrane clamped into the strain dish and covered with 15 ml of high-potassium buffer containing 108 mm NaCl, 35 mm KCl, 1 mm EDTA, 8 mm MgCl2, 1 mm NaH2PO4, 4 mm NaHCO3, 10 mm sucrose, 5 mm trehalose, 5 mm HEPES and 10 μg/ml Hoechst 33342. To securely attach the muscle filet in the open position to the center of the silicone membrane, a small amount of histoacryl blue adhesive (Tissue Seal, LLC) was applied to the four corners and to two to four additional points along the periphery of the muscle filet and then affixed to the silicone membrane. The muscle filets were then subjected to biaxial strain application as described previously for adherent cells (32,75), with minor modifications. To facilitate imaging of muscle fibers, the muscle filet was pre-stretched by applying low-level biaxial strain to the silicon membrane, resulting in taut but only lightly strained muscle fibers. Fluorescence images of 1–3 muscle fibers, containing up to 15 myonuclei, were acquired at this position. Subsequently, the silicone membrane was further stretched, resulting in larger tissue strain and induced nuclear deformations. Importantly, although the applied membrane strain is biaxial, due to the muscle physiology and attachment, the tissue strain in the muscle fibers is almost completely uniaxial in the direction of the muscle fiber. The same myonuclei were then imaged in this strained condition. To calculate tissue strain, changes in the internuclear distance within a single muscle strand were calculated using a custom-written MATLAB program. Nuclear deformations were then quantified as previously described (32,75).
In vitro lamin assembly
Recombinant human lamin A ΔC18 (‘mature’ lamin A) as well as the single-amino-acid deletion ΔK32 and the point mutations N195K and E358K were expressed using the pET24 system. A clone coding for lamin A ΔK32 was generously provided by Gisele Bonne (Inserm U582 – Institut de Myologie, Paris, France) and subcloned into pET24a. Induction with 1 mm IPTG was done for 3 h at 37°C (42). Inclusion bodies were isolated as described previously (76). For in vitro paracrystal assembly, lamin protein stored in 8 m urea at a concentration of 0.5 mg/ml was dialyzed into 10 mm Tris–HCl (pH 7.4), 300 mm NaCl, 2 mm EDTA and 1 mm DTT overnight at 4°C to obtain soluble lamin complexes. Lamins were briefly centrifuged in a benchtop centrifuge at full speed for 15 min to remove all protein at higher assembly states. Lamin proteins were diluted to 0.1 mg/ml and further dialyzed in the same buffer, but with stepwise reduction of salt concentration from 300 to 50 mm NaCl. Each dialysis step was performed for 20 min at 37°C. For in vitro filament assembly, lamin protein in 8 m urea at a concentration of 0.2 mg/ml was dialyzed into 25 mm Tris (pH 8), 250 mm NaCl and 1 mm DTT for 1 h at 37°C to obtain lamin dimers, then dialyzed into 25 mm MES, pH 6.5, 250 mm NaCl and 1 mm DTT for 50 min at 37°C. Assembled filaments, fixed with 0.1% glutaraldehyde, and paracrystals were then applied to glow-discharged, carbon-coated copper electron microscopy grids and analyzed in a Philips 410 transmission electron microscope (FEI). Images were taken with a CCD camera (Bioscan 792, Gatan).
Statistical data analysis
All measurements were performed at least three independent times. All data are expressed as mean ± standard error of the mean. Statistical analyses were performed with PRISM 3.0 (GraphPad Software, Inc.). Data were analyzed by a two-tailed Student t-test (for comparison between two groups) or one-way ANOVA with post-test for comparison of several groups. A two-tailed P-value of ≤0.05 was considered significant, with the symbols ‘*’ for P ≤ 0.05, ‘**’ for P ≤ 0.01 and ‘***’ for P ≤ 0.001.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health awards (grant numbers R01 NS059348, R01 HL082792) to J.L.; a postdoctoral fellowship from the American Heart Association to D.E.J. (AHA award 09POST2320042); a grant from the NIH/National Institutes of Health (grant number AR0600012) to L.L.W. and by postdoctoral fellowships of the American Heart Association (10POST3920014) and the Association Francaise contre les Myopathies (18336000) to G.D.
Supplementary Material
ACKNOWLEDGEMENTS
Imaging was performed with the support of the Center for Microscopy and Image Analysis, University of Zurich. We thank Dr Colin Stewart for providing Lmna+/+, Lmna+/− and Lmna−/− cell lines; Dr Howard Worman for providing plasmids and patient fibroblast cell lines; and Dr Richard C. Mulligan for providing us with the 293GPG cells. We further thank Dr Ohad Medalia and Dr Sergei Strelkov for support and advice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Stewart C.L., Roux K.J., Burke B. Blurring the boundary: the nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- 2.Dwyer N., Blobel G. A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J. Cell Biol. 1976;70:581–591. doi: 10.1083/jcb.70.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldman A.E., Maul G., Steinert P.M., Yang H.Y., Goldman R.D. Keratin-like proteins that coisolate with intermediate filaments of BHK-21 cells are nuclear lamins. Proc. Natl Acad. Sci. USA. 1986;83:3839–3843. doi: 10.1073/pnas.83.11.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKeon F.D., Kirschner M.W., Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- 5.Herrmann H., Bar H., Kreplak L., Strelkov S.V., Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat. Rev. Mol. Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- 6.Parry D.A. Microdissection of the sequence and structure of intermediate filament chains. Adv. Protein Chem. 2005;70:113–142. doi: 10.1016/S0065-3233(05)70005-X. [DOI] [PubMed] [Google Scholar]
- 7.Lin F., Worman H.J. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem. 1993;268:16321–16326. [PubMed] [Google Scholar]
- 8.Peter M., Kitten G.T., Lehner C.F., Vorburger K., Bailer S.M., Maridor G., Nigg E.A. Cloning and sequencing of cDNA clones encoding chicken lamins A and B1 and comparison of the primary structures of vertebrate A- and B-type lamins. J. Mol. Biol. 1989;208:393–404. doi: 10.1016/0022-2836(89)90504-4. [DOI] [PubMed] [Google Scholar]
- 9.Vorburger K., Kitten G.T., Nigg E.A. Modification of nuclear lamin proteins by a mevalonic acid derivative occurs in reticulocyte lysates and requires the cysteine residue of the C-terminal CXXM motif. EMBO J. 1989;8:4007–4013. doi: 10.1002/j.1460-2075.1989.tb08583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dechat T., Adam S.A., Taimen P., Shimi T., Goldman R.D. Nuclear lamins. Cold Spring Harb. Perspect. Biol. 2010;2:a000547. doi: 10.1101/cshperspect.a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Worman H.J. Nuclear lamins and laminopathies. J. Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira S., Bourgeois P., Navarro C., Esteves-Vieira V., Cau P., De Sandre-Giovannoli A., Levy N. HGPS and related premature aging disorders: from genomic identification to the first therapeutic approaches. Mech. Ageing Dev. 2008;129:449–459. doi: 10.1016/j.mad.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 13.Bertrand A.T., Chikhaoui K., Yaou R.B., Bonne G. Clinical and genetic heterogeneity in laminopathies. Biochem. Soc. Trans. 2011;39:1687–1692. doi: 10.1042/BST20110670. [DOI] [PubMed] [Google Scholar]
- 14.Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombardi M.L., Jaalouk D.E., Shanahan C.M., Burke B., Roux K.J., Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mejat A., Decostre V., Li J., Renou L., Kesari A., Hantai D., Stewart C.L., Xiao X., Hoffman E., Bonne G., et al. Lamin A/C-mediated neuromuscular junction defects in Emery-Dreifuss muscular dystrophy. J. Cell Biol. 2009;184:31–44. doi: 10.1083/jcb.200811035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burke B., Stewart C.L. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 2002;3:575–585. doi: 10.1038/nrm879. [DOI] [PubMed] [Google Scholar]
- 18.Houben F., Ramaekers F.C., Snoeckx L.H., Broers J.L. Role of nuclear lamina-cytoskeleton interactions in the maintenance of cellular strength. Biochim. Biophys. Acta. 2007;1773:675–686. doi: 10.1016/j.bbamcr.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Lammerding J., Schulze P.C., Takahashi T., Kozlov S., Sullivan T., Kamm R.D., Stewart C.L., Lee R.T. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muchir A., Pavlidis P., Decostre V., Herron A.J., Arimura T., Bonne G., Worman H.J. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J. Clin. Invest. 2007;117:1282–1293. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scaffidi P., Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat. Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Broers J.L., Peeters E.A., Kuijpers H.J., Endert J., Bouten C.V., Oomens C.W., Baaijens F.P., Ramaekers F.C. Decreased mechanical stiffness in LMNA-/- cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum. Mol. Genet. 2004;13:2567–2580. doi: 10.1093/hmg/ddh295. [DOI] [PubMed] [Google Scholar]
- 23.Lammerding J., Fong L.G., Ji J.Y., Reue K., Stewart C.L., Young S.G., Lee R.T. Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 2006;281:25768–25780. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 24.Arbustini E.A., Pasotti M., Pilotto A., Repetto A., Grasso M., Diegoli M. Gene symbol: CMD1A. Disease: dilated cardiomyopathy associated with conduction system disease. Hum. Genet. 2005;117:295. [PubMed] [Google Scholar]
- 25.Ben Yaou R., Gueneau L., Demay L., Stora S., Chikhaoui K., Richard P., Bonne G. Heart involvement in lamin A/C related diseases. Arch. Mal. Coeur Vaiss. 2006;99:848–855. [PubMed] [Google Scholar]
- 26.Decostre V., Ben Yaou R., Bonne G. Laminopathies affecting skeletal and cardiac muscles: clinical and pathophysiological aspects. Acta. Myol. 2005;24:104–109. [PubMed] [Google Scholar]
- 27.Filesi I., Gullotta F., Lattanzi G., D'Apice M.R., Capanni C., Nardone A.M., Columbaro M., Scarano G., Mattioli E., Sabatelli P., et al. Alterations of nuclear envelope and chromatin organization in mandibuloacral dysplasia, a rare form of laminopathy. Physiol. Genomics. 2005;23:150–158. doi: 10.1152/physiolgenomics.00060.2005. [DOI] [PubMed] [Google Scholar]
- 28.Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B., Gruenbaum Y., Khuon S., Mendez M., Varga R., et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl Acad. Sci. USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y., Herron A.J., Worman H.J. Pathology and nuclear abnormalities in hearts of transgenic mice expressing M371K lamin A encoded by an LMNA mutation causing Emery-Dreifuss muscular dystrophy. Hum. Mol. Genet. 2006;15:2479–2489. doi: 10.1093/hmg/ddl170. [DOI] [PubMed] [Google Scholar]
- 30.Park Y.E., Hayashi Y.K., Goto K., Komaki H., Hayashi Y., Inuzuka T., Noguchi S., Nonaka I., Nishino I. Nuclear changes in skeletal muscle extend to satellite cells in autosomal dominant Emery-Dreifuss muscular dystrophy/limb-girdle muscular dystrophy 1B. Neuromuscul. Disord. 2009;19:29–36. doi: 10.1016/j.nmd.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Fidzianska A., Hausmanowa-Petrusewicz I. Architectural abnormalities in muscle nuclei. Ultrastructural differences between X-linked and autosomal dominant forms of EDMD. J. Neurol. Sci. 2003;210:47–51. doi: 10.1016/s0022-510x(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 32.Lammerding J., Lee R.T. Mechanical properties of interphase nuclei probed by cellular strain application. Methods. Mol. Biol. 2009;464:13–26. doi: 10.1007/978-1-60327-461-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cupesi M., Yoshioka J., Gannon J., Kudinova A., Stewart C.L., Lammerding J. Attenuated hypertrophic response to pressure overload in a lamin A/C haploinsufficiency mouse. J. Mol. Cell Cardiol. 2010;48:1290–1297. doi: 10.1016/j.yjmcc.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spann T.P., Moir R.D., Goldman A.E., Stick R., Goldman R.D. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolb T., Maass K., Hergt M., Aebi U., Herrmann H. Lamin A and lamin C form homodimers and coexist in higher complex forms both in the nucleoplasmic fraction and in the lamina of cultured human cells. Nucleus. 2011;2:425–433. doi: 10.4161/nucl.2.5.17765. [DOI] [PubMed] [Google Scholar]
- 37.Herrmann H., Strelkov S.V., Burkhard P., Aebi U. Intermediate filaments: primary determinants of cell architecture and plasticity. J. Clin. Invest. 2009;119:1772–1783. doi: 10.1172/JCI38214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parry D.A., Strelkov S.V., Burkhard P., Aebi U., Herrmann H. Towards a molecular description of intermediate filament structure and assembly. Exp. Cell Res. 2007;313:2204–2216. doi: 10.1016/j.yexcr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 39.Aebi U., Cohn J., Buhle L., Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- 40.Ben-Harush K., Wiesel N., Frenkiel-Krispin D., Moeller D., Soreq E., Aebi U., Herrmann H., Gruenbaum Y., Medalia O. The supramolecular organization of the C. elegans nuclear lamin filament. J. Mol. Biol. 2009;386:1392–1402. doi: 10.1016/j.jmb.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 41.Strelkov S.V., Schumacher J., Burkhard P., Aebi U., Herrmann H. Crystal structure of the human lamin A coil 2B dimer: implications for the head-to-tail association of nuclear lamins. J. Mol. Biol. 2004;343:1067–1080. doi: 10.1016/j.jmb.2004.08.093. [DOI] [PubMed] [Google Scholar]
- 42.Foeger N., Wiesel N., Lotsch D., Mucke N., Kreplak L., Aebi U., Gruenbaum Y., Herrmann H. Solubility properties and specific assembly pathways of the B-type lamin from Caenorhabditis elegans. J. Struct. Biol. 2006;155:340–350. doi: 10.1016/j.jsb.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 43.Bank E.M., Ben-Harush K., Feinstein N., Medalia O., Gruenbaum Y. Structural and physiological phenotypes of disease-linked lamin mutations in C. elegans. J. Struct. Biol. 2012;177:106–112. doi: 10.1016/j.jsb.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Bank E.M., Ben-Harush K., Wiesel-Motiuk N., Barkan R., Feinstein N., Lotan O., Medalia O., Gruenbaum Y. A laminopathic mutation disrupting lamin filament assembly causes disease-like phenotypes in Caenorhabditis elegans. Mol. Biol. Cell. 2011;22:2716–2728. doi: 10.1091/mbc.E11-01-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dialynas G., Flannery K.M., Zirbel L.N., Nagy P.L., Mathews K.D., Moore S.A., Wallrath L.L. LMNA variants cause cytoplasmic distribution of nuclear pore proteins in Drosophila and human muscle. Hum. Mol. Genet. 2012;21:1544–1556. doi: 10.1093/hmg/ddr592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dialynas G., Speese S., Budnik V., Geyer P.K., Wallrath L.L. The role of Drosophila Lamin C in muscle function and gene expression. Development. 2010;137:3067–3077. doi: 10.1242/dev.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulze S.R., Curio-Penny B., Speese S., Dialynas G., Cryderman D.E., McDonough C.W., Nalbant D., Petersen M., Budnik V., Geyer P.K., et al. A comparative study of Drosophila and human A-type lamins. PLoS One. 2009;4:e7564. doi: 10.1371/journal.pone.0007564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duffy J.B. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 49.Koh Y.H., Popova E., Thomas U., Griffith L.C., Budnik V. Regulation of DLG localization at synapses by CaMKII-dependent phosphorylation. Cell. 1999;98:353–363. doi: 10.1016/s0092-8674(00)81964-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haque F., Mazzeo D., Patel J.T., Smallwood D.T., Ellis J.A., Shanahan C.M., Shackleton S. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J. Biol. Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roux K.J., Crisp M.L., Liu Q., Kim D., Kozlov S., Stewart C.L., Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl Acad. Sci. USA. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilhelmsen K., Litjens S.H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Q., Skepper J.N., Yang F., Davies J.D., Hegyi L., Roberts R.G., Weissberg P.L., Ellis J.A., Shanahan C.M. Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J. Cell Sci. 2001;114:4485–4498. doi: 10.1242/jcs.114.24.4485. [DOI] [PubMed] [Google Scholar]
- 54.Mejat A., Misteli T. LINC complexes in health and disease. Nucleus. 2010;1:40–52. doi: 10.4161/nucl.1.1.10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Folker E.S., Ostlund C., Luxton G.W., Worman H.J., Gundersen G.G. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc. Natl Acad. Sci. USA. 2011;108:131–136. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fidzianska A., Walczak E., Glinka Z., Religa G. Nuclear architecture remodelling in cardiomyocytes with lamin A deficiency. Folia Neuropathol. 2008;46:196–203. [PubMed] [Google Scholar]
- 57.Jahn D., Schramm S., Schnolzer M., Heilmann C.J., de Koster C.G., Schutz W., Benavente R., Alsheimer M. A truncated lamin A in the Lmna-/- mouse line: implications for the understanding of laminopathies. Nucleus. 2012;3:463–474. doi: 10.4161/nucl.21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilchrist S., Gilbert N., Perry P., Ostlund C., Worman H.J., Bickmore W.A. Altered protein dynamics of disease-associated lamin A mutants. BMC Cell Biol. 2004;5:46. doi: 10.1186/1471-2121-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Broers J.L., Kuijpers H.J., Ostlund C., Worman H.J., Endert J., Ramaekers F.C. Both lamin A and lamin C mutations cause lamina instability as well as loss of internal nuclear lamin organization. Exp. Cell Res. 2005;304:582–592. doi: 10.1016/j.yexcr.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 60.Bar H., Kostareva A., Sjoberg G., Sejersen T., Katus H.A., Herrmann H. Forced expression of desmin and desmin mutants in cultured cells: impact of myopathic missense mutations in the central coiled-coil domain on network formation. Exp. Cell Res. 2006;312:1554–1565. doi: 10.1016/j.yexcr.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 61.Bar H., Mucke N., Kostareva A., Sjoberg G., Aebi U., Herrmann H. Severe muscle disease-causing desmin mutations interfere with in vitro filament assembly at distinct stages. Proc. Natl Acad. Sci. USA. 2005;102:15099–15104. doi: 10.1073/pnas.0504568102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fatkin D., MacRae C., Sasaki T., Wolff M.R., Porcu M., Frenneaux M., Atherton J., Vidaillet H.J., Jr., Spudich S., De Girolami U., et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 63.Ostlund C., Bonne G., Schwartz K., Worman H.J. Properties of lamin A mutants found in Emery-Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J. Cell Sci. 2001;114:4435–4445. doi: 10.1242/jcs.114.24.4435. [DOI] [PubMed] [Google Scholar]
- 64.Fong L.G., Ng J.K., Lammerding J., Vickers T.A., Meta M., Cote N., Gavino B., Qiao X., Chang S.Y., Young S.R., et al. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J. Clin. Invest. 2006;116:743–752. doi: 10.1172/JCI27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davies B.S., Barnes R.H., II, Tu Y., Ren S., Andres D.A., Spielmann H.P., Lammerding J., Wang Y., Young S.G., Fong L.G. An accumulation of non-farnesylated prelamin A causes cardiomyopathy but not progeria. Hum. Mol. Genet. 2010;19:2682–2694. doi: 10.1093/hmg/ddq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coffinier C., Jung H.J., Li Z., Nobumori C., Yun U.J., Farber E.A., Davies B.S., Weinstein M.M., Yang S.H., Lammerding J., et al. Direct synthesis of lamin A, bypassing prelamin a processing, causes misshapen nuclei in fibroblasts but no detectable pathology in mice. J. Biol. Chem. 2010;285:20818–20826. doi: 10.1074/jbc.M110.128835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ory D.S., Neugeboren B.A., Mulligan R.C. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl Acad. Sci. USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muchir A., Medioni J., Laluc M., Massart C., Arimura T., van der Kooi A.J., Desguerre I., Mayer M., Ferrer X., Briault S., et al. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve. 2004;30:444–450. doi: 10.1002/mus.20122. [DOI] [PubMed] [Google Scholar]
- 69.Rowat A.C., Jaalouk D.E., Zwerger M., Ung W.L., Eydelnant I.A., Olins D., Olins A., Herrmann H., Weitz D.A., Lammerding J. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J. Biol. Chem. 2013 doi: 10.1074/jbc.M112.441535. doi:10.1074/jbc.M112.441535. http://www.ncbi.nlm.nih.gov/pubmed/23355469. (25 January 2013, date last accessted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 71.Zwerger M., Kolb T., Richter K., Karakesisoglou I., Herrmann H. Induction of a massive endoplasmic reticulum and perinuclear space expansion by expression of lamin B receptor mutants and the related sterol reductases TM7SF2 and DHCR7. Mol. Biol. Cell. 2010;21:354–368. doi: 10.1091/mbc.E09-08-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Geiger S.K., Bar H., Ehlermann P., Walde S., Rutschow D., Zeller R., Ivandic B.T., Zentgraf H., Katus H.A., Herrmann H., et al. Incomplete nonsense-mediated decay of mutant lamin A/C mRNA provokes dilated cardiomyopathy and ventricular tachycardia. J. Mol. Med. (Berl.) 2008;86:281–289. doi: 10.1007/s00109-007-0275-1. [DOI] [PubMed] [Google Scholar]
- 73.Dreger C.K., Konig A.R., Spring H., Lichter P., Herrmann H. Investigation of nuclear architecture with a domain-presenting expression system. J. Struct. Biol. 2002;140:100–115. doi: 10.1016/s1047-8477(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 74.Schulze S.R., Curio-Penny B., Li Y., Imani R.A., Rydberg L., Geyer P.K., Wallrath L.L. Molecular genetic analysis of the nested Drosophila melanogaster lamin C gene. Genetics. 2005;171:185–196. doi: 10.1534/genetics.105.043208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lombardi M.L., Lammerding J. Keeping the LINC: the importance of nucleocytoskeletal coupling in intracellular force transmission and cellular function. Biochem. Soc. Trans. 2011;39:1729–1734. doi: 10.1042/BST20110686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Herrmann H., Kreplak L., Aebi U. Isolation, characterization, and in vitro assembly of intermediate filaments. Methods Cell Biol. 2004;78:3–24. doi: 10.1016/s0091-679x(04)78001-2. [DOI] [PubMed] [Google Scholar]
- 77.Jakobs P.M., Hanson E.L., Crispell K.A., Toy W., Keegan H., Schilling K., Icenogle T.B., Litt M., Hershberger R.E. Novel lamin A/C mutations in two families with dilated cardiomyopathy and conduction system disease. J. Card. Fail. 2001;7:249–256. doi: 10.1054/jcaf.2001.26339. [DOI] [PubMed] [Google Scholar]
- 78.Bonne G., Mercuri E., Muchir A., Urtizberea A., Becane H.M., Recan D., Merlini L., Wehnert M., Boor R., Reuner U., et al. Clinical and molecular genetic spectrum of autosomal dominant Emery-Dreifuss muscular dystrophy due to mutations of the lamin A/C gene. Ann. Neurol. 2000;48:170–180. [PubMed] [Google Scholar]
- 79.Cao H., Hegele R.A. Nuclear lamin A/C R482Q mutation in Canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum. Mol. Genet. 2000;9:109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- 80.Vigouroux C., Magre J., Vantyghem M.C., Bourut C., Lascols O., Shackleton S., Lloyd D.J., Guerci B., Padova G., Valensi P., et al. Lamin A/C gene: sex-determined expression of mutations in Dunnigan-type familial partial lipodystrophy and absence of coding mutations in congenital and acquired generalized lipoatrophy. Diabetes. 2000;49:1958–1962. doi: 10.2337/diabetes.49.11.1958. [DOI] [PubMed] [Google Scholar]
- 81.Shackleton S., Lloyd D.J., Jackson S.N., Evans R., Niermeijer M.F., Singh B.M., Schmidt H., Brabant G., Kumar S., Durrington P.N., et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat. Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- 82.Bonne G., Di Barletta M.R., Varnous S., Becane H.M., Hammouda E.H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.A., et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.