Abstract
To maintain genome stability, mismatch repair of nuclear DNA replication errors must be directed to the nascent strand, likely by DNA ends and PCNA. Here we show that the efficiency of mismatch repair in Saccharomyces cerevisiae is reduced by inactivating RNase H2, which nicks DNA containing ribonucleotides incorporated during replication. In strains encoding mutator polymerases, this reduction is preferential for repair of mismatches made by leading strand DNA polymerase ε as compared to lagging strand DNA polymerase δ. The results suggest that RNase H2-dependent processing of ribonucleotides transiently present in DNA after replication may direct mismatch repair to the continuously replicated nascent leading strand.
INTRODUCTION
Mismatch repair (MMR) corrects DNA replication errors in the newly synthesized strand (Iyer et al., 2006; Kunkel and Erie, 2005). In order to enhance genome stability, MMR must be targeted to the nascent strand by one or more strand-discrimination signals. In E. coli and closely related bacteria, the nascent strand is marked for repair when, immediately after replication, MutH endonuclease nicks the DNA backbone at transiently under-methylated adenines in GATC sequences (Iyer et al., 2006). Moreover, a nick in DNA can signal for strand-specific eukaryotic MMR in vitro (Holmes et al., 1990; Thomas et al., 1991). However, the identity of the DNA ends used for strand discrimination during nuclear DNA replication in vivo is uncertain. Discontinuous lagging strand synthesis of Okazaki fragments (about 200 base pairs long in eukaryotes) produces transient 5′-DNA ends (Burgers, 2009), which have been implicated in discrimination for lagging strand replication errors (Nick McElhinny et al., 2010a; Pavlov et al., 2003). In contrast, the leading strand is likely to be replicated in a more continuous manner (Burgers, 2009). This raises the issue of the identity of the eukaryotic strand discrimination signal for MMR of leading strand replication errors.
We have suggested that ribonucleotides incorporated by DNA polymerases during replication might mark nascent strands for MMR (Nick McElhinny et al., 2010c). This hypothesis was based on several observations. S. cerevisiae replicases DNA polymerases α, δ and ε, (Pols α, δ and ε) frequently incorporate ribonucleotides into DNA in vitro (Nick McElhinny et al., 2010c). Pol ε incorporates ribonucleotides during replication in vivo (Nick McElhinny et al., 2010b), and does so preferentially into the nascent leading strand (Lujan et al., 2012). Most ribonucleotides incorporated during DNA replication are present only transiently, being efficiently repaired by Ribonucleotide Excision Repair (RER). RER is initiated when RNase H2 nicks the DNA backbone at ribonucleotides (Eder and Walder, 1991; Eder et al., 1993; Nick McElhinny et al., 2010b; Rydberg and Game, 2002; Sparks et al., 2012). Here we test whether this RNase H2-dependent processing of ribonucleotides provides a strand discrimination signal for MMR of leading strand replication errors.
RESULTS
The pol ε mutator incorporates ribonucleotides during leading strand synthesis
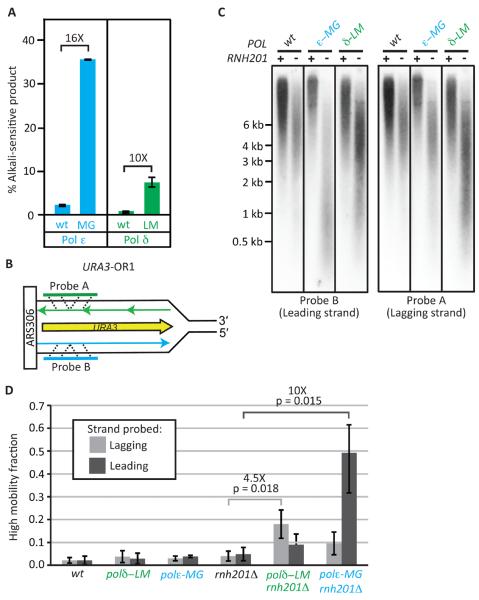
The POL2 gene encodes Pol ε, which participates in leading strand replication (Pursell et al., 2007). We began by quantifying the density of ribonucleotides in the nascent leading strand in vivo in strains encoding a pol2-M644G variant. Compared to wild type Pol ε, M644G Pol ε incorporates 16-fold more ribonucleotides into DNA during synthesis in vitro (Figure 1A, data combined from (Nick McElhinny et al., 2010b; Williams et al., 2012)). Increased ribonucleotide incorporation also occurs in vivo, where these ribonucleotides can be detected as alkali-sensitive sites in the genomic DNA of strains that are defective in RER (Nick McElhinny et al., 2010b) due to deletion of the RNH201 gene encoding the catalytic subunit of RNase H2. Consistent with Pol ε's role as a primarily leading strand replicase, strand-specific probing of DNA fragments generated by alkaline hydrolysis (Figure 1B and also see (Miyabe et al., 2011) for results in S. pombe) demonstrates that ribonucleotides are preferentially incorporated into the nascent leading strand (Figures 1C–D, and (Lujan et al., 2012)). Quantification of the DNA fragments (Figure S1) indicates that one ribonucleotide is introduced per about 950 base pairs (average of four independent experiments).
Figure 1. Ribonucleotide incorporation by Pols ε and δ.
(A) Stable incorporation of ribonucleotides into DNA was measured in vitro as described (Nick McElhinny et al., 2010c). The average percentages of alkali-sensitive product for two experiments ± the range are displayed. (B) The orientation of the URA3 reporter with respect to coding sequence is shown. The nascent leading strand synthesized by Pol ε is in blue and the nascent lagging strand synthesized by Pol δ is in green. (C) Strand-specific probing for alkali-sensitive sites in genomic DNA. DNA was subjected to alkaline hydrolysis, alkaline-agarose electrophoresis and analysis by Southern blotting using radiolabeled probes specific for nascent leading (Probe B) or nascent lagging (Probe A) strand DNA. DNA marker sizes are indicated on the left. Higher mobility (smaller) fragments reveal the presence of unrepaired ribonucleotides due to lack of RNase H2 activity. (D) Fraction of hydrolysis fragments present in the high mobility peak (see Figure S1 and Experimental Procedures). Error bars represent 95% C.I.
MMR efficiency is reduced in pol ε mutator strains lacking RNase H2
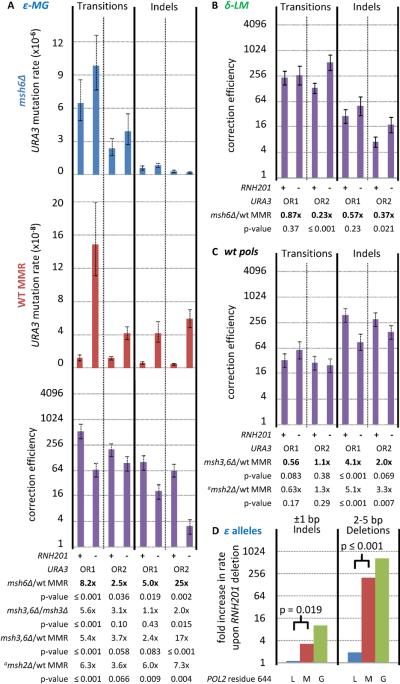
Pol ε M644G is less accurate than wild type Pol ε during DNA synthesis in vitro, and pol2-M644G strains are leading strand-specific mutators for single base substitutions and insertions/deletions (indels) (Lujan et al., 2012; Pursell et al., 2007). To determine if loss of RER reduces the efficiency of MMR of leading strand replication errors, we determined mutation rates for resistance to 5-fluoroorotic acid (5-FOA; toxic to URA3 strains) in pol2-M644G strains harboring the URA3 mutational reporter gene in each of two orientations (designated OR1 and OR2). We then determined URA3 reporter gene sequences from independent 5-FOA resistant clones (Table 1) and used the data to calculate specific rates for single base transitions and indels, the two types of mutations that are most characteristic of an MMR defect (Lujan et al., 2012; Nick McElhinny et al., 2008; Nick McElhinny et al., 2010a). Defining this signature for loss of MMR is critical, because loss of RNase H2 elevates rates for another type of mutation, 2–5 base pair deletions (Nick McElhinny et al., 2010b), which occur via nicking by Topoisomerase 1 (Top1) after replication (Kim et al., 2011) and are not subject to mismatch repair (Clark et al., 2011). We compared transition and indel rates for pol2-M644G msh6Δ strains lacking MutSα (Msh2-Msh6)-dependent MMR (Figure 2A, upper panel) to rates for isogenic pol2-M644G MSH6 MMR-proficient strains (Figure 2A, middle panel). The ratio of these rates provides MMR correction factors (Figure 2A, lower panel). We asked if the correction factors for these leading strand replication errors decreased in RER-deficient (rnh201Δ) strains, as predicted by the hypothesis that RER-dependent processing may provide a strand discrimination signal for MMR.
Table 1.
pol2-M644G mutation rates in two URA3 orientations ± MSH6 and/or RNH201.
| pol2-M644G msh6Δ/wt MMR | mutation rate(×10−7) | 95% confidence | 5-FOAR mutants | no URA3 change | URA3 mutations | Transitions | Frameshifts | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| # | rate | # | rate | ||||||||
| URA3 orientation 1 | RNH201 | wt MMR | 1.7 | (1.3–2.3) | 342 | 105 | 237 | 24 | 0.12 | 12 | 0.060 |
|
| |||||||||||
| msh6Δ | 110 | (82–150) | 219 | 26 | 205 | 130 | 65 | 12 | 6.0 | ||
|
| |||||||||||
| correction factors | 550× | 100× | |||||||||
|
| |||||||||||
| rnh201 Δ | wt MMR | 7.6 | (4.3–10) | 128 | 4 | 125 | 25 | 1.5 | 7 | 0.42 | |
|
| |||||||||||
| msh6Δ | 170 | (130–220) | 141 | 10 | 133 | 82 | 99 | 7 | 8.4 | ||
|
| |||||||||||
| correction factors | 67× | 20× | |||||||||
|
| |||||||||||
| RNH201/rnh201Δ correction factor ratios | 8.2 | 5.0 | |||||||||
| p-values | ≤ 0.001 | 0.019 | |||||||||
|
| |||||||||||
| URA3 orientation 2 | RNH201 | wt MMR | 0.83 | (0.71–0.96) | 246 | 123 | 124 | 36 | 0.12 | 14 | 0.047 |
|
| |||||||||||
| msh6Δ | 54 | (40–75) | 181 | 38 | 149 | 79 | 24 | 10 | 3.0 | ||
|
| |||||||||||
| correction factors | 190× | 63× | |||||||||
|
| |||||||||||
| rnh201 Δ | wt MMR | 14 | (9.8–20) | 138 | 0 | 138 | 5 | 0.51 | 7 | 0.71 | |
|
| |||||||||||
| msh6Δ | 81 | (57–110) | 134 | 20 | 126 | 65 | 39 | 3 | 1.8 | ||
|
| |||||||||||
| correction factors | 77× | 2.6× | |||||||||
|
| |||||||||||
| RNH201/rnh201Δ correction factor ratios | 2.5 | 25 | |||||||||
| p-values | 0.036 | 0.002 | |||||||||
For comparison, all rates are listed by strain in Table S6. All rates are multiplied by 107. Correction factors are compared in Figure 2A, listed as “msh6Δ/wt MMR”. Strain origins and related references may be found in Table S5. Among the mutants with no sequence change in the URA3 open reading frame, some may have arisen due to sequence changes in the URA3 promoter or in another gene that affects resistance to 5-FOA. Because these mutants contribute to the overall mutation rate, they are included in the calculation of rates for individual mutation classes.
Figure 2. The effect of RNH201 deletion on mutation rates and correction factors.
Error bars represent 95% C.I. (A) URA3 mutation rates are shown for pol2-M644G strains with MSH6 deleted (msh6Δ; blue bars) or wild type mismatch repair (WT MMR; red bars). Correction factors (correction efficiency; purple bars), are ratios of mutation rates (msh6Δ : MSH6). Rates and correction factors are shown for transitions (left) and indels (right). RNase H2 status (RNH201 = “+”; rnh201Δ = “−”) and URA3 orientation are indicated below the charts. Ratios of correction factors (RNH201 : rnh201Δ) and the corresponding p-values (indicating confidence that correction factors differ) are shown at bottom, beginning with ratios derived from correction factors shown (“msh6Δ/wt MMR”; Table 1) and continuing for strains with different MMR deficiencies (see Tables S1–S2). aNo measurements were made in an msh2Δ rnh201Δ strain (see Table S2). (B) As per the bottom of panel A, but for the pol3-L612M mutator background (δ-LM; see Table S3). (C) As per panel B, but for the wild type polymerase background (wt pols; see Table S4). (D) Fold increases in single-base indel and 2–5 bp tandem repeat deletion rates - due to RNH201 deletion - are shown for three Pol ε variants: M644L Pol ε (blue bars; see Table S7), wild type Pol ε (red bars), and M644G Pol ε (green bars).
In the absence of RNase H2, correction factors in OR1 strains were reduced by 8.2-fold and 5.0-fold for transitions and indels respectively (Figure 2A, values below lower panel). To determine if this reduction was reproducible, an independent set of measurements (Table 1) was made in pol2-M644G strains in which the location of URA3 was preserved (adjacent to replication origin ARS306) but the URA3 gene orientation was reversed (OR2). In these strains, Pol ε M644G replicates the complementary strand, providing completely different sequence contexts for incorporating ribonucleotides and for generating and repairing mismatches. Correction factors for transitions and indels in these strains were again reduced in the absence of RNase H2, this time by 2.5-fold and 25-fold, respectively (Figure 2A). These reductions are statistically significant (p < 0.05).
Similar reductions in MMR correction were observed when the experiments were repeated with msh3Δ msh6Δ strains that lack both MutSα-dependent and MutSβ (Msh2-Msh3)-dependent MMR. This included comparing msh3Δ msh6Δ strains to msh3Δ strains (Table S1 and Figure 2A bottom, with one exception for indels in OR1, where the p values was 0.43), as well as comparing msh3Δ msh6Δ strains to MMR-proficient strains (Table S2, upper panel). Because rates in msh2Δ strains are similar to rates in msh3Δ msh6Δ strains, similar reductions in MMR efficiency were observed using rates from msh2Δ strains (Table S2, lower panel) that also lack MutSα-dependent and MutSβ-dependent MMR.
MMR efficiency is not reduced in Pol δ mutator strains lacking RNase H2
Genetic evidence in S. cerevisiae (Nick McElhinny et al., 2010a; Pavlov et al., 2002) led us to previously suggest that the 5′-DNA ends of Okazaki fragments, perhaps in conjunction with PCNA, may be used as signals for strand discrimination during MMR. These 5′ ends are transiently present about every 200 base pairs ((Burgers, 2009; Smith and Whitehouse, 2012) and references therein). This density is about 5 to 10 -fold higher than for ribonucleotides in the nascent leading strand in the pol2-M644G rnh201Δ strains (Figures 1A and 1D). This led us to hypothesize that loss of RNase H2-dependent processing of ribonucleotides will have a lesser role in strand discrimination during lagging strand replication. To test this, we used the L612M mutator variant of Pol δ. Pol δ L612M is a mutator polymerase (Nick McElhinny et al., 2007) with a base substitution and indel error signature characteristic of lagging strand replication (Nick McElhinny et al., 2008). We now report that Pol δ L612M also incorporates 10-fold more ribonucleotides into DNA during synthesis in vitro than does wild type Pol δ (Figure 1A), and that this property is manifested in vivo as an increased number of strand-specific alkali-sensitive sites in DNA isolated from pol3-L612M rnh201Δ strains (Figures 1B–C). However in this case, the ribonucleotides are preferentially observed in the nascent lagging strand (Figures 1C–D,). This supports the interpretation, based earlier on mutational signatures (Larrea et al., 2010; Nick McElhinny et al., 2008), that Pol δ is primarily a lagging strand replicase. The density of alkali-sensitive sites in the pol3-L612M rnh201Δ strain (Figure S1) further indicates that one ribonucleotide is present per about 3400 base pairs. At this density, incisions by RNase H2 are expected to have little effect on the total number of transient DNA ends in the nascent strand compared to those already resulting from Okazaki fragment synthesis. Consistent with this, mutational analyses with pol3-L612M strains (Table S3), analogous to those described above for pol2-M644G strains, indicated that loss of RNase H2 did not decrease the efficiency of MutSα-dependent MMR of transition and indel mismatches generated by L612M Pol δ (Figure 2B). Instead, there are small increases in MMR efficiency in the pol3-L612M rnh201Δ strains. These increases are statistically significant in one URA3 orientation (OR2, p ≤ 0.021), but not in the other (OR1, p > 0.23).
Indel MMR efficiency is reduced in RNase H2-defective strains with wild type replicases
Does RNase H2 status affect MMR efficiency in strains with wild type Pols δ and ε? To test this, we examined the number of alkali-sensitive sites in DNA isolated from rnh201Δ strains encoding wild type polymerases. The results (Figures 1C–D) suggest that one ribonucleotide is present in the nascent leading strand per about 6500 deoxyribonucleotides. When mutational analyses similar to those described above were performed in strains encoding wild type polymerases (Table S4), the results showed that loss of RNase H2 decreased the efficiency of MutSα-dependent MMR of indel mismatches (Figure 2C) by 4.1-fold in OR1 strain (p ≤ 0.001) and by 2-fold in OR2 strains (p = 0.069), with no significant effect on MMR of transitions. The decrease in MMR efficiency for indels but not transitions is consistent with many studies showing that indels are more sensitive biomarkers for a eukaryotic MMR deficiency than are transitions. This applies to our own studies in yeast strains with wild type replicases, where the majority of uncorrected replication errors are single base deletions in mononucleotide runs (7) that are corrected at least 10 times more efficiently than transitions.
MMR efficiency is largely unaffected in a strain containing fewer ribonucleotides in DNA
The hypothesis that ribonucleotides direct MMR to the nascent leading strand predicts that reducing the number of ribonucleotides incorporated into DNA will reduce the effect of deleting RNH201 on MMR efficiency. To test this prediction, we measured spontaneous mutation rates and mutational specificity in pol2-M644L strains that contain fewer genomic ribonucleotides than POL2 and pol2-M644G strains (Nick McElhinny et al., 2010b). The mutation data (Table S7) were then used to calculate the difference in single-base indel rate in rnh201Δ versus RNH201 strains. This difference in the pol2-M644L strain is a negligible 1.1-fold (Figure 2D, blue bar on left, and Table S7), indicating that RNase H2 status is irrelevant in the pol2-M644L strain, which exhibits diminished genomic ribonucleotide incorporation. This 1.1-fold difference is significantly lower than the 3.2-fold difference in the POL2 strain (red bar; p = 0.019). An even greater difference (10-fold; green bar) is observed in the pol2-M644G strain, which contains the greatest number of ribonucleotides in the genome. Importantly, RNH201 deletion has no significant effect on single-base indel rates in MMR-deficient strains in the pol2-M644G background (Figure 2A and Table S1). Assuming this is also true in the POL2 and pol2-M644L strains, the results for single-base indels in Figure 2D strongly support the hypothesis that ribonucleotides in the nascent leading strand are important for repairing replication errors in that strand. Serving as a positive control in these experiments are results for 2–5 base pair deletions from direct repeats in the same three strains. The effect of RNH201 deletion on the rate of these events, which are due to topoisomerase 1-dependent cleavage of unrepaired ribonucleotides incorporated into DNA (Kim et al., 2011), is highest in the pol2-M644G rnh201Δ strain, intermediate in the POL2 rnh201Δ strain, and lowest in the pol2-M644L rnh201Δ strain (Figure 2D, right). The latter result confirms recent observations (Cho et al.).
DISCUSSION
The results in the M644G Pol ε background (Figure 2A), and to a lesser extent in the wild type (Figure 2C) and M644L Pol ε backgrounds (Figure 2D), are consistent with the idea that RNase H2-dependent processing of ribonucleotides incorporated by Pol ε may direct MMR to correct mismatches made during continuous leading strand replication. Thus RNase H2 action on ribonucleotides transiently present in the nascent leading strand may generate strand discrimination signals in a manner akin to transiently under-methylated adenines in GATC sequences and MutH endonuclease in E. coli. A simple explanation is that RNase H2 nicks the DNA backbone at positions of newly incorporated ribonucleotides, allowing these nicks to serve as strand discrimination signals, and possibly as entry points for removing mismatches.
Removal could occur by excising the nascent strand containing the mismatch, e.g., by exonuclease 1, or removal could occur by DNA synthesis to displace the nascent strand containing the mismatch followed by cleavage of the 5′-DNA flap (Kadyrov et al., 2009). In fact, the latter mechanism operates during ribonucleotide excision repair (Sparks et al., 2012). An additional attractive feature of the RNase H2-dependent signaling model is that the non-catalytic B subunit of RNase H2 contains a PCNA binding motif (Chon et al., 2009). PCNA directs RNase H2 to replication foci and imposes strand specificity on its function (Bubeck et al., 2011). PCNA is required for RER (Sparks et al., 2012), and it also required for early steps in MMR (Umar et al., 1996), where it has been implicated in signaling for strand discrimination (see (Pluciennik et al., 2010) and references therein) Thus, in addition to initiating RER, RNase H2 may deliver PCNA in an orientation-dependent manner to the nascent leading strand after replication in order to participate in signaling for MMR.
Our estimates of the density of ribonucleotides in the leading strand template are based on probing the URA3 locus, which is only a tiny fraction of the yeast genome (Figure 1B). Because ribonucleotide incorporation by Pol ε varies considerably with sequence context and ribonucleotide identity (Nick McElhinny et al., 2010b), ribonucleotide density in vivo could differ elsewhere in the genome. Nonetheless, in the URA3 reporter gene adjacent to ARS306, ribonucleotide density is sufficient to introduce nicks at intervals consistent with excision between the signal and the mismatch, which can cover a distance of 1,000 base pairs (Fang and Modrich, 1993). Moreover, a nick introduced by RNase H2 need not necessarily be the starting point for excision per se, but could rather be the signal needed for directing the endonuclease activity of the hPms2/yPms1 subunit of MutLα to introduce nicks that can be used for excision (Kadyrov et al., 2006; Kadyrov et al., 2007).
It is also important to note that mutation rates for transitions and indels in the pol2-M644G background are much higher when MMR is completely inactivated, as in msh3Δ msh6Δ strains (Table S2, upper panel) and msh2Δ strains (Table S2, lower panel), than are rates in rnh201Δ strains lacking RNase H2 (Table 1). Thus, while nicking by RNase H2 may contribute to signaling for MMR of leading strand errors, one or more other signals are likely to exist. Given the importance of MMR to genome stability, redundant signals are to be expected. Candidate signals include the 3′-DNA ends used for chain elongation and/or PCNA (e.g., (Hombauer et al., 2011; Kleczkowska et al., 2001; Kunkel and Erie, 2005; Modrich and Lahue, 1996; Pavlov et al., 2003; Pluciennik et al., 2010; Tran et al., 1999; Umar et al., 1996)).
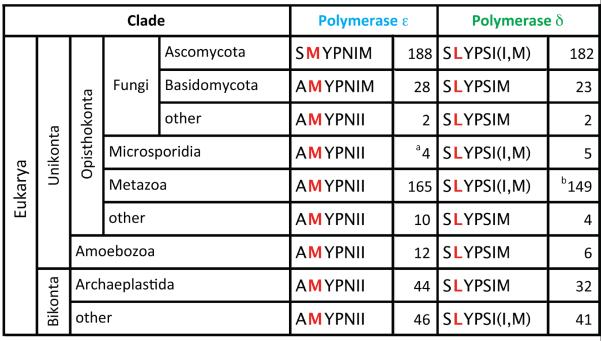
Sequence alignments (Figure 3) show that the amino acid in yeast Pol δ that was changed in this study, Leu612, is invariant among all Pols δ. When changed to methionine, L612M Pol δ becomes promiscuous for ribonucleotide incorporation (Figure 1). Thus, this lagging strand replicase conserves a leucine that helps to prevent ribonucleotide incorporation. In contrast, the opposite is true for Pol ε, which has an invariant methionine at the same position (Met644 in yeast). When Met644 is changed to leucine, M644L Pol ε becomes more like Pol δ, in that it incorporates fewer ribonucleotides (Nick McElhinny et al., 2010b). In other words, wild type Pol ε is only one amino acid away from being able to more effectively exclude ribonucleotides from DNA, yet Met644 is strictly conserved in Pol ε from many organisms (Figure 3). This suggests that there may be positive selective advantages to Pol ε incorporation of ribonucleotides into the nascent leading strand. Among several possibilities we discussed previously (Nick McElhinny et al., 2010c), this study provides evidence for one advantage, i.e., using the transient presence of a ribonucleotide to stabilize the genome by signaling for MMR, followed by ribonucleotide removal (Nick McElhinny et al., 2010b; Sparks et al., 2012) to prevent short deletions via a post-replication pathway initiated by endonucleolytic cleavage by topoisomerase 1 (Kim et al., 2011).
Figure 3. Conservation in the polymerase active site of Pols ε and δ.
Organisms are grouped by clade with organism counts to the right of each sequence. aPutative Polymerase ε from Nosema ceranae [SLYTDII]. bPolymerases δ from Trichoplax ahaerens [SLYP(TII,SII,SIM,SVM)] and Loxodonta africana [SMYPSIV].
EXPERIMENTAL PROCEDURES
Strains used in this study
All Saccharomyces cerevisiae strains used in this study may be found in Table S5, along with associated references.
Stable rNMP incorporation in vitro measurements
Assays were performed as in (Nick McElhinny et al., 2010a). The average percentages of alkali-sensitive product for each indicated DNA polymerase was calculated using data from two experiments (error bars indicate the range). Data for four-subunit wild-type and M644G Pol ε are from (Nick McElhinny et al., 2010b; Williams et al., 2012). Wild-type and L612M Pol δ were purified as described (Burgers and Gerik, 1998).
Probing for ribonucleotides in yeast genomic DNA in vivo
Genomic DNA was isolated from asynchronously growing strains in the exponential phase of growth using the Epicentre Yeast DNA purification kit. Five μg of DNA were subjected to alkaline-lysis by treatment with 0.3 M KOH for 2 hours at 55 °C. Alkaline-agarose electrophoresis was performed as described (Nick McElhinny et al., 2010b) and the DNA was transferred to a charged nylon membrane by capillary action and subjected to Southern analysis. Probing was performed using single-strand radiolabeled probes that anneal to either the nascent leading or nascent lagging strand of the URA3 reporter gene inserted in orientation 1 (OR1) at the AGP1 locus on chromosome III.
Quantitative analysis of strand-specific alkaline-hydrolysis fragmentation patterns
Fluorescence intensity was quantified for each alkali gel lane (Figure 1C) in 0.1 mm increments in order to create a curve – I(di) – of intensity versus migration distance (examples in Figure S1A–B). DNA ladders were used to build a standard curve for each gel. The migration distance of each standard, d, was related to the standard size, s (in base pairs), by the relation:
| Equation 1. |
This function was fit to each standard curve via least squares regression. Each point, i, in I(di) was transformed via the following function:
| Equation 2. |
This resulted in curves – I(si) – of intensity versus hydrolysis fragment size (examples in Figure S1C–D). Due to the logarithmic transformation, the width of bins represented by each point differ by up to 100-fold. This effect is corrected by first finding the bin width, w, for each point via
| Equation 3. |
and then converting to the average intensity per minimum bin width, , for each point via
| Equation 4. |
is then converted to the fraction of fragments of a given size, f(si) (examples in Figure S1E–F and I–J), by normalizing versus total lane intensity, IT.
| Equation 5. |
| Equation 6. |
In many cases observed in this study, f(si) is clearly bimodal. Monte Carlo simulations of Saccharomyces cerevisiae chromosome 3 fragmentation were performed, assuming probe location at the AGP1 locus, 1.5 kbp from ARS306 (Figure 1B; Supplemental Methods: Alkaline hydrolysis). The simulated f(si) curves each fit to a Weibull distribution, Wi(s):
| Equation 7. |
The bimodal observed f(si) curves each fit to a sum of two Weibull distributions (examples in Figure S1G–H), representing a high-mobility population (H; small fragments) and a low-mobility population (L; large fragments). The high-mobility fraction, fH, may be calculated from the scaling parameters, AH and AL via:
| Equation 8. |
High-mobility fractions are shown in Figure 1E.
Forward mutation assays
Mutation rates for each strain were estimated from URA3 mutation frequencies that were collected as in (Nick McElhinny et al., 2008; Pursell et al., 2007).
URA3 mutation spectra
URA3 mutation spectra were collected as per (Nick McElhinny et al., 2008; Pursell et al., 2007).
Mutation rates and correction factor calculations
Mutation rates and mismatch repair correction factors for specific mutation types (i.e. transitions and indels) mutation were calculated for each spectrum URA3 as per (Lujan et al., 2012).
Statistical analysis of mismatch repair efficiencies
In order to show differences in mismatch repair between RNH201 and rnh201Δ strains, p-values for comparison of correction factors were assigned (Figure 2). These were estimated as the proportion of at least 1000 Monte Carlo simulated data sets (both fluctuation and spectral results) that resulted in correction factors exceeding the relevant comparators. For purposes of Monte Carlo simulations, forward mutation rates were assumed to follow log-normal distributions and mutation spectrum counts to follow binomial distributions, as per (Lujan et al., 2012).
Supplementary Material
Highlights
Deletion of RNH201 causes a decrease in mismatch repair efficiency.
This effect is specific to errors made on the nascent leading strand.
Acknowledgements
We thank Jeffrey Stumpf and Kin Chan for helpful comments on the manuscript. We acknowledge the Molecular Genetics Core Facility at the National Institute of Environmental Health Sciences for sequence analysis of 5-FOA-resistant mutants. This work was supported by Project Z01 ES065070 to T.A.K. from the Division of Intramural Research of the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bubeck D, Reijns MA, Graham SC, Astell KR, Jones EY, Jackson AP. PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 2011;39:3652–3666. doi: 10.1093/nar/gkq980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. The Journal of biological chemistry. 2009;284:4041–4045. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- Cho JE, Kim N, Li YC, Jinks-Robertson S. Two distinct mechanisms of Topoisomerase 1-dependent mutagenesis in yeast. DNA repair. doi: 10.1016/j.dnarep.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H, Vassilev A, DePamphilis ML, Zhao Y, Zhang J, Burgers PM, Crouch RJ, Cerritelli SM. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 2009;37:96–110. doi: 10.1093/nar/gkn913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AB, Lujan SA, Kissling GE, Kunkel TA. Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase epsilon. DNA Repair (Amst) 2011;10:476–482. doi: 10.1016/j.dnarep.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder PS, Walder JA. Ribonuclease H from K562 human erythroleukemia cells. Purification, characterization, and substrate specificity. J Biol Chem. 1991;266:6472–6479. [PubMed] [Google Scholar]
- Eder PS, Walder RY, Walder JA. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- Fang WH, Modrich P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]
- Holmes J, Jr., Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci U S A. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci U S A. 2009;106:8495–8500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov FA, Holmes SF, Arana ME, Lukianova OA, O'Donnell M, Kunkel TA, Modrich P. Saccharomyces cerevisiae MutLalpha is a mismatch repair endonuclease. J Biol Chem. 2007;282:37181–37190. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Huang SN, Williams JS, Li YC, Clark AB, Cho JE, Kunkel TA, Pommier Y, Jinks-Robertson S. Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science (New York, N.Y. 2011;332:1561–1564. doi: 10.1126/science.1205016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Larrea AA, Lujan SA, Nick McElhinny SA, Mieczkowski PA, Resnick MA, Gordenin DA, Kunkel TA. Genome-wide model for the normal eukaryotic DNA replication fork. Proc Natl Acad Sci U S A. 2010;107:17674–17679. doi: 10.1073/pnas.1010178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan SA, Williams JS, Pursell ZF, Abdulovic-Cui AA, Clark AB, Nick McElhinny SA, Kunkel TA. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012;8:e1003016. doi: 10.1371/journal.pgen.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabe I, Kunkel TA, Carr AM. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kissling GE, Kunkel TA. Differential correction of lagging-strand replication errors made by DNA polymerases {alpha} and {delta} Proc Natl Acad Sci U S A. 2010a;107:21070–21075. doi: 10.1073/pnas.1013048107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, Johansson E, Chabes A, Kunkel TA. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010b;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Stith CM, Burgers PM, Kunkel TA. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 2007;282:2324–2332. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts B, Kumar D, Watt DL, Lundström E-B, Burgers PMJ, Johansson E, Chabes A, Kunkel TA. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010c;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13:744–748. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Newlon CS, Kunkel TA. Yeast origins establish a strand bias for replicational mutagenesis. Mol Cell. 2002;10:207–213. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- Pluciennik A, Dzantiev L, Iyer RR, Constantin N, Kadyrov FA, Modrich P. PCNA function in the activation and strand direction of MutLalpha endonuclease in mismatch repair. Proc Natl Acad Sci U S A. 2010;107:16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science (New York, N.Y. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydberg B, Game J. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc Natl Acad Sci U S A. 2002;99:16654–16659. doi: 10.1073/pnas.262591699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Whitehouse I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature. 2012;483:434–438. doi: 10.1038/nature10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, Burgers PM. RNase H2-Initiated Ribonucleotide Excision Repair. Mol Cell. 2012;47:980–986. doi: 10.1016/j.molcel.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC, Roberts JD, Kunkel TA. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- Tran HT, Gordenin DA, Resnick MA. The 3'-->5' exonucleases of DNA polymerases delta and epsilon and the 5'-->3' exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- Williams JS, Clausen AR, Nick McElhinny SA, Watts BE, Johansson E, Kunkel TA. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase epsilon. DNA Repair (Amst) 2012;11:649–656. doi: 10.1016/j.dnarep.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.