1. DISEASE CHARACTERISTICS
1.1 Name of the disease (synonyms)
Pseudohypoparathyroidism (PHP), Pseudohypoparathyroidism type Ia (PHP-Ia), Pseudohypoparathyroidism type Ib (PHP-Ib), Pseudohypoparathyroidism type Ic (PHP-Ic), Pseudopseudohypoparathyroidism (PPHP), Albright's Hereditary Osteodystrophy (AHO).
1.2 OMIM# of the disease
PHP-Ia (MIM 103580), PHP-Ib (MIM 603233), PHP-Ic (MIM 612462), PPHP (MIM 612463), AHO (MIM 103580).
1.3 Name of the analyzed genes or DNA/chromosome segments
GNAS/20q13.
1.4 OMIM# of the gene(s)
139320.
1.5 Mutational spectrum
In ≈70–80% of cases, PHP-Ia is caused by haploinsufficiency because of maternally-inherited heterozygous inactivating mutations of GNAS gene1, 2 (personal data). In very few cases, large deletions including part or the whole gene have been reported,3, 4 while in a further subset of patients cytosine methylation defects of the GNAS promoters have been identified, a pattern typically found in PHP-Ib.5, 6, 7, 8
Patients affected with PHP-Ib share a loss of methylation on the maternal A/B exon of GNAS. Those with the autosomal dominant form of PHP-Ib (AD-PHP-Ib) display an isolated loss of methylation at exon A/B associated with a recurrent 3-kb deletion in the STX16 gene, although a 4.4-kb deletion has been described in one family with AD-PHP-Ib.9, 10 In four families with AD-PHP-Ib, NESP55 and NESPAS deletions have also been described leading to the loss of all maternal GNAS imprints (epimutations).11, 12, 13
In most cases, PHP-Ib is sporadic, and is characterized by complete loss of methylation at the NESPas, XLαs and A/B promoters, and no other changes in cis- or trans-acting elements have been found to explain this loss of methylation. In some cases (2–20% of the patients with PHP-Ib) paternal 20q disomies have been described.14, 15, 16, 17
1.6 Analytical methods
Bi-directional DNA sequencing of exons 1–13 and their flanking intronic sequences is employed for the detection of point mutations and small insertions and deletions. Sequencing is recommended when PHP-Ia is suspected.
Methylation specific-multiplex ligation-dependant probe amplification (MS-MLPA) with SALSA kit ME031 (MRC-Holland, Amsterdam, The Netherlands) can be applied to detect methylation defects,16, 18 reported STX16 deletions, and deletions encompassing GNAS. Pyrosequencing with specific primers after bisulfite treatment of the genomic DNA, allows the detection of methylation defects,19 although it cannot uncover deletions of GNAS.
Fluorescent PCR of microsatellites D20S102, D20S1063, D20S496, D20S459, D20S443, D20S171 and D20S173 is recommended to confirm/exclude paternal 20q disomy. Analysis of the trio (parents and index) is essential for definitive conclusions.
Analysis of deletions causing methylation defects should be run by long-PCR.11, 12, 13
1.7 Analytical validation
Sequencing validation: confirmation of mutation in an independent biological sample of the index case. Special care is required in the interpretation of variants of unknown clinical significance (non-previously reported missense mutations). The gold standard is functional analysis (view 3.1.1) but this is usually not possible in a diagnostic set up. The variants should therefore be interrogated using conservation analysis, segregation analysis in the relatives of the index patient, analysis of at least 300 chromosomes from normal ethnically matched controls, and, if possible, predictive protein analysis. Thus, a statement must indicate the analysis that has been undertaken.
MS-MLPA validation:
Parallel analysis of negative and positive controls
Deletions encompassing a single probe should always be confirmed by alternative methods. Initially, sequencing should be undertaken to exclude the presence of a mutation or polymorphism in the probe-binding sequence, resulting in a false positive result. If no variant is observed, quantitative PCR,19 quantitative multiple PCR of short fragments20 or high density array CGH3 should be undertaken to confirm the presence of the deletion.
Methylation defects should be validated by pyrosequencing, methylation specific-PCR or combined bisulfite resctriction analysis19 in the absence of associated deletions.
Pyrosequencing validation: Accuracy in quantification by pyrosequencing technology is primarily limited by variation in PCR amplification, so it is recommended to run analysis in triplicate with internal controls for fully methylated and/or unmethylated DNAs. Confirmation of methylation defect in an independent biological sample of the index case.
1.8 Estimated frequency of the disease
(Incidence at birth (‘birth prevalence') or population prevalence)
If known to be variable between the ethnic groups, please report:
0.79/100.000 (according to Orphanet Report Series, November 2011)
1.9 Diagnostic setting

Comment:
PHP and AHO (short stature, rounded face, brachydactyly, ectopic ossifications and mental retardation) are rare, related, highly heterogeneous disorders with proven genetic component. The two main subtypes of PHP, PHP-Ia and PHP-Ib are caused by molecular alterations within or upstream of the GNAS locus.1, 21 In particular, most PHP-Ia patients, who show AHO associated with resistance toward multiple hormones (parathyroid hormone (PTH)/TSH/GHRH/gonadotrophins/calcitonin), are affected by heterozygous, maternally-derived mutations in GNAS exons 1–13. The same mutations inherited from the father lead to PPHP, in which AHO occurs in the absence of endocrine abnormalities.
On the other hand, the majority of PHP-Ib patients, who classically display hormone resistance limited to PTH and sometimes TSH with no AHO, display methylation defects in the imprinted GNAS cluster. Recent data on both clinical and molecular aspects of these complex disorders have challenged the distinction of different GNAS-related diseases. As stated in point 1.2, in a subset of patients with PHP and variable degrees of AHO, epigenetic defects of GNAS similar to those classically found in PHP-Ib have been detected, suggesting a molecular overlap between PHP-Ia and Ib.
Overall, the phenotypes are highly variable. If an alternative diagnosis is excluded, GNAS mutation/epimutation screening should be performed in all the subjects with hormone resistance with or without AHO signs.
Prenatal diagnosis is technically feasible but it is only recommended when one of the parents carries a coding mutation or deletions/alterations in the regulatory regions of GNAS.
2. TEST CHARACTERISTICS

2.1 Analytical sensitivity
(proportion of positive tests if the genotype is present)
The sensitivity of MS-MLPA approaches 100% for deletion detection, but errors can be made when a polymorphism is present in the probe-binding site, thus resulting in a false allele dropout as described by the manufacturer. For methylation defects low-grade mosaics16, 19 might not be detected.
The sensitivity for genomic sequencing also approaches 100% for mutation detection, but errors can be made because of polymorphisms causing allele dropout. Mutations outside the coding exons in promoters or enhancers are likely to be missed.
2.2 Analytical Specificity
(proportion of negative tests if the genotype is not present)
Nearly 100%.
2.3 Clinical Sensitivity
(proportion of positive tests if the disease is present)
The clinical sensitivity can be dependent on variable factors such as age or family history. In such cases a general statement should be given, even if a quantification can only be made case by case.
The presence of ectopic ossifications in the context of normal kidney function is associated with haploinsufficiency of GNAS in the great majority of patients.
The proportion of identified loss-of-function mutations of GNAS is close to 70% when the phenotype is present (ie AHO and hormone resistance).
The proportion of identified loss of imprinting at GNAS is close to 80–90% when the phenotype is present (ie isolated hormone resistance).
2.4 Clinical specificity
(proportion of negative tests if the disease is not present)
The clinical specificity can be dependent on variable factors such as age or family history. In such cases a general statement should be given, even if quantification can only be made case by case.
Nearly 100% (the unaffected carriers of STX16 and NESP55/NESPas deletions in PHP-Ib families will have the test positive, but are not predicted to develop the disease at all).
2.5 Positive clinical predictive value
(life time risk to develop the disease if the test is positive)
Nearly 100%.
So far, the identification of the mutation has been always associated with development of the disease (personal experience,22), excluding the unaffected carriers of the regulatory mutations in PHP-Ib families.
2.6 Negative clinical predictive value
(probability not to develop the disease if the test is negative)
Assume an increased risk based on family history for a non-affected person. Allelic and locus heterogeneity may need to be considered.
Index case in that family had been tested:
If a pathogenic GNAS alteration is identified in the index case, the negative predictive value is close to 100%.
Index case in that family had not been tested:
Genetic heterogeneity with undiscovered gene(s) or alternative (epi)genetic mechanisms might account for ≈30% of PHP individuals with negative testing for GNAS but who still have the condition.
3. CLINICAL UTILITY
3.1 (Differential) diagnosis: The tested person is clinically affected
(To be answered if in 1.10 ‘A' was marked)
3.1.1 Can a diagnosis be made other than through a genetic test?
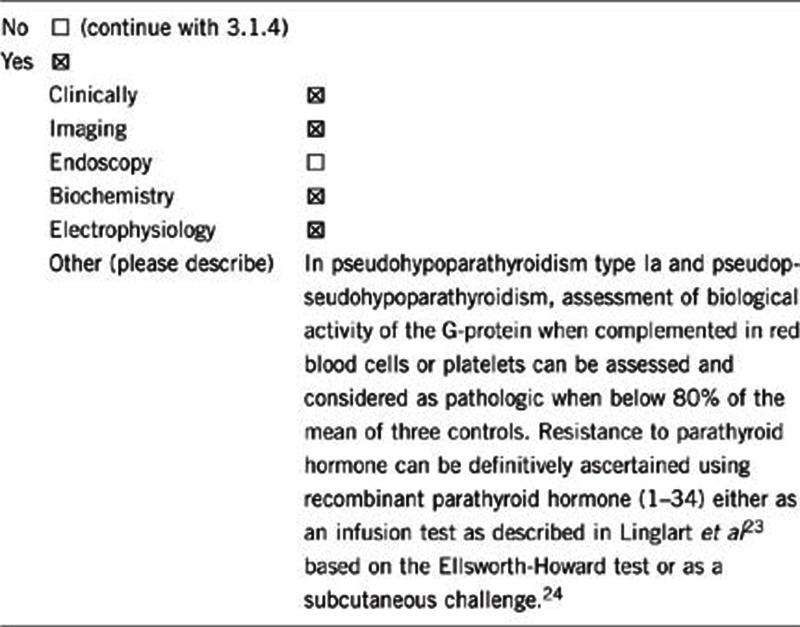
Overall, careful clinical examination, together with appropriate endocrine testing, can suggest the diagnosis even if genetic testing cannot confirm it.
3.1.2 Describe the burden of alternative diagnostic methods to the patient
Diagnosis of PHP is defined by the co-existence of hypocalcemia and hyperphosphatemia with elevated PTH levels in the presence of normal vitamin D values, normal renal function and the absence of hypercalciuria.
Beside calcium metabolism, thyroid and gonadal function, GH secretion and calcitonin should be measured in all the patients with suspected hormone resistance. In difficult cases, resistance to PTH can be definitively ascertained using recombinant PTH (1–34) either as an infusion test as described in Linglart et al23 based on the Ellsworth-Howard test or as a subcutaneous challenge.24
For PHP-Ia diagnosis, diagnostic procedures should also include evaluation of AHO: X-ray analysis revealing brachydactyly of all digits or affecting mainly the fourth and fifth digits is usually considered very suggestive. CT scan often reveals intracranial calcifications of the basal ganglia as signs of chronic hypocalcemia. Careful physical examination to detect and/or follow-up ectopic ossifications should be carried out regularly. However, AHO features are highly variable, depending upon the age of the patient and patient-to-patient variability. Genetic test remains useful to confirm clinical diagnosis and to help prognosis evaluation.
3.1.3 How is the cost effectiveness of alternative diagnostic methods to be judged?
X-rays and repeated lab work approximate the cost of the genetic analysis, yet do not formally establish the diagnosis, as there is no specific bio- or radio-marker of the disease.
On the other hand, confirmation of the diagnosis by genetic testing does not prevent the use of radiological and regular biochemical analysis as the onset of the different clinical manifestations is highly variable and unpredictable.
3.1.4 Will disease management be influenced by the result of a genetic test?

3.2 Predictive Setting: The tested person is clinically unaffected but carries an increased risk based on family history
(To be answered if in 1.10 ‘B' was marked)
3.2.1 Will the result of a genetic test influence lifestyle and prevention?
If the test result is positive (please describe)
Forecasts the development of the disease, and therefore induces the clinical and biochemical follow-up:
For patients with maternal haploinsufficiency of GNAS, we recommend vitamin D analogs (calcitriol, alfacalcidol) when PTH and phosphate increase, before the occurrence of hypocalcemia, cognitive support and physiotherapy, dietary monitoring and follow-up of growth and, if the growth hormone secretion is insufficient, growth hormone treatment.
For patients with methylation defects, we recommend vitamin D analogs (calcitriol, alfacalcidol) when PTH and phosphate increase, before the occurrence of hypocalcemia.
For patients with PPHP and paternal haploinsufficiency of GNAS, we recommend a regular clinical follow-up. In case of extensive or progressive ossifications, patients should be managed in collaboration with surgeons/orthopedics in expert centers. Surgery to remove ectopic ossifications is often not recommended because of the risk of recurrence; medical treatments are not evaluated.
If the test result is negative (please describe)
Patients with AHO and PTH resistance negative for GNAS (mutation and methylation) should be tested for PRKAR1A.23, 25
3.2.2 Which options in view of lifestyle and prevention does a person at-risk have if no genetic test has been done (please describe)?
Regular clinical (growth, weight) and biochemical follow-up, at least yearly
Recurrent information about the disease symptoms and risks
3.3 Genetic risk assessment in family members of a diseased person
(To be answered if in 1.10 ‘C' was marked)
3.3.1 Does the result of a genetic test resolve the genetic situation in that family?
This is only possible if genetic mutation/alteration (mutations in the coding sequence of GNAS or deletions of regulatory regions, including STX16, NESP/NESPas deletions) is found in the affected proband. Otherwise, relatives are advised to undergo detailed biochemical testing.
3.3.2 Can a genetic test in the index patient save genetic or other tests in family members?
Yes, it will be cost effective, if a GNAS alteration is identified in the index patient, it can reduce the need for testing for other genetic conditions in family members by providing a diagnosis and, on the other hand, a positive genetic test result implies the possibility of predictive genetic testing in relatives. Relatives being non-carrier of the pathogenic mutation can be excluded from clinical follow-up and this will mean saving the costs of repeated biochemical analysis.
A negative test result in the index patient does not mean the disease is not hereditary so all relatives are advised to continue regular clinical follow-up.
3.3.3 Does a positive genetic test result in the index patient enable a predictive test in a family member?
Yes, if genetic mutations/deletions are found.
In particular:
The identification of a heterozygous inactivating mutation of the GNAS gene (including deletion in NESP55, NESPas or STX16) in patients with PHP predicts that mothers carry the mutation, except for de novo mutation. Conversely, fathers will not be affected.
The identification of a heterozygous inactivating mutation of the GNAS gene (including deletion in NESP55, NESPas or STX16) in female patients with PHP or PPHP predicts that children of the index case are at 50% risk of developing PHP. Conversely, the identification of a heterozygous inactivating mutation of the GNAS gene (including deletion in NESP55, NESPas or STX16) in male patients with PHP or PPHP predicts that children of the index case are at 50% risk of developing PPHP and/or extensive ectopic ossifications yet no hormone resistance.
No, if apparently sporadic epigenetic alterations are found, with the exception of patients with 20q disomy who will not transmit the disease.
3.4 Prenatal diagnosis
(To be answered if in 1.10 ‘D' was marked)
3.4.1 Does a positive genetic test result in the index patient enable a prenatal diagnosis?
Depending of the laws of the Country, prenatal diagnosis may be requested when one of the parents is affected and/or carries a genetic mutation of the GNAS gene (including deletion in NESP55, NESPas or STX16).
Prenatal screening of epigenetic aberrations has not been performed for this condition.
4. IF APPLICABLE, FURTHER CONSEQUENCES OF TESTING
Please assume that the result of a genetic test has no immediate medical consequences. Is there any evidence that a genetic test is nevertheless useful for the patient or his/her relatives? (Please describe)
The genetic diagnosis of PHP has clinical validity for both index cases and their relatives. Genetic counseling and appropriate predictive genetic testing of family members should establish their risk for the condition. Analysis of structural-defects carrier status could be requested owing to the psychosocial implications of short stature and be useful to plan appropriate endocrine testing and consequent specific treatment.
Acknowledgments
This work was supported by EuroGentest2 (Unit 2: ‘Genetic testing as part of health care'), a Coordination Action under FP7 (Grant Agreement Number 261469) and the European Society of Human Genetics. GPN is partially funded by the I3SNS Program of the Spanish Ministry of Health (CP03/0064; SIVI 1395/09), IG is supported by FIS-program I3SNS-CA10/01056. This work was also partially supported by a grant from the Italian Ministry of Health to GM (GR-2009-1608394), the French center of reference for rare disorders of calcium and phosphorus metabolism to AL and Fundacion Eugenio Rodriguez Pascual and the Instituto de Salud Carlos III (PI10/0148) to GPN.
The authors declare no conflict of interest.
References
- Mantovani G. Clinical review: pseudohypoparathyroidism: diagnosis and treatment. J Clin Endocrinol Metab. 2011;96:3020–3030. doi: 10.1210/jc.2011-1048. [DOI] [PubMed] [Google Scholar]
- Linglart A, Carel JC, Garabedian M, Le T, Mallet E, Kottler ML. GNAS1 lesions in pseudohypoparathyroidism Ia and Ic: genotype phenotype relationship and evidence of the maternal transmission of the hormonal resistance. J Clin Endocrinol Metab. 2002;87:189–197. doi: 10.1210/jcem.87.1.8133. [DOI] [PubMed] [Google Scholar]
- Fernandez-Rebollo E, Garcia-Cuartero B, Garin I, et al. Intragenic GNAS deletion involving Exon A/B in pseudohypoparathyroidism type 1A resulting in an apparent loss of exon A/B methylation: potential for misdiagnosis of pseudohypoparathyroidism type 1B. J Clin Endocrinol Metab. 2010;95:765–771. doi: 10.1210/jc.2009-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui T, Nagasaki K, Takagi M, Narumi S, Ishii T, Hasegawa T.A family of pseudohypoparathyroidism type Ia with an 850-kb submicroscopic deletion encompassing the whole GNAS locus Am J Med Genet A 2011. e-pub ahead of print 2 December 2011 doi: 10.1002/ajmg.a.34393 [DOI] [PubMed]
- Mantovani G, de SL, Barbieri AM, et al. Pseudohypoparathyroidism and GNAS epigenetic defects: clinical evaluation of albright hereditary osteodystrophy and molecular analysis in 40 patients. J Clin Endocrinol Metab. 2010;95:651–658. doi: 10.1210/jc.2009-0176. [DOI] [PubMed] [Google Scholar]
- Perez de Nanclares G, Fernandez-Rebollo E, Santin I, et al. Epigenetic defects of GNAS in patients with pseudohypoparathyroidism and mild features of Albright's hereditary osteodystrophy. J Clin Endocrinol Metab. 2007;92:2370–2373. doi: 10.1210/jc.2006-2287. [DOI] [PubMed] [Google Scholar]
- Unluturk U, Harmanci A, Babaoglu M, et al. Molecular diagnosis and clinical characterization of pseudohypoparathyroidism type-Ib in a patient with mild Albright's hereditary osteodystrophy-like features, epileptic seizures, and defective renal handling of uric acid. Am J Med Sci. 2008;336:84–90. doi: 10.1097/MAJ.0b013e31815b218f. [DOI] [PubMed] [Google Scholar]
- Mariot V, Maupetit-Mehouas S, Sinding C, Kottler ML, Linglart A. A maternal epimutation of GNAS leads to Albright osteodystrophy and PTH resistance. J Clin Endocrinol Metab. 2008;93:661–665. doi: 10.1210/jc.2007-0927. [DOI] [PubMed] [Google Scholar]
- Bastepe M, Frohlich LF, Hendy GN, et al. Autosomal dominant pseudohypoparathyroidism type Ib is associated with a heterozygous microdeletion that likely disrupts a putative imprinting control element of GNAS. J Clin Invest. 2003;112:1255–1263. doi: 10.1172/JCI19159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linglart A, Gensure RC, Olney RC, Juppner H, Bastepe M. A novel STX16 deletion in autosomal dominant pseudohypoparathyroidism type Ib redefines the boundaries of a cis-acting imprinting control element of GNAS. Am J Hum Genet. 2005;76:804–814. doi: 10.1086/429932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastepe M, Frohlich LF, Linglart A, et al. Deletion of the NESP55 differentially methylated region causes loss of maternal GNAS imprints and pseudohypoparathyroidism type Ib. Nat. Genet. 2005;37:25–27. doi: 10.1038/ng1487. [DOI] [PubMed] [Google Scholar]
- Chillambhi S, Turan S, Hwang DY, Chen HC, Juppner H, Bastepe M. Deletion of the noncoding GNAS antisense transcript causes pseudohypoparathyroidism type Ib and biparental defects of GNAS methylation in cis. J. Clin. Endocrinol. Metab. 2010;95:3993–4002. doi: 10.1210/jc.2009-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard N, Abeguile G, Coudray N, et al. A new deletion ablating NESP55 causes loss of maternal imprint of A/B GNAS and autosomal dominant pseudohypoparathyroidism type Ib. J Clin Endocrinol Metab. 2012;97:E863–E867. doi: 10.1210/jc.2011-2804. [DOI] [PubMed] [Google Scholar]
- Bastepe M, Lane AH, Juppner H. Paternal uniparental isodisomy of chromosome 20q--and the resulting changes in GNAS1 methylation—as a plausible cause of pseudohypoparathyroidism. Am J Hum Genet. 2001;68:1283–1289. doi: 10.1086/320117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastepe M, Altug-Teber O, Agarwal C, Oberfield SE, Bonin M, Juppner H. Paternal uniparental isodisomy of the entire chromosome 20 as a molecular cause of pseudohypoparathyroidism type Ib (PHP-Ib) Bone. 2010;48:659–662. doi: 10.1016/j.bone.2010.10.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Rebollo E, Lecumberri B, Garin I, et al. New mechanisms involved in paternal 20q disomy associated with pseudohypoparathyroidism. Eur J Endocrinol. 2010;163:953–962. doi: 10.1530/EJE-10-0435. [DOI] [PubMed] [Google Scholar]
- Lecumberri B, Fernandez-Rebollo E. Coexistence of two different pseudohypoparathyroidism subtypes (Ia and Ib) in the same kindred with independent Gs{alpha} coding mutations and GNAS imprinting defects. J Med Genet. 2010;47:276–280. doi: 10.1136/jmg.2009.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsum Z, Abu SL, Nygren AO, Al-Hamed MA, Alkuraya FS. Methylation-specific multiplex-ligation-dependent probe amplification as a rapid molecular diagnostic tool for pseudohypoparathyroidism type 1b. Genet Test Mol Biomarkers. 2010;14:135–139. doi: 10.1089/gtmb.2009.0092. [DOI] [PubMed] [Google Scholar]
- Maupetit-Mehouas S, Mariot V, Reynes C, et al. Quantification of the methylation at the GNAS locus identifies subtypes of sporadic pseudohypoparathyroidism type Ib. J Med Genet. 2011;48:55–63. doi: 10.1136/jmg.2010.081356. [DOI] [PubMed] [Google Scholar]
- Perez-Nanclares G, Romanelli V, Mayo S, et al. Detection of hypomethylation syndrome among patients with epigenetic alterations at the GNAS locus. J Clin Endocrinol Metab. 2012;97:E1060–E1067. doi: 10.1210/jc.2012-1081. [DOI] [PubMed] [Google Scholar]
- Bastepe M. The GNAS locus and pseudohypoparathyroidism. Adv Exp Med Biol. 2008;626:27–40. doi: 10.1007/978-0-387-77576-0_3. [DOI] [PubMed] [Google Scholar]
- Linglart A, Bastepe M, Juppner H. Similar clinical and laboratory findings in patients with symptomatic autosomal dominant and sporadic pseudohypoparathyroidism type Ib despite different epigenetic changes at the GNAS locus. Clin Endocrinol (Oxf) 2007;67:822–831. doi: 10.1111/j.1365-2265.2007.02969.x. [DOI] [PubMed] [Google Scholar]
- Linglart A, Menguy C, Couvineau A, Auzan C, Gunes Y. Recurrent PRKAR1A mutation in acrodysostosis with hormone resistance. N Engl J Med. 2011;364:2218–2226. doi: 10.1056/NEJMoa1012717. [DOI] [PubMed] [Google Scholar]
- Todorova-Koteva K, Wood K, Imam S, Jaume JC. Screening for parathyroid hormone resistance in patients with non-phenotypically evident pseudohypoparathyroidism. Endocr Pract. 2012;11:1–21. doi: 10.4158/EP12007.OR. [DOI] [PubMed] [Google Scholar]
- Silve C, Clauser E, Linglart A.Acrodysostosis Horm Metab Res 2012. e-pub ahead of print 19 July 2012. [DOI] [PubMed]


