Recently, Hedberg et al1 identified a DES mutation (p.P419S) in a Swedish family, suffering from myofibrillar myopathy (MFM) in combination with arrhythmogenic right ventricular cardiomyopathy (ARVC), by next-generation sequencing. Originally, a linkage analysis indicated that the genetic defect is located on chromosome 10q22.3 in this family.2 The analysis of muscle biopsies of affected patients demonstrated an aggregation of desmin and further proteins.
The same desmin mutation (p.P419S) was identified before by Olivé et al3 in patients suffering from skeletal myopathy or hypertrophic cardiomyopathy, respectively. However, Hedberg et al1 demonstrated that this mutation did not completely co-segregate within the Swedish family, raising the questions on pathogenesis or penetrance, respectively. Of note, in both studies the desmin mutant p.P419S was not functionally characterized. Hence, it is currently difficult to judge the pathological potential of this variant. Especially, it is unclear whether the DES mutation p.P419S is a sufficient molecular trigger for aggregate formation.
For this reason, we introduced this mutation by site-directed mutagenesis into a desmin construct (pEYFP-N1-Desmin) using appropriate oligonucleotides and transfected H9c2, C2C12 and SW-13 cells with mutant and wild-type desmin–eYFP constructs. The filament or aggregate formation was investigated in cell culture and the filament formation of purified recombinant mutant desmin was analysed in vitro by atomic force microscopy (AFM), as previously described.4
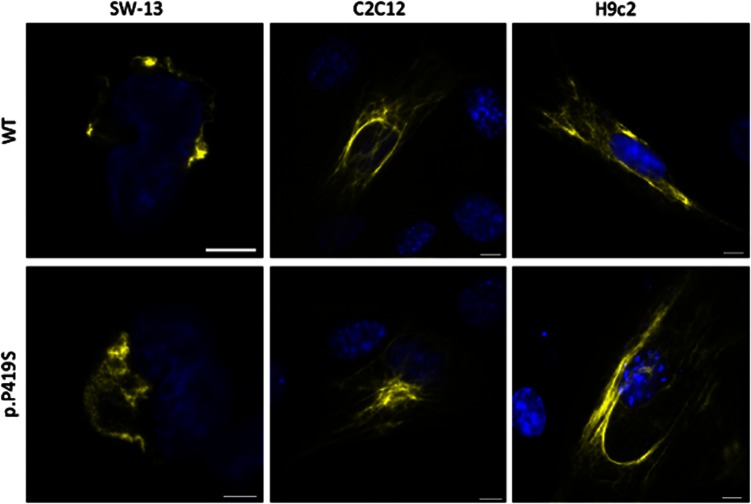
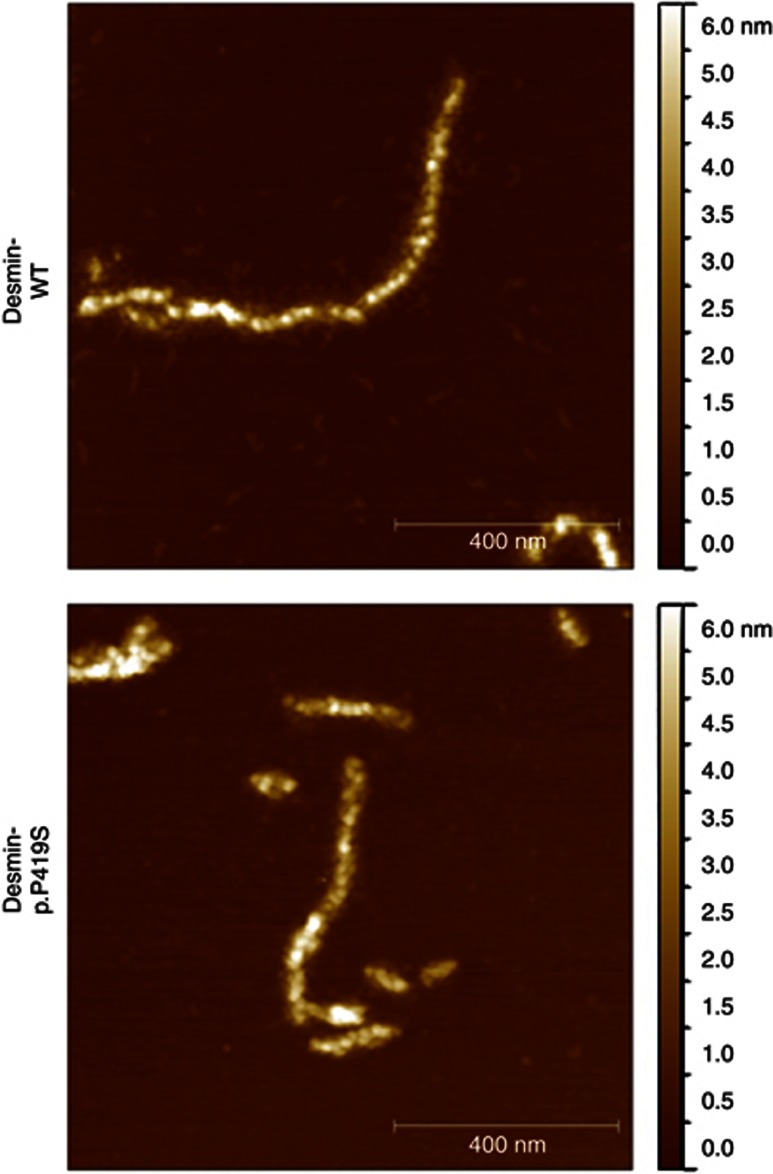
To our surprise, the expression of desmin-p.P419S does not induce an aggregation in either cell line as recently described for other ARVC-related desmin mutants4, 5, 6 (Figure 1). The cell culture data were also supported by the AFM analysis virtually yielding undistinguishable desmin filaments between wild-type and desmin-p.P419S in vitro (Figure 2). Thus, our data reveal that the desmin mutant p.P419S published by Hedberg et al1 forms filaments in vitro and in transfected cells. Consequently, it might be important to look for further molecular triggers, which induce or influence the protein aggregation in the Swedish patients suffering from MFM/ARVC. From our point of view, the next-generation sequencing data of Hedberg et al1 might provide an important basis for further studies, identifying modifier genes or other molecular abnormalities responsible for desmin aggregate formation.
Figure 1.
Filament formation of wild-type and mutant (p.P419S) desmin in transfected cells. Representative fluorescence images of transfected SW-13, C2C12 and H9c2 cells expressing desmin–eYFP constructs (yellow) were shown. The nuclei were stained with DAPI. Scale bars represent 10 μm.
Figure 2.
In vitro filament formation of wild-type and mutant (p.P419S) desmin. The desmin molecules were expressed in Escherichia coli and purified by ion-exchange and affinity chromatography. Filament assembly was initiated by the addition of sodium chloride (100 mℳ) and was analysed by AFM. The height is colour coded in the representative topography images.
The authors declare no conflict of interest.
References
- Hedberg C, Melberg A, Kuhl A, Jenne D, Oldfors A. Autosomal dominant myofibrillar myopathy with arrhythmogenic right ventricular cardiomyopathy 7 is caused by a DES mutation. Eur J Hum Genet. 2012;20:984–985. doi: 10.1038/ejhg.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melberg A, Oldfors A, Blomstrom-Lundqvist C, et al. Autosomal dominant myofibrillar myopathy with arrhythmogenic right ventricular cardiomyopathy linked to chromosome 10q. Ann Neurol. 1999;46:684–692. doi: 10.1002/1531-8249(199911)46:5<684::aid-ana2>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Olivé M, Armstrong J, Miralles F, et al. Phenotypic patterns of desminopathy associated with three novel mutations in the desmin gene. Neuromuscul Disord. 2007;17:443–450. doi: 10.1016/j.nmd.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodehl A, Hedde PN, Dieding M, et al. Dual-color photoactivation localization microscopy of cardiomyopathy associated desmin mutants. J Biol Chem. 2012;287:16047–16057. doi: 10.1074/jbc.M111.313841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauke B, Kossmann S, Gaertner A, et al. De novo desmin-mutation N116S is associated with arrhythmogenic right ventricular cardiomyopathy. Hum Mol Genet. 2010;19:4595–4607. doi: 10.1093/hmg/ddq387. [DOI] [PubMed] [Google Scholar]
- Vernengo L, Chourbagi O, Panuncio A, et al. Desmin myopathy with severe cardiomyopathy in a Uruguayan family due to a codon deletion in a new location within the desmin 1A rod domain. Neuromuscul Disord. 2010;20:178–187. doi: 10.1016/j.nmd.2010.01.001. [DOI] [PubMed] [Google Scholar]