Abstract
In this paper we present an automated system based mainly on the computed tomography (CT) images consisting of two main components: the midline shift estimation and intracranial pressure (ICP) pre-screening system. To estimate the midline shift, first an estimation of the ideal midline is performed based on the symmetry of the skull and anatomical features in the brain CT scan. Then, segmentation of the ventricles from the CT scan is performed and used as a guide for the identification of the actual midline through shape matching. These processes mimic the measuring process by physicians and have shown promising results in the evaluation. In the second component, more features are extracted related to ICP, such as the texture information, blood amount from CT scans and other recorded features, such as age, injury severity score to estimate the ICP are also incorporated. Machine learning techniques including feature selection and classification, such as Support Vector Machines (SVMs), are employed to build the prediction model using RapidMiner. The evaluation of the prediction shows potential usefulness of the model. The estimated ideal midline shift and predicted ICP levels may be used as a fast pre-screening step for physicians to make decisions, so as to recommend for or against invasive ICP monitoring.
Keywords: Medicine, Issue 74, Biomedical Engineering, Molecular Biology, Neurobiology, Biophysics, Physiology, Anatomy, Brain CT Image Processing, CT, Midline Shift, Intracranial Pressure Pre-screening, Gaussian Mixture Model, Shape Matching, Machine Learning, traumatic brain injury, TBI, imaging, clinical techniques
Introduction
Every year there are about 1.4 million traumatic brain injuries (TBI) related emergency department cases in the United States, of which, over 50,000 result in death1. Severe TBI is usually accompanied by an increase in intracranial pressure (ICP) with symptoms such as hematomas and swelling brain tissue. These result in reduced cerebral perfusion pressure and cerebral blood flow, placing the injured brain in additional risk. Severe ICP increase can be fatal, so monitoring ICP for patients with TBI is crucial. This typically requires placement of indwelling catheters directly into the brain for monitoring of pressure, a risky procedure for patients that can only be performed at specialized medical centers. The procedure also involves risk such as infection. However, some signs of elevated ICP may be noticeable in medical imaging. In particular, midline shift is often associated with an increase in the ICP and can be captured from the brain computed tomography (CT) images. As such, these images provide an opportunity for non-invasive detection of elevated ICP which can be used as a pre-screening step before cranial trepanation. CT imaging is still the gold standard for initial TBI assessment among all other imaging modalities, e.g. MRI, because of its high speed and relative low cost2. Furthermore, a CT examination does not require strict patient immobility, and has advantage in revealing severe abnormalities such as bone fractures and hematomas. While CT is commonly used for detection of injuries in the brain, based on the current technology, midline shift is not automatically measured and therefore physicians must assess this important factor by visual inspection. Inaccurate or inconsistent CT interpretation is often associated with the nature of the human visual system and the complex structure of the brain. While small midline shifts are elusive, they are often invaluable for assessment of brain injury, in particular at early stages of injury before a patient's condition becomes more severe. On the other side of the spectrum, large midline shift suggests highly elevated ICP and more severe TBI. However, it is a very challenging task for humans to visually inspect CT images and predict the level of ICP quantitatively. Due to advances in automated computational techniques, features extracted from CT images, such as midline shift, hematoma volume, and texture of brain CT images, can be measured accurately and automatically using advanced image processing methods. However, the relationship between ICP and midline shift as well as other features such as degree of bleeding, the texture from CT images has not been explored. In this paper, a computational framework has been proposed to measure the midline shift measurement as well as other physiological / anatomical features on brain CT images and then predict the degree of ICP non-intrusively using machine learning techniques.
Protocol
1. Methodology Overview
The proposed framework processes the brain CT images of traumatic brain injury (TBI) patients to automatically calculate midline shift in pathological cases and use it as well as other extracted information to predict intracranial pressure (ICP). Figure 1 shows the schematic diagram of the entire framework. The automated midline shift measurement can be divided into three steps. First, the ideal midline of the brain, i.e. the midline before injury, is found via a hierarchical search based on skull symmetry and tissue features3. Secondly, the ventricular system is segmented for each brain CT image4. Thirdly, the actual midline is estimated from the segmented deformed ventricular system using a shape matching method5. The horizontal shift of the ventricular system is then estimated based on the estimation of the ideal midline and the actual midline. After the midline shift is successively estimated, features including midline shift, texture information of CT images, as well as other demographic information are used to predict ICP. Machine learning algorithms are used to model the relationship between the ICP and the extracted features6.
2. Ideal Midline Estimation
This step detects the approximate ideal midline using symmetry of the skull. First, using grayscale thresholding, the skull needs to be segmented from the rest of the content in the CT image. After which, perform an exhaustive search to identify the rotation angles around the mass center of the skull. The optimal rotation angle is defined as the angle that maximizes the symmetry of the resulting halves of the skull. The approximate ideal midline is the line passing through the mass center point and has the optimal rotation angel with respect to the original vertical direction of the CT image.
This step detects the posterior falx cerebri and anterior falx attachment to the margins of the sagittal sulcus. This step is performed to refine the identified approximate ideal midline. First define two searching rectangles centered at the two intersection points between the approximate ideal midline and the calvarium. Next, choose the sizes of the rectangles empirically so that they cover the anatomical features to be detected as explained below. The anterior falx attachment is detected as the peak point of the ridge on the calvarium and the falx cerebri is detected as the gray line in the posterior region3.
This step uses those features detected above to refine the ideal midline position. Once the peak point of the anterior falx attachment and the furthest point in the posterior falx cerebri from the calvarum are specified, the refined ideal midline is the line connecting the two points.
3. Ventricle Segmentation
First apply a low-level segmentation using a Gaussian mixture model (GMM) for each CT slice4,7. CT images can be divided into 4 types of tissue: bone/blood, cerebrospinal fluid (CSF), gray matter and white matter. For the initialization of the Gaussian Mixture Model, the parameters are estimated based on an iterative K-means segmentation result of the CT image. Then use the expectation-maximization (EM) method to optimize GMM iteratively to better represent the CT image. The hard segmentation result can be obtained by partitioning the CT image into regions based on the maximum probability of the membership that each pixel belongs to different region types.
After the low-level segmentation of GMM, apply further constraints on the segmentation result in order to recognize ventricular regions. Only ventricular regions with size above a certain threshold are retained. Apply constraints on the location of ventricular regions as well using the brain bounding box and the set of ventricle templates. Extract the set of ventricle templates from a standard brain MR images and then enlarge it using the morphological dilation to accommodate variations among different subjects and pathological cases.
4. Actual Midline Estimation
Specify the feature points on the ventricle templates extracted from MR.
Do the Multiple Regions Shape Matching5,8 between segmented ventricles and the MR template.
Estimate the actual midline based on the identified feature points on the ventricle shapes using shape matching. Then use the average of left side mean and right side mean of the x-coordinates of those feature points to define the x-coordinate of the midline.
5. More Feature Extractions
Measure the intracranial hematoma/bleeding volume based on the Gaussian mixture model (GMM) segmentation results obtained from the CT images. The segmented result may include small regions including venous sinus blood and falx cerebri, but they can usually be neglected in comparison to the regions of hematoma. Then count the number of pixels classified as blood for each slice and sum them up. The final sum quantifies the extravasated blood volume in the CT examination.
Extract texture features. First select six windows in each CT image which captures the gray and white matter but avoid the blood and ventricle structures in the CT image, see Figure 6. Then extract the corresponding texture features by using the following methods: Gray Level Run length9, Histogram analysis, Fourier analysis, Dual Tree Complex Wavelet analysis10.
Demographic information and injury severity score are also collected.
All extracted features of each CT image are aggregated to represent the entire CT examination. Specifically, min(f), max(f), median(f), mean(f), std(f) are calculated among all the selected features belonging to the particular CT examination, e.g. a feature ƒ of the midline shift or a texture feature. For the intracranial hemorrhage amount feature, besides the 5 operators listed above, sum(f) is also added to record the total blood volume.
6. ICP Estimation
The main idea of ICP estimation is to apply machine learning techniques to build a model based on a set of training samples. Then the built model is evaluated on the remaining test samples. Because of the high dimension of extracted features including those from the CT scans and demographic information, feature selection is important to remove unrelated features for a relatively simple thus stable model. Therefore there are two steps to be performed for ICP estimation/prediction. First, select the relative features which are informative in predicting ICPs. The second step is to use Support Vector Machines (SVM) as the learning algorithm to develop and evaluate the training model. Software such as RapidMiner11 is ideal for this task because it is a very well developed tool for most of machine learning algorithms and provides very powerful interfaces to train and evaluate models.
Perform the feature selection in two stages. First use the information gain ratio criterion in the first stage to select the top 50 features. In the second stage, use a genetic algorithm incorporating SVM to further optimize the feature selection. Then use the final set of selected features to build the model for ICP prediction in the following step.
Perform ICP classification and evaluation through machine learning techniques. The top level diagram of the training and testing are shown in RapidMiner in Figure 8. Apply a 10 fold cross-validation scheme as the outmost layer for evaluation. In order to build a model with better generalization and avoid over-fitting to the training data, nest another layer of cross-validation inside each training fold. In these modules, first apply the feature selection process described above and then use a SVM for classification with its own parameter selection module6.
Representative Results
The testing CT datasets were provided by the Carolinas Healthcare System (CHS) under Institutional Review Board approval. All subjects were diagnosed with mild to severe TBI when first admitted to hospital. For each patient, the ICP value was recorded every hour using ICP probes inside the ventricle region both before and after CT scans were obtained. To associate the ICP value with each CT scan, average the two closest measurements of ICP to the time of CT scan, both of which are within an hour of the CT scan. Then assign the average as the estimated ICP value at the time of the CT scan. Group the ICP values into two classes: elevated ICP if ICP >12 mm Hg and normal ICP if ICP ≤ 12 mm Hg. The datasets contain 17 patients. From this set, 391 axial CT scan images are selected that show ventricles or regions that should have contained ventricles. Figure 2 shows the result of the ideal midline detection. In Figure 3, the ventricles are segmented. Figure 4 shows the estimated actual midline. Figure 5 shows the estimation of midline shift. Figure 7 shows the segmentation of blood using GMM. A quantitative evaluation of the performance is also performed. In most slices (over 80%), the error between the ideal midline estimated by the framework and the manual annotation are around 2 pixels, which is about 1mm. For the actual midline, above 80% has less than 2.25 mm difference provided that the quality of the ventricular segmentation is relatively good (it is defined as "relatively good" if the quality of the segmentation result can be used for estimation of the actual midlines manually). There are 57 CT scans in the evaluation of ICP prediction. The result of the proposed method is evaluated using the following three measures: sensitivity, specificity and accuracy. Sensitivity is defined as
sensitivity = #(true positives) / #(positives).
The specificity is defined as
specificity = #(true negatives) / #(negatives).
The predication accuracy is defined as
accuracy = #(corrected predicted) / #(total samples).
An accuracy of about 70% was achieved in our study using 10 fold cross validation. The sensitivity was found to be about 65% and specificity of about 73%. This may suggest a certain predictive power of the proposed method on this data set. The following step would be replication of similar results on other independent data sets.
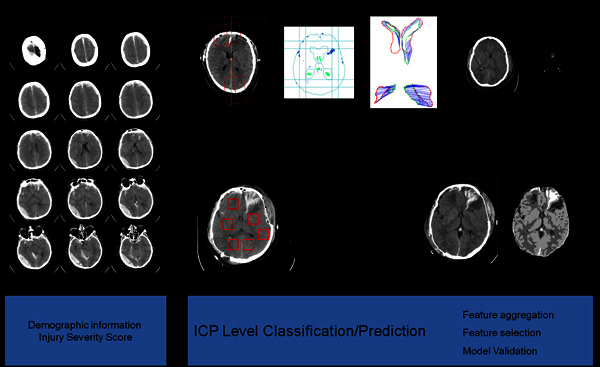 Figure 1. The top level framework of the method. There are three feature extraction modules for the raw CT images: midline shift measurement, texture analysis and blood amount estimation. All the extracted features and other recorded information, such as demographic information are fed into the classification module to predict the ICP levels.
Figure 1. The top level framework of the method. There are three feature extraction modules for the raw CT images: midline shift measurement, texture analysis and blood amount estimation. All the extracted features and other recorded information, such as demographic information are fed into the classification module to predict the ICP levels.
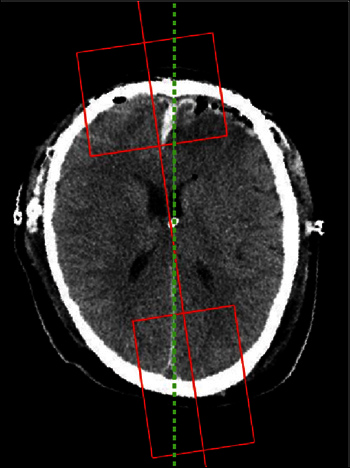 Figure 2. The result of the ideal midline detection. The red line is the approximate ideal midline. The two rectangular boxes cover the bone protrusion and the lower falx cerebri respectively. These boxes are used to reduce the regions of interest. The green dash line is the final detected ideal midline, which captures the bone protrusion and the lower falx cerebri accurately.
Figure 2. The result of the ideal midline detection. The red line is the approximate ideal midline. The two rectangular boxes cover the bone protrusion and the lower falx cerebri respectively. These boxes are used to reduce the regions of interest. The green dash line is the final detected ideal midline, which captures the bone protrusion and the lower falx cerebri accurately.
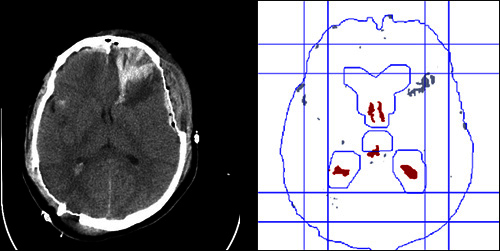 Figure 3. The result of the ventricle segmentation. The left image is the original CT image. The right image consists of the following lines: the interior edge of the skull, the inner bounding box formed by four lines, the outer bounding box formed by another four lines, the edge of the matched enlarged ventricle template, the red regions representing the detected ventricular regions, the gray regions representing other regions rejected being ventricular regions after applying several constraints on recognizing ventricles.
Figure 3. The result of the ventricle segmentation. The left image is the original CT image. The right image consists of the following lines: the interior edge of the skull, the inner bounding box formed by four lines, the outer bounding box formed by another four lines, the edge of the matched enlarged ventricle template, the red regions representing the detected ventricular regions, the gray regions representing other regions rejected being ventricular regions after applying several constraints on recognizing ventricles.
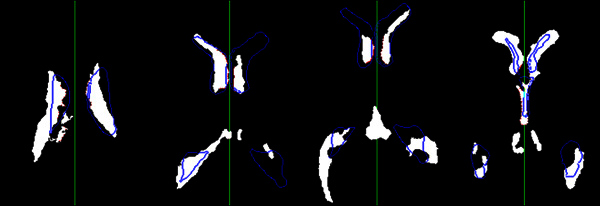 Figure 4. The result of the actual midline estimation. This figure shows the estimation results in different CT images with different ventricular shapes. The white regions are segmented ventricular regions. The blue contours are the edge of the matched ventricular templates. The red points are detected feature points representing the inner edges of the ventricles used to calculate the actual midline. The green line is the final estimated actual midline. Click here to view larger figure.
Figure 4. The result of the actual midline estimation. This figure shows the estimation results in different CT images with different ventricular shapes. The white regions are segmented ventricular regions. The blue contours are the edge of the matched ventricular templates. The red points are detected feature points representing the inner edges of the ventricles used to calculate the actual midline. The green line is the final estimated actual midline. Click here to view larger figure.
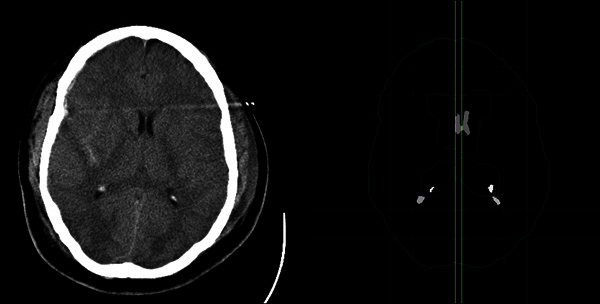 Figure 5. The result of the midline shift estimation. The left image shows the input CT slice. The right image shows the processed result similar to Figure 3. The left vertical green line represents the estimated ideal midline, and the right vertical green line represents the estimated actual midline. The distance between the two lines is the estimated midline shift.
Figure 5. The result of the midline shift estimation. The left image shows the input CT slice. The right image shows the processed result similar to Figure 3. The left vertical green line represents the estimated ideal midline, and the right vertical green line represents the estimated actual midline. The distance between the two lines is the estimated midline shift.
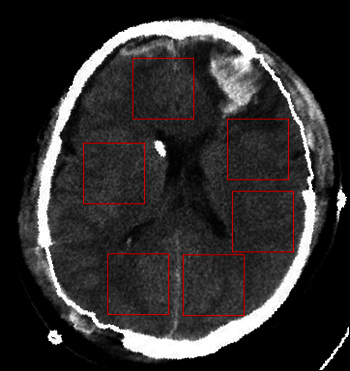 Figure 6. The six selected windows for texture analysis. The selected windows are the red rectangles avoiding the ventricles.
Figure 6. The six selected windows for texture analysis. The selected windows are the red rectangles avoiding the ventricles.
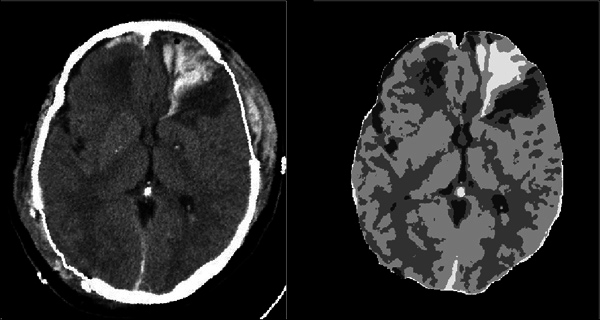 Figure 7. The blood segmentation. The left image is the input CT image. The right image shows the segmentation map produced by the GMM method. The brightest region corresponds to the blood region.
Figure 7. The blood segmentation. The left image is the input CT image. The right image shows the segmentation map produced by the GMM method. The brightest region corresponds to the blood region.
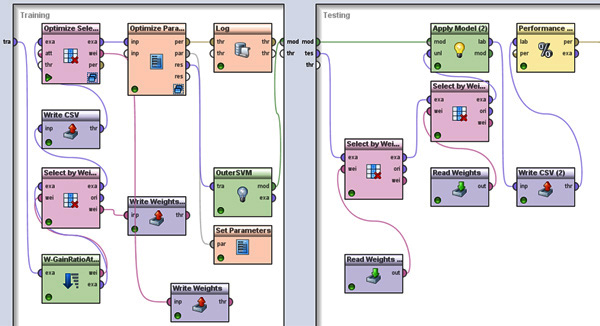 Figure 8. The top level cross-validation in RapidMiner. The left window shows the modules in the training process and the right window shows the modules in the testing process. In the training process, first we use the information gain module and the genetic algorithm module to select features. Then a SVM is used to do classification. The parameter tuning of the SVM is done through a nested cross-validation in the Optimize Parameter process. The final full trained model is the output from the OuterSVM process using all the training data. In the testing module, the selected features are used and the trained model is evaluated. Click here to view larger figure.
Figure 8. The top level cross-validation in RapidMiner. The left window shows the modules in the training process and the right window shows the modules in the testing process. In the training process, first we use the information gain module and the genetic algorithm module to select features. Then a SVM is used to do classification. The parameter tuning of the SVM is done through a nested cross-validation in the Optimize Parameter process. The final full trained model is the output from the OuterSVM process using all the training data. In the testing module, the selected features are used and the trained model is evaluated. Click here to view larger figure.
Discussion
In this study, an intuitive and flexible framework is proposed to address two challenging problems: the estimation of the midline shift in CT images and ICP level prediction based on extracted features. The evaluation results show the effectiveness of the proposed method. As far as we know, this is the first time of a systematic study in addressing these two problems. We notice that based on the general framework, there are many potential improvements that can be achieved. For example, in the proposed segmentation, the low level segmentation and high level recognition are separated and currently there is no feedback from the high level to the low level segmentation. This is different from human visual inspection, which has interactions between the low level vision and high level recognition. One potential approach to combine these two levels together is the so-called "model based low-level segmentation". In this method, the low-level segmentation is guided by the high level atlas models of the target structure. For example, a registration algorithm can be applied in the first step to align the CT images to a standard CT image. This may further improve the accuracy of ventricle recognition as well as the estimation of the midline shift because it may provide more accurate mapping in the brain structures between the CT images and the standard CT image. For the ICP prediction, even though the result is promising based on the dataset tested, we have to notice that the size of the samples is limited. It will be more persuasive to validate the results based on another independent dataset. In the application of machine learning for ICP prediction, the sample size is a very important factor. A larger dataset of CT examinations, which may contain more different patterns in both CT images and ICP signals, may give a more informative evaluation of the proposed framework.
Disclosures
No conflicts of interest declared.
Acknowledgments
The material is based upon work partially supported by the National Science Foundation under Grant No. IIS0758410. The data was supplied by Carolinas Healthcare System.
References
- Langlois JA, Rutland-Brown W, Thomas KE. Traumatic brain injury in the united states: emergency department visits, hospitalizations, and deaths. Atlanta, GA: Centers for Disease Control and Prevention, National Center for Injury Prevention and Control; 2006. [Google Scholar]
- Moore EE, Feliciano DV, Mattox KL. Trauma. 5th. McGraw-Hill Professional; 2003. [Google Scholar]
- Chen W, Smith R, Ji SY, Ward KR, Najarian K. Automated Ventricular Systems Segmentation in Brain CT Images by Combining Low-level Segmentation and High-level Template Matching. BMC Medical Informatics and Decision Making. 2009;9(1) doi: 10.1186/1472-6947-9-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Najarian K, editors. Segmentation of Ventricles in Brain CT Images Using Gaussian Mixture Model Method. 2009 IEEE/ICME International Conference on Complex Medical Engineering (ICME2009); 2009. pp. 15–20. [Google Scholar]
- Chen W, Ward KR, Najarian K, editors. Actual Midline Estimation from Brain CT Scan Using Multiple Regions Shape Matching. International Conference on Pattern Recognition; 2010. pp. 2552–2555. [Google Scholar]
- Chen W, Cockrell C, Ward KR, Najarian K, editors. Intracranial Pressure Level Prediction in Traumatic Brain Injury by Extracting Features from Multiple Sources and Using Machine Learning Methods. IEEE International Conference on Bioinformatics & Biomedicine; 2010. pp. 510–515. [Google Scholar]
- Greenspan H, Ruf A, Goldberger J. Constrained Gaussian mixture model framework for automatic segmentation of MR brain images. IEEE Trans. Med. Imaging. 2006;25(9):1233–1245. doi: 10.1109/tmi.2006.880668. [DOI] [PubMed] [Google Scholar]
- Belongie S, Malik J, Puzicha J. Shape matching and object recognition using shape contexts. IEEE Trans. Pattern Anal. Mach. Intell. 2002;24:509–522. [Google Scholar]
- Weszaka JS, Dyer CR, Rosenfeld A. A comparative study of texture measures for terrain classification. IEEE Trans. on Syst., Man, Cyber. 1976.
- Kingsbury N. Complex wavelets for shift invariant analysis and filtering of signals. Applied and Computational Harmonic Analysis. 2002;10(3):234–253. [Google Scholar]
- Mierswa I, Wurst M, Klinkenberg R, Scholz M, Euler T, editors. Yale: Rapid prototyping for complex data mining tasks. KDD '06: Proceedings of the 12th ACM SIGKDD international conference on Knowledge discovery and data mining; 2006. pp. 935–940. [Google Scholar]


