Abstract
Despite the importance of the immune system in many diseases, there are currently no objective benchmarks of immunological health. In an effort to identifying such markers, we used influenza vaccination in 30 young (20–30 years) and 59 older subjects (60 to >89 years) as models for strong and weak immune responses, respectively, and assayed their serological responses to influenza strains as well as a wide variety of other parameters, including gene expression, antibodies to hemagglutinin peptides, serum cytokines, cell subset phenotypes and in vitro cytokine stimulation. Using machine learning, we identified nine variables that predict the antibody response with 84% accuracy. Two of these variables are involved in apoptosis, which positively associated with the response to vaccination and was confirmed to be a contributor to vaccine responsiveness in mice. The identification of these biomarkers provides new insights into what immune features may be most important for immune health.
Keywords: aging, apoptosis, influenza, systems immunology, vaccinology
Introduction
The role of the immune system in human health is widely recognized. This is particularly evident in aging, where the immune response is often attenuated as observed in the decreased protection following vaccination and high prevalence and severity of infectious diseases in the elderly. Of the current estimated 36 000 influenza deaths in the United States each year, 90% occur in people older than 65 years of age (Thompson et al, 2003), and in this age group influenza alone contributes to 64% of the estimated total economic annual burden of $10.4 billion (Molinari et al, 2007). Moreover, the most common causes of death in the older population, including coronary heart disease, stroke and cancer, may also be due, at least in part, to immune system dysfunction (Hansson, 2005; de Visser et al, 2006).
The changes that occur with age affect both the innate and the adaptive components of the immune system. These include major alterations in cell subset composition, partly due to thymic involution (Fagnoni et al, 2000), defects in antigen presentation, reduced cytotoxic function, a restricted T-cell repertoire and defects in antibody responses (Pawelec and Larbi, 2008; Weiskopf et al, 2009).
Because of the complexity of the immune system, the majority of the studies to date have measured a relatively small number of variables. However, recent advances in technology enable many parameters to be measured quickly and at low cost, allowing much broader immune profiling. This ‘systems immunology’ approach to vaccination has been pioneered in the studies of Sékaly and colleagues (2008) and Pulendran and colleagues (2009) in analyses of yellow fever vaccine, and more recently in the study of Nakaya et al (2011) in influenza vaccination, who used gene expression data from post-vaccination samples to identify differences between responders and non-responders. Here we have used pre-vaccination samples to determine whether it is feasible to find baseline biomarkers from blood that could predict the serological response to the trivalent, inactivated seasonal influenza vaccine (TIV) in advance of its administration. We comprehensively characterized baseline young and older individuals’ blood using whole-genome DNA microarrays, peptide microarrays for detection of influenza-specific antibodies to linear epitopes, multiplexed serum protein assays comprising 50 cytokines and chemokines, and high-resolution immune phenotyping for 15 immune cell subsets. We also measured the antibody responses to TIV and the signaling responses to cytokines in various cell subsets from peripheral blood, using antibodies to phosphorylated STAT proteins, also known as phosphoFlow (pFlow).
A strong negative effect of pre-existing anti-influenza antibodies with hemagglutinin (HA) inhibition (HAI) activity on the fold-change HAI titer following vaccination has been reported in several studies (Beyer et al, 1996; He et al, 2008). We characterized the reactivity of influenza-specific antibodies recognizing HA peptides from baseline serum samples with the aim of (1) investigating the HA linear epitope specificities in pre-vaccine antibodies and (2) finding the minimal set of pre-existing antibodies that could potentially explain the negative effect of baseline HAI titers on the response to the influenza vaccine. We found a set of pre-existing antibodies recognizing viral HA peptides that can distinguish individuals with high versus low pre-vaccine HAI titers. We also identified antibodies against HA peptides that associate with the HAI response to the vaccine. Using a machine learning approach to identify other relevant predictors of vaccine response, we found age-related and -independent variables, which correlated with and predicted the serological response with 84% accuracy. Two of these markers were involved in apoptosis, which was confirmed to be important in influenza vaccine responses in mice.
The immune predictors found here may also be important in the response to other vaccines and help to define metrics of immunological health (Davis, 2008).
Results
Antibody responses to influenza vaccination in young versus older vaccine recipients
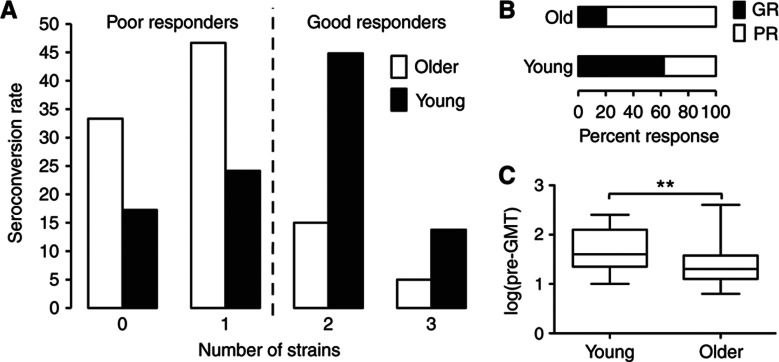
We carried out an influenza vaccine study in 91 young and older ambulatory subjects to better characterize immune parameters from peripheral blood that associate with vaccine responsiveness. Eighty-nine individuals’ serum specimen sets (pre- and post-vaccine) were complete for testing the vaccine response. The number of previous influenza immunizations, history of influenza infection, demographic data and other characteristics of the subjects are depicted in Table I. We measured a comprehensive set of immune parameters in pre-vaccine samples (day 0) (Figure 1) and the antibody responses by the HAI assay at day 0 and 28±7 days after vaccination. We used the HAI assay, because it is a direct measure of antibodies produced in vivo against viral HA proteins in response to infection or vaccination and is generally accepted as the gold standard for gauging the effectiveness of influenza vaccines. Seroconversion (defined as an HAI titer post-/pre-vaccine ⩾4) was computed for each strain in the vaccine. The individuals were classified according to the breadth and magnitude of the response as poor responders (PR), if seroconversion was achieved for one strain or none, or good responders (GR), if seroconversion was achieved for two or all three strains in the vaccine (Figure 2A). By this definition, 80% of young individuals were GRs and only 38% were GRs in the older cohort (Figure 2B). These results are in agreement with several previous studies.
Table 1. Subjects’ baseline characteristics.
| Baseline characteristic | Young | Older |
|---|---|---|
| Age range (median) | 20–30 (24.5) | 61 to >89 (78) |
| Gender | ||
| Male | 16 (53%) | 21 (66%) |
| Female | 14 (47%) | 40 (34%) |
| Ethnicity | ||
| White | 21 (70%) | 59 (97%) |
| Asian | 9 (30%) | 2 (3%) |
| Body mass index (range) | 19–44 (23.1) | 18–47 (25.1) |
| Cytomegalovirus (+) | 57% | 59% |
| Epstein Barr virus (+) | 53% | 67% |
| Previous immunizations | 1–20 (4) | 2–30 (13) |
| Ever diagnosed with Flu | 2 (7%) | 4 (7%) |
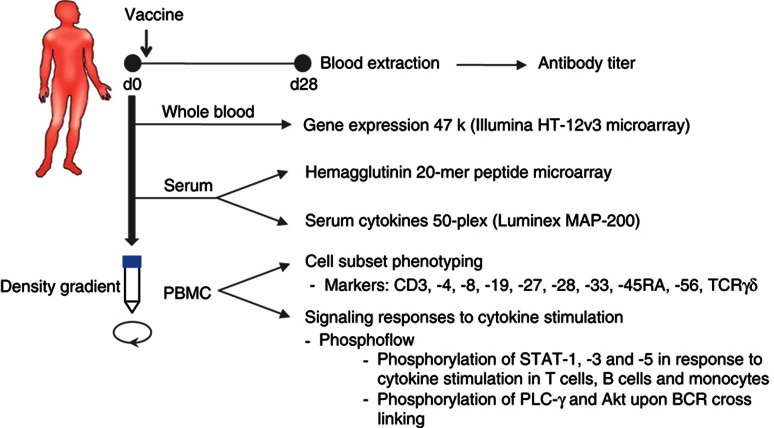
Figure 1.
Study design. Blood samples are obtained before (d0) and 28±7 days (d28) after a single intramuscular inoculation of the seasonal inactivated influenza vaccine. Samples from d0 are used for gene expression analysis, hemagglutinin 20-mer peptide microarrays, determination of serum cytokines and chemokines, cell subset phenotyping and signaling responses to cytokine stimulations on CD4+ and CD8+ T cells, B cells and monocytes, as well as the phosphorylation of PLC-γ and Akt upon BCR crosslinking on B cells. Serum samples from d0 and d28 are utilized for determination of anti-influenza antibody titers by the HAI assay. Measurements from d0 are processed and subjected to computational modeling for prediction of the antibody response.
Figure 2.
Antibody responses to the influenza vaccine in young and older subjects. Serum samples obtained from 89 vaccine recipients before and ∼3 weeks after vaccination are assayed for HAI against each vaccine strain of the influenza virus to determine the antibody response to the vaccine. Seroconversion (⩾4-fold increase) is calculated and the number of strains that subjects seroconverted against is used to define response categories; seroconversion to 0 or 1 strain is considered as a poor response (PR), whereas seroconversion to 2 or all 3 strains is considered as a good response (GR) to the vaccine (A). (B) Percentage of PR and GR for young and older subjects. (C) Pre-vaccine geometric mean titer for all strains in young versus older subjects. **P-value <0.01.
An important feature of the immune response to influenza in humans is the presence of pre-existing HAI antibodies, the titer of which negatively correlates with responsiveness (fold increase) to the vaccine (Beyer et al, 1996; He et al, 2008). Pre-existing flu-specific memory CD4+ T cells seem to have an important role by activating (CD56dim)-NK cells that are able to inhibit antigen presentation by dendritic cells, thereby suppressing the subsequent CD4+ T-cell help to B cells (He et al, 2008). Therefore, we calculated for each subject the pre-vaccine HAI geometric mean titer for all three strains in the vaccine (pre-GMT) and compared pre-vaccine titer in young individuals with older individuals. The older cohort had significantly less pre-GMT than the young cohort (P=0.0017; Figure 2C), which suggests that pre-GMT is a stronger determinant of the vaccine response in young than in older adults. To address this in detail, we compared the pre-GMT levels in PRs versus GRs in young or older individuals. The differences between PRs and GRs were more pronounced for the young group (P=0.005) than for the older group (P=0.039; Supplementary Figure 1).
Pre-existing antibodies to HA peptides and pre-vaccine HAI titer
To characterize the pool of pre-existing HAI antibodies with more precision, we developed peptide microarrays for profiling the antibody repertoire directed against viral HA peptides. These studies were restricted to 76/89 subjects because of limitations in the availability of serum from some subjects. Our peptide microarray technology is able to cover ∼40% of the HA protein sequence for each vaccine strain. Thus, to maximize the number of positive hits, we selected an HA region found in an H5N1 influenza strain containing epitopes for neutralizing antibodies against different influenza strains (broadly neutralizing antibodies; Khurana et al, 2009).
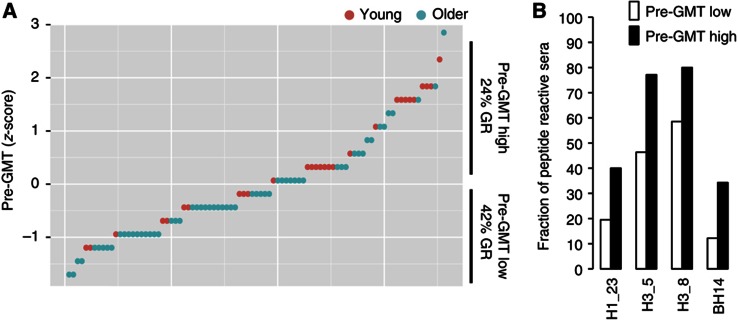
To determine whether the reactivity against these HA peptides at baseline was associated with the presence of pre-existing HAI antibodies, we compared individuals with high versus low pre-GMT (above versus below the group average, respectively). As anticipated, there were significantly more GR within the subjects with low pre-GMT compared with those with high pre-GMT (P<0.01 by Fisher’s exact test; Figure 3A). We identified four peptides to which serum reactivity was significantly more prevalent in individuals with high compared with low pre-GMT (P<0.05, false discovery rate (FDR) Q<0.1; Figure 3B). This result indicated that individuals with a high pre-vaccine HAI titer, associated with poor responses here and elsewhere, are more likely to have antibodies directed against peptides H1_23, H3_5, H3_8 and BH14 corresponding to potential linear HA epitopes from each of the virus strains in the vaccine.
Figure 3.
Pre-existing antibodies to HA peptides that correlate with pre-vaccine HAI antibody titers (pre-GMT). Individuals were sorted by pre-GMT levels and divided into high or low pre-GMT (A) and reactivities against HA peptides were compared (B). A significantly higher fraction of samples reactive to peptides H1_23, H3_5, H3_8 and BH14 were found in individuals with high pre-GMT compared with low pre-GMT (P<0.05 by Student’s t-test). Red circles=young, green circles=older.
Pre-existing antibodies to HA peptides and prediction of the HAI response
We next addressed the possibility that the pre-existing antibodies to HA peptides were also associated with and could predict the HAI titer in response to the vaccine. To do so, we used the ‘elastic net’ (Friedman et al, 2010) with fivefold cross validation, a machine learning procedure that enables the discovery of relevant features from high-dimensional data and estimates the performance of the resulting prediction model, such that the most informative factors are selected out of the pool of available parameters in an automated manner, to generate the model with the lowest error. In the fivefold cross validation, the samples are randomly partitioned into five sets, of which four sets are used to ‘train’ the algorithm and blindly predict the outcome of the fifth set (test set). This process is repeated iteratively five times until all sets are tested. Then a cross-validated area under the ROC curve (cvAUROC) can be computed to assess the goodness of the model without the known problem of overfitting.
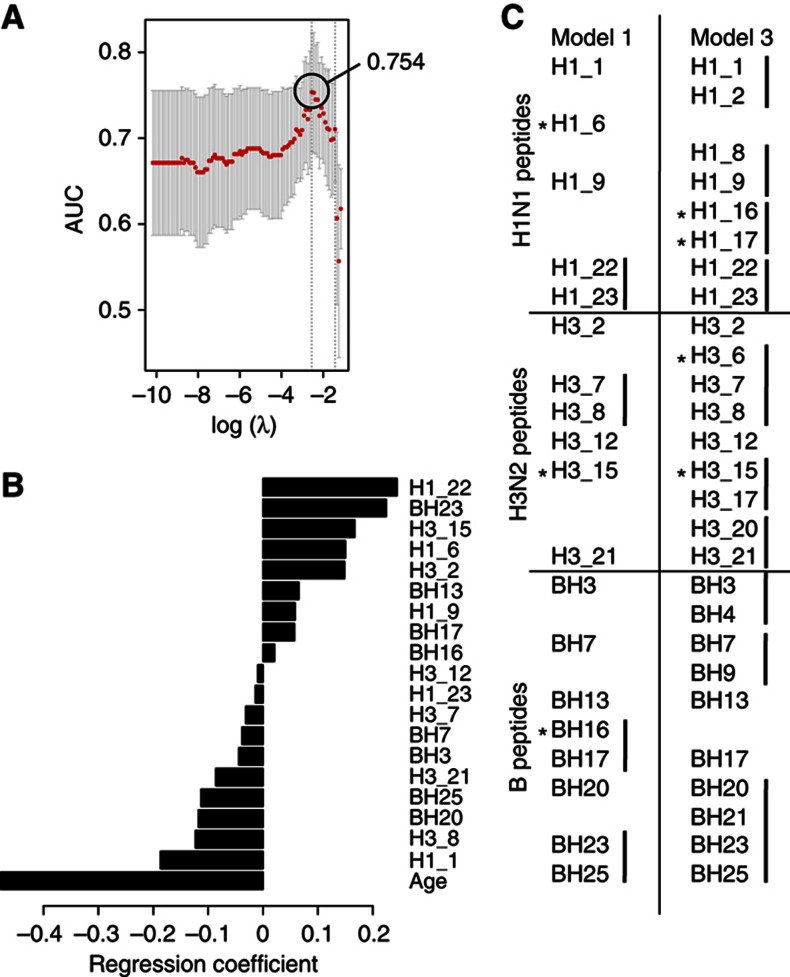
By this procedure, we built a first model (Model 1) that included age, and identified a total of 19 potential epitopes recognized by pre-vaccine antibodies, resulting in an cvAUROC=0.754 (Figure 4A), which is significantly better than the model including age alone (Model 2), which yielded an cvAUROC=0.686 (P<0.0001 by likelihood-ratio test). Not surprisingly, in Model 1 age had the largest regression coefficient (contribution to the model; Figure 4B). Of note, two of the peptide predictors identified in Model 1 (H1_23, NH2-FALSRGFGSGIINSNAPMD-COOH and H3_8, NH2-SSGTLEFNNESFNWTGVTQ-COOH) were significantly more prevalent in subjects with high pre-GMT (Figure 3B) and neither of these peptide predictors were age-associated (see below). Thus, we identified two HA peptides that contribute to distinguish individuals with a high versus low pre-vaccine HAI titer and to which high antibody levels are negatively associated with the HAI activity in response to influenza vaccination.
Figure 4.
Hemagglutinin peptides targeted by pre-existing antibodies that predict the antibody response to the influenza vaccine. Pre-vaccine serum reactivity to HA peptides for each of the three strains was subjected to feature selection and prediction of the HAI response. The crossvalidated area under the ROC curve (cvAUROC) (A) and regression coefficients for each feature (B) from Model 1 were obtained from the cross-validation procedure. Excluding age from the learning procedure results in incorporation of adjacent peptide reactivities to Model 3 (C); however, the accuracy of the prediction drops to 0.711. Asterisks in (C) indicate significant age associations.
We then tested the predictability and changes in the selection of predictors when age is removed from the model (Model 3) and only peptide variables are used to predict the HAI response. By performing this procedure, 26 peptides were selected, yielding a cvAUROC=0.711, only moderately better than the model where age is used as the only predictor (P<0.001, by likelihood-ratio test). Intriguingly, pre-vaccine reactivity to several sets of adjacent peptides was computationally selected for the prediction model when the contribution of age was removed (Figure 4C). The fact that clusters of adjacent peptides (both by sequence and within the three-dimensional structure of HA) were selected in these models highlights potential HA epitopes important for inhibiting the binding of influenza virus to cells.
Age-related pre-existing antibodies against HA peptides
To investigate whether reactivities to HA peptides were different in young versus older individuals, we performed simple regression analysis on each peptide variable and age, and corrected for multiple testing using permutation tests (Benjamini and Hochberg, 1995). Older subjects showed differences in the pre-existing reactivity to 11/72 peptides at an FDR<20% (P <0.05, FDR Q<0.2; Supplementary Table 1); three of which were selected as predictors of vaccine response in Model 1 and four in Model 3 (Figure 4C, asterisks).
These findings demonstrate that the levels of some pre-existing antibodies targeting viral HA peptides correlate with pre-vaccine HAI titers and with the magnitude of the HAI antibody response to each of the three strains in TIV. The fact that the antibody levels to some of these peptides are lower in older subjects might reflect differences in influenza strain exposure or changes in their antibody repertoire with age (Yager et al, 2008).
Baseline parameters and prediction of the HAI response: Models 1 and 2
We next tested the hypothesis that the baseline levels of immune parameters and subjects’ characteristics are indicative of an ability to respond to vaccination. To do so, we first generated gene modules from whole-blood gene expression data and estimated their corresponding regulatory programs using a method similar to what has previously been described (Segal et al, 2003). Of a total of 48 771 gene probes in the microarray per sample, we derived 109 gene modules and their corresponding regulatory programs (Supplementary Table 2 and Methods). In addition to these gene modules, we also explored other immune parameters, including serum proteins, immune cell subsets, signaling events and the subjects’ characteristics, such as demographics and CMV and EBV serological status. A total of 241 variables were used for feature selection and prediction of the antibody response. As with the peptide microarray data, we performed cross validation using the elastic net algorithm (Friedman et al, 2010), which is well suited for scenarios having more features than samples.
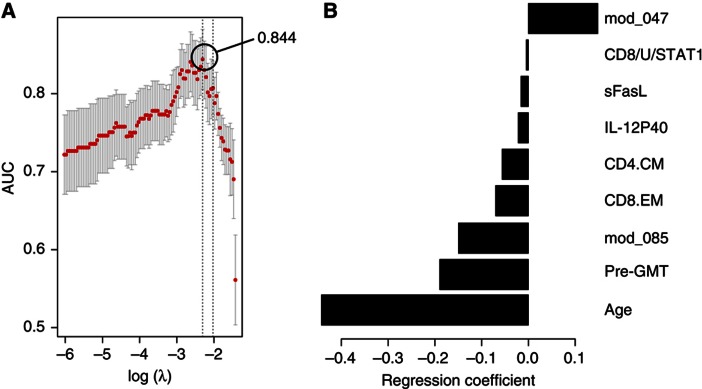
We first generated two models as with the peptide array data: Model 1 (including all features) that selected nine relevant features, which correlated with the HAI response and resulted in a cvAUROC=0.844 (Figure 5A), and Model 2 in which the immune features are excluded and only age is used to predict the HAI response. The latter model with age alone predicted the HAI response with an accuracy of cvAUROC=0.694, which is significantly less accurate than our full model (P=0.000012 by likelihood-ratio test).
Figure 5.
Baseline blood measurements that predict the antibody response to the influenza vaccine. Diverse measurements from pre-vaccine samples were used for feature selection and prediction of the HAI response. The cross-validated area under the ROC curve (cvAUROC) for Model 1 (A) and regression coefficients for each feature (B) were obtained from the cross-validation procedure. mod_047, gene module APO; CD8/U/STAT1, pSTAT1 in unstimulated CD8+ T cells; CD4.CM, central-memory CD4+ T cells; CD8.EM, effector-memory CD8+ T cells; mod_085, gene module PROL; pre-GMT, geometric mean titer for all three virus strains.
Only one of the nine features identified in Model 1 (gene module 47) had a positive regression coefficient (Figure 5B). This gene module is enriched in genes involved in apoptosis (P<0.01; module ‘APO’), including (1) GSTP1, which suppresses the anti-apoptotic protein JNK (Gate et al, 2004; Yu et al, 2004), (2) the STAT4 inhibitor PIAS4, which stimulates transcriptional activity of p53 and increases Rb-dependent corepression through recruitment to E2F-responsive promoter (Bischof et al, 2006; Zhang et al, 2004), (3) IL17D, which acts by downregulating T cell-mediated immune hypereactivity and proinflammatory cytokine production (Hamano et al, 2003; Villarino et al, 2003; Owaki et al, 2008; Yoshimoto et al, 2008) and (4) ZNF-148 (also known as ZBP-89), which encodes for a protein that binds to p53 and p21 (waf1), enhancing their transcriptional activity (Bai and Merchant, 2001; Zhang et al, 2010). Our regulatory program analysis indicated that the gene module APO is likely repressed by IRF9, XBP-1 and Bcl-3, among other transcription factors (Supplementary Figure 2) that promote cell proliferation and survival in diverse cell types, and negatively associate with apoptosis (Takaoka et al, 2003; Tsuno et al, 2009; Thorpe and Schwarze, 2010). Of note, XBP-1 also controls expression of IL-6, which in turn, promotes plasma cell growth and survival (Iwakoshi et al, 2003).
Age and pre-GMT were selected as negative predictors of the HAI response in Model 1, which is consistent with previous reports (Beyer et al, 1996, 1999; Beyer et al, 2004; He et al, 2008). Gene module 85 had a negative regression coefficient as well, and is composed of genes associated with the regulation of B-cell proliferation (P<0.0001; module ‘PROL’), such as CDK6, BCS1L, PBX4, CD320 and CD40L. The regulatory program of the PROL module indicated that it is likely repressed by TSC-22d1 (TGF-stimulated clone 22), implicated in the TGF-mediated apoptosis (Uchida et al, 2000) and the androgen receptor (AR) coregulator ARA70 (NCOA4), which is known to be repressed by Akt and has been found to act as a tumor suppressor by inducing AR-dependent apoptosis (Ligr et al, 2010). Activators of this gene module include the following: (1) PHB2, an anti-apoptotic factor upregulated by Myc (O'Connell et al, 2003) (2) ID3, a target of Blimp-1 and essential for B-cell proliferation (Pan et al, 1999) and (3) STAT4. Of note, PIAS4, a suppressor of STAT4, was found in the APO module (Supplementary Table 2), a positive predictor of vaccine response (see above).
This suggested that upregulation of genes in the APO module and the concomitant downregulation of genes found in the PROL module is beneficial for cell renewal and optimal antibody production in response to immunization, which is consistent with the notion that the limited niche availability restricts the number of cells to respond to antigenic challenges (Discussion and Franceschi et al, 2000). Indeed, interesting data from Xiang et al (2007) demonstrated that crosslinking the IgG receptor FcγRII induces apoptosis of bone marrow plasma cells in mice and of plasmablasts in humans likely contributing to cell turnover (Xiang et al, 2007). This prompted us to compare the expression of the FcγRIIB gene (FCGR2B) in PRs versus GRs. However, there was not sufficient variation in the expression levels of FCGR2B for its selection and gene module construction (Methods). Therefore, we focused on FcγRIIA (FCGR2A), as in mice FcγRIIB and FcγRIIIA exert similar pro-apoptotic characteristics (Fang et al, 2012), and mouse FcγRIIIA is predicted to be the counterpart of human FcγRIIA in terms of functionality and sequence similarity in the extracellular portion.
Strikingly, FCGR2A clustered with a number of genes participating in cell apoptosis in module 54 (Supplementary Table 2), with differential expression between PRs and GRs (Supplementary Figure 3) and selected for prediction of the HAI response in Model 3 (see below). This suggests that diverse pathways implicated in apoptosis must function properly for a high cell turnover and the establishment of new antibody-producing cells.
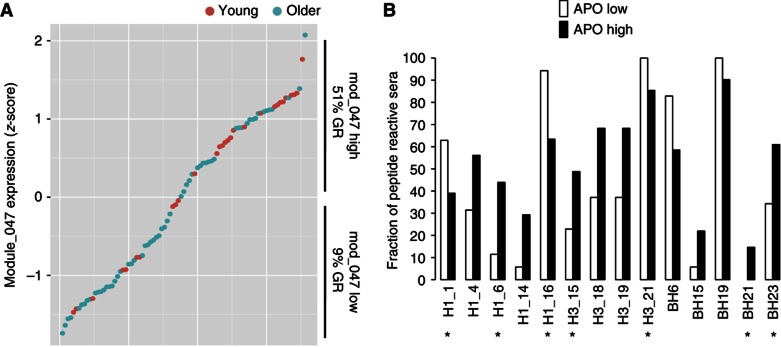
To determine whether apoptosis genes were also linked to the presence of pre-existing antibodies to the potential linear epitopes identified using the peptide array technology, we divided the subjects according to their gene expression levels of the module APO into high or low, if they were above or below the group average, respectively (Figure 6A). As anticipated, the fraction of GRs was much higher in subjects with high expression of genes in the module APO (P=0.00007). Significant differences were also observed in many of the pre-existing antibodies to linear determinants (P<0.05, FDR Q<0.1; Figure 6B). Of note, some of these antibodies (Figure 6B, asterisks) were identified to be predictive of the HAI response in our previous analyses (Figure 4).
Figure 6.
Pre-existing antibodies to HA peptides that correlate with expression of apoptosis module. Individuals were sorted and divided by expression levels of genes in the APO module (A) and reactivity to HA peptides was compared between individuals with high or low gene expression (B). Various HA peptides’ reactivities were significantly different between the APO high and low groups. Peptides H1_1, H1_6, H1_16, H3_15, H3_21, BH21 and BH23 (asterisks) were found to be predictive of the HAI titer in response to the vaccine. Red circles=young, Green circles=older.
For serum cytokines, we found that soluble Fas ligand (sFasL) and IL-12p40 were negative predictors (Figure 5B). sFasL prevents membrane-bound FasL from binding to Fas, thereby decreasing susceptibility to apoptosis (Knox et al, 2003). This strengthens our previous findings and indicates that the presence of soluble apoptosis inhibitors could affect the antibody response to vaccination.
Additional immune features that were selected in Model 1 were the frequency of central-memory CD4+ and effector-memory CD8+ T cells, and the baseline STAT1 levels in CD8+ T cells, which are elevated in the elderly and negatively correlate with the pSTAT response to diverse cytokine stimuli.
Baseline parameters and prediction of the HAI response: Model 3
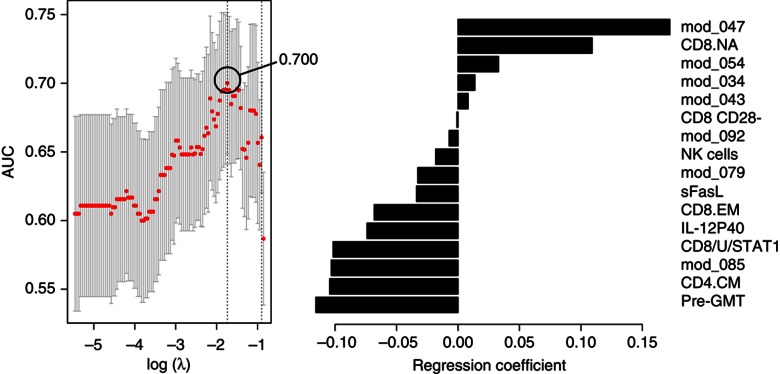
We next tested the possibility that new immune features may emerge from the classification model when age is excluded from the list of candidate predictors (Model 3). Indeed, excluding age resulted in the identification of a new set of features in addition to those previously discovered (Figure 7A), and yielded a cvAUROC=0.700, which is similar to Model 2 (age alone). Model 3 included four positive and four negative additional associations (Figure 7A and B). The positive predictors were the CD8+ naive T-cell frequency, gene module 34 (cell-to-cell signaling and interaction, P<0.01), gene module 43 (RNA post-transcriptional modification, P<0.01) and gene module 54 (cell death by apoptosis, P<0.01). The negative predictors incorporated in Model 3 were the frequency of CD8+ CD28− cells and of NK cells, and modules 79 and 92 both enriched for genes associated with metabolism of carbohydrates (P<0.01). In addition to age and pre-GMT, the frequency of CD8+ CD28− cells was the only feature identified in previous studies to associate with poor vaccine response in the elderly (Goronzy et al, 2001).
Figure 7.
Immune features from Model 3 that associate with and predict the HAI response to TIV. The cross-validated area under the ROC curve (cvAUROC) for Model 3 is depicted in A and the selected immune features that associate with and predict the HAI response to TIV in B. mod_047, gene module APO; CD8/U/STAT1, pSTAT1 in unstimulated CD8+ T cells; CD4.CM, central-memory CD4+ T cells; CD8.EM, effector-memory CD8+ T cells; mod_085, gene module PROL; pre-GMT, geometric mean titer for all three virus strains; CD8.NA, naive CD8 T cell frequency; mod_054, gene module 54 (cell death by apoptosis; P<0.01); mod_034, gene module 34 (cell-to-cell signaling and interaction; P<0.01); mod_043, gene module 43 (RNA post-transcriptional modification; P<0.01); CD8 CD28-, CD8+ CD28− cell frequency; NK cells, NK cell frequency; mod_079 and mod_092, gene modules 79 and 92 (metabolism of carbohydrates (P<0.01)).
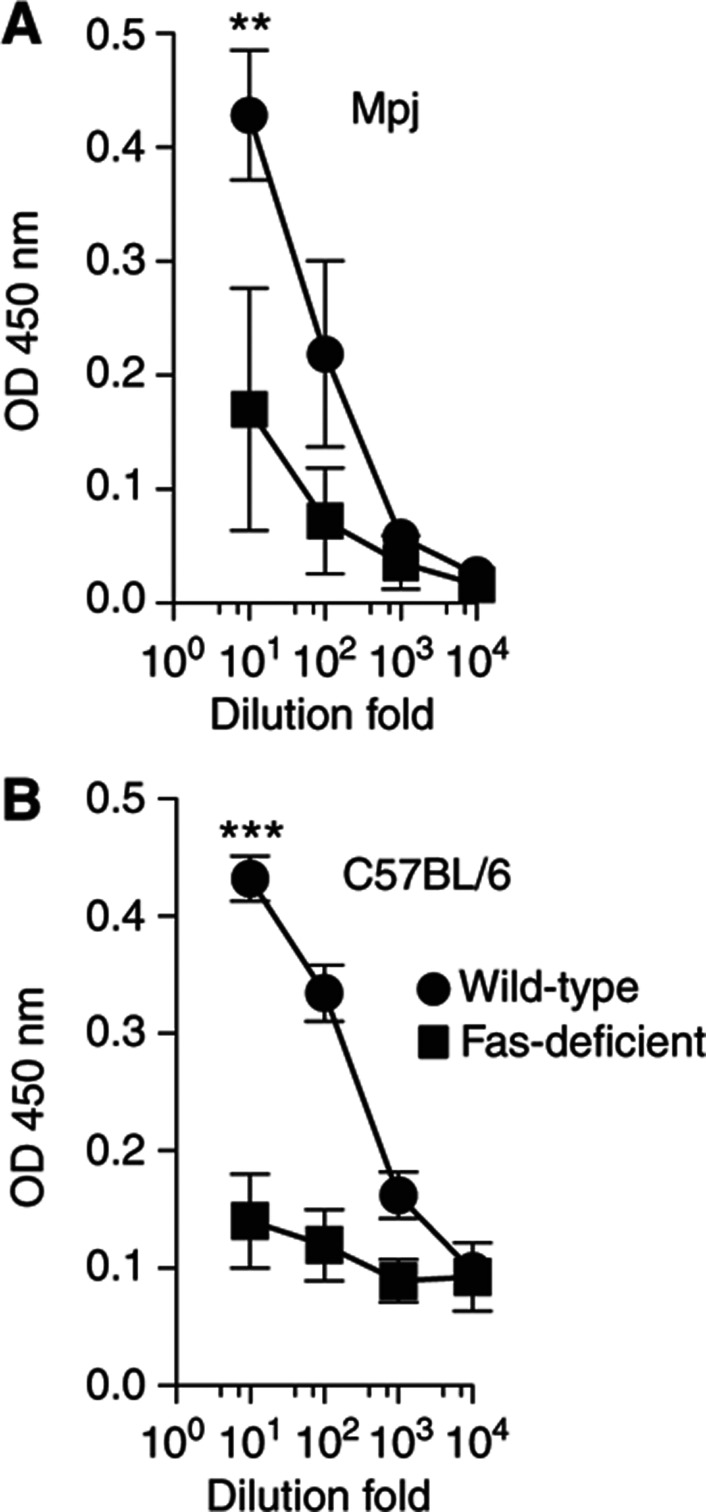
The fact that three apoptosis-related features (module 47, module 54 and sFasL) correlated with a strong vaccine response motivated us to investigate these findings in a mouse model. Thus, we tested two strains of mice lacking CD95 (Fas); MRL/Mpj-Faslpr/J) and B6.MRL-Faslpr/J. In addition to the lack of Fas, plasma cells from these mice do not express FcγRIIB and, therefore, they are also protected from FcγRII-mediated apoptosis (Xiang et al, 2007). After vaccination with TIV, mice lacking Fas on either genetic background showed a significant reduction in their ability to produce influenza-specific antibodies (P<0.05 by t-test) (Figure 8A and B), which suggests a critical role of the apoptosis system in the antibody response in vivo.
Figure 8.
Antibody responses to vaccination in apoptosis-deficient mice. Fas-deficient (squares) or control (circles) mice were vaccinated with a single intramuscular injection of the seasonal trivalent inactivated influenza vaccine and specific anti-vaccine IgG levels were measured by ELISA before and ∼4 weeks after vaccination. Results from four MRL/Mpj-Faslpr/J (Mpj/lpr) and four MRL/MpJ (Mpj) mice (A); and from six B6.MRL-Faslpr/J (B6/lpr) and three C57BL/6 (B6) (B) mice are shown. **P=0.006, ***P=0.0001 by Student’s t-test.
Age-dependent and independent predictors of vaccine response
It has been well established that many older individuals fail to generate protective antibody titers following vaccination with TIV. Therefore, given our model for feature selection where features are not necessarily orthogonal, i.e., they can correlate with one another (Methods); some of the predictors could be correlated with age. To identify age-dependent and -independent features in our data, for each predictor we performed a multiple regression analysis on all samples by age, gender and CMV status (Supplementary Table 3). We used an FDR lower than 5% (P<0.05; FDR Q<0.05) as a cut-off value for significance. Of the 16 immune predictors selected in Model 3, only 2 were not associated with age (Supplementary Figure 3, asterisks). These are the levels of sFasL and expression of the PROL module (module 85). The elderly expressed low levels of positive predictors and high levels of negative predictors (Supplementary Table 3), with the exception of pre-GMT, which were reduced in older compared with young subjects. This suggests that the effect of age on the antibody response overcomes the effect of pre-existing immunity to the vaccine.
Discussion
In this study, we comprehensively characterized the immune system of young and older subjects across multiple technological platforms and identified new candidate traits that associate with and partially predict the humoral response to vaccination in both a young and an old cohort.
We first characterized the reactivity of influenza-specific antibodies recognizing HA peptides from baseline serum samples with the aims of (1) examining the HA-linear epitope specificities in the pre-vaccine antibodies and (2) finding the minimal set of pre-existing antibodies that could potentially explain the negative effect of baseline HAI titers on the response to the influenza vaccine observed here and elsewhere (Beyer et al, 1996; He et al, 2008). We found a set of four peptides, the reactivities of which correlated with the pre-vaccination HAI titer. In two of these, robust activity negatively correlated with the HAI response to influenza vaccination. Thus, our system can rapidly identify GRs from PRs based on their pre-vaccination antibody repertoire against HA protein regions with known neutralization activity (Khurana et al, 2009), as conserved sequences are characteristic for neutralization epitopes on HA across different influenza virus strains (Yamashita et al, 2010). The negative effect of pre-existing antibodies on the response to vaccination has been suggested to be due to pre-existing flu-specific memory CD4+ T cells that inhibit antigen-presentation by dendritic cells, thereby suppressing the subsequent CD4+ T-cell help to B cells (He et al, 2008). Therefore, it is possible that such memory CD4+ T cells are able to maintain a background T-cell-dependent antibody production represented by the reactivities against the linear epitopes we found here. Further studies are needed to clarify the mechanisms underlying these observations.
We identified both negative and positive peptide predictors of the HAI response, many of which were age-associated. These may partially explain the overall poor response observed in aging. However, neither of the reactivities to peptides associated with pre-vaccine HAI changed with age, although the levels of baseline HAI titers were lower in older subjects. This could be due to the limitations of the assay that (1) does not detect conformational epitopes and (2) does not allow for the identification of antibodies that depend on glycosylation for binding. Several previous studies have compared the pre-GMT (baseline titers) with influenza in elderly versus young individuals with no general consensus (Clark et al, 2009; Vajo et al, 2010; Chen et al, 2011; Ehrlich et al, 2012). These discrepancies have been examined in detail by Sasaki et al (2011) who used plasmablast-derived polyclonal antibodies and found greater influenza-specific broad reactivity of antibodies from older versus young vaccine recipients. Thus, the pre-GMT in the elderly is more dependent on their previous exposure history than that of young subjects. This is relevant for vaccinology, as it suggests differential strategies in young versus older subjects based on the historical findings of circulating strains.
Additional predictors of vaccine response included various immune features connected with apoptosis function. Consistent with this, apoptosis-deficient (lpr) mice exhibited poor serologic responses to the influenza vaccine. It is known that the lpr mice have defects in T-cell and B-cell development. Thus, our results might not be only due to apoptosis defects in these mice (although this is suggested by the apoptosis gene modules and sFasL), but these other factors. However, there is no agreement as to whether the germinal center (GC) formation and B-cell responses in these mice are normal (Smith et al, 1995; Takahashi et al, 2001). In particular, our results argue against a previous study that reported memory and antibody-forming cell populations appear to be normal in lpr mice (Smith et al, 1995). However, more recent studies have shown that these mice have defects in clonal selection and the establishment of the memory B-cell repertoire (Takahashi et al, 2001).
Apoptosis has a fundamental role in lymphocyte development and in the termination of the immune response. In the GC reaction, an optimal apoptotic machinery ensures the survival of only high-affinity plasmablasts (Smith et al, 2000; Takahashi et al, 1999), which we found to decrease with age in a recent study (Sasaki et al, 2011). In addition, a functional Fas-mediated pro-apoptotic program is required for clearance of reactive T and B cells after an immune response takes place (Nagata, 1999), as well as for augmentation and maintenance of naive cells, which ultimately results in an optimal balance between the naive and memory cell pools (Zhou et al, 1995). When an immune response ends, the expanded clones of effector cells must be reduced in size. This allows the immune system to cope with new influenza virus challenges, such as those originating from antigenic drift and/or found in a new influenza vaccine preparation. Thus, the appropriate regulation of apoptosis and rapid removal of reactive memory cells may improve immune responses to newly encountered antigenic challenges as observed in the study by Haynes et al (2005), in which new functional CD4+ naive T cells developed after CD4+ T-cell depletion in aged and young mice, restoring optimal responses to antigen (Haynes et al, 2005). This has a number of implications for vaccination and suggests possible ways to overcome, at least partially, the immune system defects observed here. This would also suggest competition and a restricted immunological niche largely occupied by memory cells that, unless cross-reactive, cannot respond optimally to novel antigens.
We also find an age-dependent reduction in the gene modules associated with apoptosis. However, at the level of the serum cytokine sFasL, there was no difference between young and older subjects. This indicates that aging results in the accumulation of factors contributing to less cell renewal and complements previous observations of decreased susceptibility to apoptosis in cells from older subjects (Zhou et al, 1995; Salvioli et al, 2001; Hsu and Mountz, 2010). Indeed, Hsu et al (2006) showed that T cells from nonagenarians have much higher levels of Fas and increased activation-induced cell death than subjects 65 to 89 years old (Hsu et al, 2006), suggesting that apoptosis is beneficial in maintaining good immune health and attaining a longer life span. Although the role of apoptosis in the maintenance of immune tolerance has been well established, to the best of our knowledge, its association with a robust antibody response to a vaccine has not been shown previously.
The accumulation of memory cells in those older subjects who do not respond to vaccination could also be due to chronic antigenic stimulation. Thus, it has been postulated that the constant activation of clonal T-cell populations may lead to increasing numbers of memory cells filling the immunological space and compromising responses to newly encountered antigens (De Martinis et al, 2005). Our results indicate that the increasing size of the memory compartment in older individuals that do not respond well could result from a loss of pro-apoptotic activity and increased levels of cell survival factors. Indeed, recent investigations have shown that memory CD4+ T cells are more resistant to apoptosis than naive CD4+ T cells (Grayson et al, 2002; Jaleco et al, 2003) and, at the gene expression level, memory CD4+ T cells express lower levels of pro-apoptotic-related genes and higher levels of cell survival and proliferation-related genes (Liu et al, 2001).
Our study also identifies a link between cell survival and proliferation, and defective antibody responses. We found that the CD40L gene clustered in a module enriched for proliferation genes (module PROL), which is a negative indicator of the vaccine response. Although molecules in the gene module PROL (e.g., CD40L, CD320) are critical for plasmablast growth and survival, results from different studies suggest that elevated proliferation signals from these molecules have a detrimental effect. For example, heightened CD40 signaling causes B cells to shunt into an extrafollicular plasma cell fate and this prevents the generation of long-lived bone-marrow plasma cells. This has consequences for the B-cell response, including the premature termination of the humoral immunity and the disruption of GC formation in vivo (Erickson et al, 2002). Therefore, it is appealing to hypothesize that weaker responses to the vaccine in individuals with augmented CD40L expression result from the preferential generation of extrafollicullar plasma cells that compromise the accumulation of somatic mutations in the GC B cells, dampening late-appearing high-affinity antibodies, as demonstrated in transgenic CD40L mice (Kishi et al, 2010). An interesting finding in support of this hypothesis is the impairment in GC formation found in old mice with poor responses to vaccine (Eaton et al, 2004). Further studies in humans could potentially address this.
Our results also suggest the presence of other genes in the PROL module with similar functions to that of CD40L, which could contribute to the diminished antibody responses. For example, the gene CD320 has been shown to participate in GC differentiation by directing B cells to mature into plasma cells (Zhang et al, 2001).
The large variation in the responses to vaccines in humans creates a number of challenges. We have utilized a ‘systems biology’ approach that allows us to embrace this variation and to integrate diverse measurements in the same individuals to generate hypothesis on how a system functions. Systems biology approaches, which have been extensively applied in the study of metabolism networks and genetics, have only recently been utilized in vaccinology (Gaucher et al, 2008; Querec et al, 2009; Nakaya et al, 2011). For instance, a recent study by Nakaya et al (2011) has been successful in finding features of the innate immune response, 3 and 7 days after influenza vaccination, which partially predict the subsequent antibody response in young, healthy adults. One of the genes the authors found in their predictive signatures and confirmed in a mouse model, encoding for the Ca2+/calmodulin kinase IV (CaMKIV), was inversely correlated with the HAI response to the influenza vaccine. This kinase has been implicated in the regulation of CRE-dependent transcription (Sato et al, 2006) and, consistent with our findings, a recent study has shown the requirement of CaMKIV for the accumulation of the anti-apoptotic molecules Bcl-2 and Bcl-x and the survival of DCs (Illario et al, 2008). Despite the fact that we did not find CaMKIV in our predictive gene modules, we hypothesize that CaMKIV could also prevent apoptosis in other immune cells, resulting in an overall low cell renewal and poor immune response. The study of Nakaya et al (2011) is therefore complementary to the work described here, which identifies biomarkers before vaccination in different compartments of the human immune system, and across age groups. Being able to predict whether or not an individual will respond productively to a given vaccine is important clinically, because if the prognosis is poor, one might choose not to vaccinate or choose a more potent formulation. For example, in seasonal influenza vaccines, there is a 4 × formulation that has been reported to be more efficacious than the standard one used here (DiazGranados et al, 2013), and adjuvanted influenza vaccines, although not yet licensed in the United States, are available in many other countries.
In summary, we have used immune aging as a model for impaired immunity and identified biomarkers that point to what factors have the greatest role in the response to a seasonal influenza vaccine, now recommended annually for all persons older than 6 months of age with few exceptions. An important feature of our approach is that each individual is profiled for many facets of the immune system. This enables us to observe within single individuals or across groups of individuals the relationship between multiple factors as well as the effects these factors have on the immune response. Ultimately, this methodology, applied to further studies of influenza or other vaccines and infections, should enable us to gain a more complete understanding of immune system function and dysfunction, as well as identifying key variables and mechanisms of immunological health (Davis, 2008).
Materials and methods
Subjects and sample collection
Ninety-one healthy donors (ages 20 to >89 years) were enrolled in an influenza-vaccine study at the Stanford-LPCH Vaccine Program during the fall of 2008 of which eighty-nine completed the study. The protocol of this study was approved by the Institutional Review Board of the Research Compliance Office at Stanford University. Informed consent was obtained from all subjects in the study. All individuals were ambulatory and generally healthy as determined by clinical records. Females of childbearing potential were tested for pregnancy by a urine sample. Volunteers had no acute systemic or serious concurrent illness, no history of immunodeficiency, nor any known or suspected impairment of immunologic function, including clinically significant liver disease, diabetes mellitus treated with insulin, moderate to severe renal disease, blood pressure >150/95 at screening, chronic hepatitis B or C, and recent or current use of immunosuppressive medication. In addition, none of the volunteers were recipients or donors of blood or blood products within the past 6 months and 6 weeks, respectively, nor showed any signs of febrile illness on the day of enrollment and baseline blood draw. A total of ∼40 ml per visit peripheral blood were obtained at day 0 (pre-vaccine) and 28±7 days after receiving a single intramuscular dose of trivalent seasonal influenza vaccine Fluzone (Sanofi Pasteur). Each dose of the vaccine contained 15 μg HA each of H1N1, H3N2 and B strains of the virus. Whole blood was used for gene expression analysis (below). Peripheral blood mononuclear cells (PBMCs) were obtained by density gradient centrifugation (Ficoll-Paque) and frozen at −80°C for 24–48 h before transferring to LN2. Serum was separated by centrifugation of clotted blood and was stored at −80°C before use. Whole blood, PBMCs or serum from the first visit (baseline: day 0) were processed and used for determination of gene expression, leukocyte subset frequency, signaling responses to stimulation, serum cytokine and chemokine levels, and CMV and EBV serostatus by ELISA (Calbiotech, San Diego, CA). Serum samples from day 0 and day ∼28 were used for HAI titer determination.
HA microarray design and fabrication
We printed streptavidin-surface glass slides (Arrayit, Sunnyvale, CA) with biotinylated 19-mer peptides (Sigma Aldrich, St Louis, MO) at 0.5 mg/ml in PBS plus 2.5% glycerol (Sigma) in triplicate using a VersArray ChipWriter Compact robotic microarrayer and ChipWriter Pro software (BioRad). We then blocked printed peptide arrays with biotin at 0.1 mg/ml (Sigma) in PBS for 10 min at room temperature (RT) with 500 r.p.m. orbital agitation. After several washes with peptide-binding buffer (PBB): 50 mM Tris pH 7.5, 150 mM NaCl, 0.05% NP-40 plus 2.5% FCS, we incubated the arrays with the subject’s sera diluted in PBB. Following primary incubation for 2.5 h at RT with light rocking agitation, we rinsed the arrays with PBB, then with PBST (PBS, pH 7.5 with 0.05% Tween-20), before incubating with goat anti-mouse IgG or goat anti-human IgG conjugated to Cy3 (Jackson ImmunoResearch) diluted to 1.5 μg/ml in PBST plus 20% FCS for 30 min at RT. We then rinsed arrays several times in PBST, then with PBS and finally with diH20, and dried arrays by centrifugation at 300 g for 5 min at RT. We immediately scanned processed microarrays with an Axon digital scanner and analyzed scanned images with Genepix Pro 6.1 software (Molecular Devices, Sunnyvale, CA).
Whole-blood microarray analysis of gene expression
Total RNA was extracted from PAXgene blood RNA tubes (PreAnalytiX, USA) using the QIAcube automation RNA extraction procedure according to the manufacturer’s protocol (Qiagene, Valencia, CA). The amount of total RNA, and A260/A280- and A260/A230-nm ratios were assessed using the NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, DE). RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). For each sample, 750 ng of total RNA were hybridized to Beadchips (HumanHT-12v3 Expression Bead Chip, Illumina, San Diego, CA) that contains 48 771 probes for around 25 000 annotated genes. The hybridized Beadchips were scanned on an Illumina BeadScan confocal scanner and were analyzed by Illumina’s GenomeStudio software, version 2.0. After checking the quality of each individual array, the Feature Extraction Files were imported into R Bioconductor and analyzed using the Beadarray package for probe filtering, quantile normalization, replicate probe summarization and log2 transformation. The original microarray probe-level data files were entered into the GEO repository under accession number GSE41080.
Leukocyte subset frequency determination
PBMCs were thawed in warm media, washed twice and stained with three separate anti-human antibody cocktails containing: (1) anti-CD3 AmCyan, CD4 Pacific Blue, CD8 APCH7, CD28 APC; (2) CD3 AmCyan, CD4 Pacific Blue, CD8 APCH7, CD27 PE, CD45RA PE-Cy5; and (3) CD3 AmCyan, CD19 Alexa Fluor700, CD56 PE, CD33 PE-Cy7, TCR APC, all reagents from BD Biosciences. Incubation with antibodies was performed for 40 min at 4°C. Cells were washed with FACS buffer (PBS supplemented with 2% FBS and 0.1% Na Azide) and resuspended in 200 μl FACS buffer. Data were collected using DIVA software in an LRSII instrument (BD Biosciences). Data analysis was performed using FlowJo 8.8.6 by gating on live cells based on forward versus side-scatter profiles, then using double gating for singlet discrimination, followed by cell subset-specific gating.
Phosphorylation of intracellular proteins by pFlow
Cells were thawed in warm media and rested at 37°C in RPMI with 10% FBS. Cells were then distributed in 96 deep-well blocks (2 ml) and stimulated with IFN-γ, IL-2, IL-6, IL-7, IL-10 or IL-21 at 50 ng/ml or with 104 U/ml IFN-α for 15 min. After stimulation, cells were immediately fixed with 1.5% PFA for 10 min at RT, washed with an excess of plain PBS and permeabilized with 95% ice-cold methanol for 20 min on ice. Different stimuli conditions were barcoded using a 3 × 3 matrix with Pacific Orange and Alexa Fluor 750 (Invitrogen Corp.) at 0.03 and 0.04 μg/ml for low and 0.2 and 0.3 μg/ml for high staining, respectively. Incubation with barcoding dyes was performed at 4°C for 30 min. After several washes with FACS buffer, stimulated and barcoded cells were pooled into single tubes and stained for 30 min at 4°C, with an antibody cocktail containing anti-pSTAT1 Alexa Fluor 488, pSTAT3 Alexa Fluor 647, pSTAT5 PE, CD3 Pacific Blue, CD4 PerCP-Cy5.5, CD20 PerCP-Cy5.5 and CD33 PE-Cy7 (all from BD Phosflow). After washing, cells were resuspended in FACS buffer and acquisition was performed on an LSRII instrument (BD Biosciences). Data were collected using DIVA software. Data analysis was performed using FlowJo 8.8.6. by gating on live cells, then using double gating for singlet discrimination, followed by cell subset-specific gating. Phosphorylation of STAT1, 3 and 5 proteins in B cells, CD4− or CD4+ CD3+ T cells, or monocytes was analyzed by deconvolution of stimuli-specific gating. Baseline levels and fold increase between stimulated and unstimulated conditions were calculated using the 90th percentile fluorescence intensity of the pSTAT1, 3 or 5 signals.
Phosphorylation of Akt and PLC-γ was assessed in B cells by crosslinking of the B-cell receptor. After resting PBMC samples at 37°C (as conducted for cytokine stimulations), cells were distributed in V-bottom 96-well plates at 0.5 × 106 cells per well, and incubated for 4 min at 37°C in CO2 incubator with anti-IgM and anti-IgG both at 10 μg/ml (BD Biosciences) and 3% H2O2 for phosphatase inhibition. Cells were then fixed with 1.5% PFA for 10 min at RT. After washing twice with plain PBS, cells were permeabilized by 20 min incubation in 95% ice-cold methanol. Cells were then washed with FACS buffer and stained with an antibody cocktail containing: CD3 Pacific Blue, CD20 PerCp Cy5.5, CD27 PE Cy7, PLGγ2 (BD Biosciences) and pAkt-S473 Alexa Fluor 488 (Cell Signaling Technologies, Danvers MA). After 30 min incubation at 4°C, cells were washed in FACS buffer and analyzed by flow cytometry (as for cytokine stimulation). Median fluorescence intensity was recorded and used for the calculation of baseline levels of phosphorylated proteins and fold increase after BCR stimulation.
Serum cytokine levels determination
Cytokines were measured by a Luminex system (Luminex Corp., Austin, TX). The 50-plex kits were purchased from Millipore and were used according to manufacturer’s recommendations, with modifications as described below. Briefly, serum samples were mixed with antibody-linked polystyrene beads on 96-well filter plates and incubated at RT for 2 h followed by overnight incubation at 4°C. Plates were then vacuum filtered and washed twice before 2 h incubation with biotinylated detection antibody. Samples were filtered as above, washed twice and incubated with streptavidin-PE for 40 min, then filtered and washed twice again before resuspending in reading buffer. Each sample was measured in duplicate. Plates were read using a Luminex LabMap200 instrument with a lower bound of 100 beads per sample per measured cytokine. The Luminex LabMap200 outputs the fluorescence intensity of each bead measured for a given cytokine in a sample. For each well, we considered the median fluorescence intensity (MFI) of all beads measured for a given cytokine and averaged the MFI of the two replicates. Values were normalized to a control sample ran in each of the plates.
HAI assay
The HAI assay was performed on sera from day 0 and day 28 using a standard technique (Webster et al, 2002); serially diluted 25-μl aliquots of serum samples in PBS were mixed with 25-μl aliquots of virus, corresponding to 4 HA units, in V-bottom 96-well plates (Nunc, Rochester, NY, USA) and incubated for 30 min at RT. At the end of the incubation, 50 μl of 0.5% chicken red blood cells were added and incubated for a minimum of 45 min before reading for HAI activity. The HAI titer of a given sample was defined as the reciprocal of the last serum dilution with no HA activity. A titer of five was assigned to all samples in which the first dilution (1:10) was negative. Seroconversion has been defined as an increase of at least fourfold in the antibody titer from pre- to post-vaccination. Categories of vaccine PRs, if seroconversion was achieved for 0 or 1 strain, and vaccine GRs, if seroconversion was achieved for 2 or all 3 strains were utilized to account for both the strength and the breadth of the response.
Vaccination of CD95−/ − mice and ELISA for detection of influenza-specific antibodies
Mutant mice MRL/Mpj-Faslpr/J (Mpj/lpr) and B6.MRL-Faslpr/J (B6/lpr), and MRL/MpJ (Mpj) and C57BL/6L (B6) control mice were obtained from Jackson Laboratories (Bar Harbor, ME). Young mice (5–7 weeks of age) were housed at the Stanford Animal Facility until the protocol was completed. All mice were handled in accordance with APLAC and Stanford University animal care guidelines. Four male mice per group in the Mpj background, six male B6/Lpr and three B6 mice were given a single intramuscular dose (1/50 dilution, ∼1 μg HA) of the trivalent 2008 seasonal influenza vaccine Fluzone (Sanofi Pasteur). Samples were obtained from tail blood draws before and ∼4 weeks after vaccination. Serum was separated by centrifugation and stored at −80°C until collection of all samples. Vaccine-induced influenza-specific antibodies levels were determined by ELISA as follows. Ninety-six-well Vinyl Microtiter Microplates were coated with 2008 TIV (Fluzone) at 1 mg/ml in PBS. Plates were incubated overnight at 4°C, washed with wash buffer (0.1% of Tween 20 in PBS) and blocked with blocking buffer (3% BSA in PBS) for 1 h at 37°C. Serum samples were serially diluted with blocking buffer, added to the wells of coated/blocked plates and incubated for 1 h at 37°C. Wells incubated with blocking buffer only served as negative control. The plates were then washed, incubated for 1 h at 37°C with peroxidase-conjugated goat anti-IgG (KPL) diluted 1:2000 with blocking buffer. After washing, the plates were developed with TMB substrate (KPL) and the OD450nm of each well determined with an ELISA plate reader.
pFlow normalization scheme
We analyzed 89 samples across 13 different plates. Fold change due to stimulation was computed as the ratio of the cell, cytokine stimulation, phospho-protein measure to the raw, unnormalized, cell phospho-protein matching baseline that was measured on the same plate. Fold-change values were then normalized by the median fold-change difference of a given cell cytokine stimulation phospho-protein measure within a given plate. We tested each assay for plate-dependent differences and no significant differences between plates were detected post-day normalization.
Gene module construction
Of a total of 48 771 gene probes in the microarray per sample, we first selected 6234 (s.d. cutoff value of 0.24) and subsequently normalized their expression by centering and scaling the expression so that each gene’s expression across all subjects had euclidean norm equal to 1 for purposes of clustering. We utilized hierarchical agglomerative clustering with average linkage, euclidean distance and a height cutoff value of 1.5 to derive 109 modules. For each gene module, we assigned a set of regulatory genes (regulatory program), based on regression analysis of genes in the modules onto expression of transcription factors using a Akaike Information Criterion (AIC;Akaike, 1974). To do so, we used a set of candidate regulators composed of known signaling and transcription factors, 394 in total (Supplementary Table 4), of which 188 met the s.d. cutoff value. Briefly, we performed linear regression with elastic net penalty of each module’s expression onto the set of regulators using LARS-EN algorithm with l2 penalty weighted by 0.01. The LARS-EN algorithm provides fits of increasing number of predictors. To select the best model among the outputs of LARS-EN, we assessed quality of the resulting models by AIC, with sample specific terms weighted by within-module variance. The fit with the best AIC score was selected for each module. Gene modules and their regulatory programs can be accessed at http://www.cs.unc.edu/~vjojic/fluy2/.
Cross validation and feature selection for prediction of antibody response to vaccination
An integral part of our training algorithm is a procedure for fitting a logistic regression model with l1 and l2 penalties, the elastic net penalty, a regularization algorithm that uses cyclical coordinate descent in a pathwise fashion as described (Friedman et al, 2007, 2010). The optimization cost can be stated as:
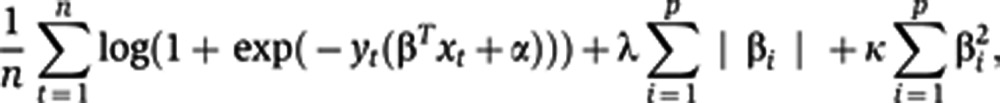 |
where n is the number of donors in the sample, p is the number of predictors, xt denotes a vector of predictor values for subject t and yt is the observed outcome (PRs versus GRs based on the seroconversion to 0–1 or 2–3 strains, respectively). We assume all of our predictors are standardized to mean 0 and s.d. 1. The result of our fitting procedure is the set of predictor weights β and intercept α for the logistic regression model. In practice, penalty weights λ and κ in Eq. 1 are set by a data-driven procedure, such as cross validation. The minimum λ was chosen to yield the highest AUC with the minimum set of features. For prediction of vaccine response using peptide predictors, we applied sevenfold cross validation with the aim of including at least 10 subjects per set (these experiments were conducted in 76 subjects only). For prediction of HAI response using blood measurements (gene module expression levels, serum proteins, pFlow conditions, immune cell subsets), all 89 subjects were included. Hence, eightfold cross validation was performed to include at least 10 subjects per test set.
Gene enrichment analysis
Genes from each module were explored by using Ingenuity Pathway Analysis for function enrichments. Data was imported and Core Analysis was performed with the following setting: Data Source: Ingenuity Expert Findings; Confidence: Experimentally Observed, TarBase, Protein–protein Interactions, Additional interactions; Species: Human. Most significant function enrichment for each module were explored in selected modules and were used for quality control. As an example, regression of each module by age and gender (Supplementary Table 3) identified a gene module (106) highly correlated with gender (r=0.714, P-value=7.9 × 10−14) of which 17 genes (42% of total genes in module) are Y chromosome-linked genes.
Function enrichment with highest significance, cell death and cell cycle, is reported from predictive modules APO and PROL, respectively. In addition, all genes in these modules and module regulators with highest regression coefficient or with known function were manually curated using a variety of sources including PubMed, IPA and BIOBASE Knowledge Library.
Data availability
Gene modules and their regulatory programs can be accessed at http://www.cs.unc.edu/~vjojic/fluy2/. Probe-level expression data was submitted to the GEO repository under accession number GSE41080.
Supplementary Material
Metadata, exposure history and antibody titer pre- and post-vaccine
Acknowledgments
We are grateful to all our volunteers for agreeing to participate in this study. We also thank Yueh-hsiu Chien, Rob Tibshirani, Jerry Friedman, Karen Sachs, Harry Greenberg, Janet McElhaney (University of British Columbia, Canada) and Fleur Tynan for illuminating discussions, and/or help and comments on the manuscript; Patty Lovelace and Garry Nolan for invaluable help with the pFlow assays; Yael Rosenberg-Hasson and Iris Herschmann for their great help in cytokine level determination; and the Stanford-LPCH Vaccine Program staff: Sally Mackey MS, Sue Swope RN, Cynthia Walsh RN, Kyrsten Spann, Thu Quan and Michele Ugur who enthusiastically enrolled subjects into the study and obtained samples from the participants. This work has been supported by grants from the Ellison Medical Foundation (AG-SS-1788), the Howard Hughes Medical Institute and the National Institutes of Health (U19 AI057229 and U19 AI090019) to MMD, and grants for the Stanford CTRU (NIH contract M01 RR00070). PJU is the recipient of a Donald E. and Delia B. Baxter Foundation Career Development Award, and is supported by the National Institutes of Health HHSN288201000034C, U19-AI050864 and U19 AI090019; and FP Grant number 261, Euro Commission, European Union Seventh Framework Program (FP7/2007-2013) under grant agreement number 261382. DF was supported by the Stanford Center on Longevity.
Author contributions: DF performed pFlow experiments; DF, TT and PL performed antibody titer studies; JP and JJ performed peptide microarray assays; DF and HH performed vaccination experiments in mice; HTM coordinated gene expression, luminex and phenotyping experiments; DF, VJ, BK and SS analyzed data; PJU and BK provided support and guidance in peptide microarray experiments and data analysis, respectively; CLD coordinated and organized the clinical study and contributed to study design and manuscript review; DF and MMD wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akaike H (1974) A new look at the statistical model identification. IEEE Trans Auto Control 19: 716–723 [Google Scholar]
- Bai L, Merchant JL (2001) ZBP-89 promotes growth arrest through stabilization of p53. Mol Cell Biol 21: 4670–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B-57: 289–300 [Google Scholar]
- Beyer WE, de Bruijn IA, Palache AM, Westendorp RG, Osterhaus AD (1999) Protection against influenza after annually repeated vaccination: a meta-analysis of serologic and field studies. Arch Intern Med 159: 182–188 [DOI] [PubMed] [Google Scholar]
- Beyer WE, Palache AM, Luchters G, Nauta J, Osterhaus AD (2004) Seroprotection rate, mean fold increase, seroconversion rate: which parameter adequately expresses seroresponse to influenza vaccination? Virus Res 103: 125–132 [DOI] [PubMed] [Google Scholar]
- Beyer WE, Palache AM, Sprenger MJ, Hendriksen E, Tukker JJ, Darioli R, van der Water GL, Masurel N, Osterhaus AD (1996) Effects of repeated annual influenza vaccination on vaccine sero-response in young and elderly adults. Vaccine 14: 1331–1339 [DOI] [PubMed] [Google Scholar]
- Bischof O, Schwamborn K, Martin N, Werner A, Sustmann C, Grosschedl R, Dejean A (2006) The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol Cell 22: 783–794 [DOI] [PubMed] [Google Scholar]
- Chen WH, Cross AS, Edelman R, Sztein MB, Blackwelder WC, Pasetti MF (2011) Antibody and Th1-type cell-mediated immune responses in elderly and young adults immunized with the standard or a high dose influenza vaccine. Vaccine 29: 2865–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TW, Pareek M, Hoschler K, Dillon H, Nicholson KG, Groth N, Stephenson I (2009) Trial of 2009 influenza A (H1N1) monovalent MF59-adjuvanted vaccine. N Engl J Med 361: 2424–2435 [DOI] [PubMed] [Google Scholar]
- Davis MM (2008) A prescription for human immunology. Immunity 29: 835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Martinis M, Franceschi C, Monti D, Ginaldi L (2005) Inflamm-ageing and lifelong antigenic load as major determinants of ageing rate and longevity. FEBS Lett 579: 2035–2039 [DOI] [PubMed] [Google Scholar]
- de Visser KE, Eichten A, Coussens LM (2006) Paradoxical roles of the immune system during cancer development. Nat Rev Cancer 6: 24–37 [DOI] [PubMed] [Google Scholar]
- DiazGranados CA, Dunning AJ, Jordanov E, Landolfi V, Denis M, Talbot HK (2013) High-dose trivalent influenza vaccine compared to standard dose vaccine in elderly adults: safety, immunogenicity and relative efficacy during the 2009–2010 season. Vaccine 31: 861–866 [DOI] [PubMed] [Google Scholar]
- Eaton SM, Burns EM, Kusser K, Randall TD, Haynes L (2004) Age-related defects in CD4 T cell cognate helper function lead to reductions in humoral responses. J Exp Med 200: 1613–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich HJ, Muller M, Kollaritsch H, Pinl F, Schmitt B, Zeitlinger M, Loew-Baselli A, Kreil TR, Kistner O, Portsmouth D, Fritsch S, Maritsch F, Aichinger G, Pavlova BG, Barrett PN (2012) Pre-vaccination immunity and immune responses to a cell culture-derived whole-virus H1N1 vaccine are similar to a seasonal influenza vaccine. Vaccine 30: 4543–4551 [DOI] [PubMed] [Google Scholar]
- Erickson LD, Durell BG, Vogel LA, O'Connor BP, Cascalho M, Yasui T, Kikutani H, Noelle RJ (2002) Short-circuiting long-lived humoral immunity by the heightened engagement of CD40. J Clin Invest 109: 613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagnoni FF, Vescovini R, Passeri G, Bologna G, Pedrazzoni M, Lavagetto G, Casti A, Franceschi C, Passeri M, Sansoni P (2000) Shortage of circulating naive CD8(+) T cells provides new insights on immunodeficiency in aging. Blood 95: 2860–2868 [PubMed] [Google Scholar]
- Fang Y, Larsson L, Bruhns P, Xiang Z (2012) Apoptosis of mouse mast cells is reciprocally regulated by the IgG receptors FcgammaRIIB and FcgammaRIIIA. Allergy 67: 1233–1240 [DOI] [PubMed] [Google Scholar]
- Franceschi C, Bonafe M, Valensin S (2000) Human immunosenescence: the prevailing of innate immunity, the failing of clonotypic immunity, and the filling of immunological space. Vaccine 18: 1717–1720 [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R (2010) Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 33: 1–22 [PMC free article] [PubMed] [Google Scholar]
- Gate L, Majumdar RS, Lunk A, Tew KD (2004) Increased myeloproliferation in glutathione S-transferase pi-deficient mice is associated with a deregulation of JNK and Janus kinase/STAT pathways. J Biol Chem 279: 8608–8616 [DOI] [PubMed] [Google Scholar]
- Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR 3rd, Castro E, Akondy R, Rinfret A, Yassine-Diab B, Said EA, Chouikh Y, Cameron MJ, Clum R, Kelvin D, Somogyi R, Greller LD et al. (2008) Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med 205: 3119–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goronzy JJ, Fulbright JW, Crowson CS, Poland GA, O'Fallon WM, Weyand CM (2001) Value of immunological markers in predicting responsiveness to influenza vaccination in elderly individuals. J Virol 75: 12182–12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson JM, Harrington LE, Lanier JG, Wherry EJ, Ahmed R (2002) Differential sensitivity of naive and memory CD8+ T cells to apoptosis in vivo. J Immunol 169: 3760–3770 [DOI] [PubMed] [Google Scholar]
- Hamano S, Himeno K, Miyazaki Y, Ishii K, Yamanaka A, Takeda A, Zhang M, Hisaeda H, Mak TW, Yoshimura A, Yoshida H (2003) WSX-1 is required for resistance to Trypanosoma cruzi infection by regulation of proinflammatory cytokine production. Immunity 19: 657–667 [DOI] [PubMed] [Google Scholar]
- Hansson GK (2005) Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med 352: 1685–1695 [DOI] [PubMed] [Google Scholar]
- Haynes L, Eaton SM, Burns EM, Randall TD, Swain SL (2005) Newly generated CD4 T cells in aged animals do not exhibit age-related defects in response to antigen. J Exper Med 201: 845–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XS, Holmes TH, Sasaki S, Jaimes MC, Kemble GW, Dekker CL, Arvin AM, Greenberg HB (2008) Baseline levels of influenza-specific CD4 memory T-cells affect T-cell responses to influenza vaccines. PLoS One 3: e2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HC, Mountz JD (2010) Metabolic syndrome, hormones, and maintenance of T cells during aging. Curr Opin Immunol 22: 541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HC, Scott DK, Zhang P, Zhou J, Yang P, Wu Q, Schroeder HW Jr, Gerald LB, Ravussin E, Jazwinski SM, Mountz JD (2006) CD8 T-cell immune phenotype of successful aging. Mech Ageing Dev 127: 231–239 [DOI] [PubMed] [Google Scholar]
- Illario M, Giardino-Torchia ML, Sankar U, Ribar TJ, Galgani M, Vitiello L, Masci AM, Bertani FR, Ciaglia E, Astone D, Maulucci G, Cavallo A, Vitale M, Cimini V, Pastore L, Means AR, Rossi G, Racioppi L (2008) Calmodulin-dependent kinase IV links Toll-like receptor 4 signaling with survival pathway of activated dendritic cells. Blood 111: 723–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH (2003) Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol 4: 321–329 [DOI] [PubMed] [Google Scholar]
- Jaleco S, Swainson L, Dardalhon V, Burjanadze M, Kinet S, Taylor N (2003) Homeostasis of naive and memory CD4+ T cells: IL-2 and IL-7 differentially regulate the balance between proliferation and Fas-mediated apoptosis. J Immunol 171: 61–68 [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Höfling Holger, Tibshirani Robert (2007) Pathwise coordinate optimization. Ann Appl Stat 1: 302–332 [Google Scholar]
- Khurana S, Suguitan AL Jr, Rivera Y, Simmons CP, Lanzavecchia A, Sallusto F, Manischewitz J, King LR, Subbarao K, Golding H (2009) Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med 6: e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi Y, Aiba Y, Higuchi T, Furukawa K, Tokuhisa T, Takemori T, Tsubata T (2010) Augmented antibody response with premature germinal center regression in CD40L transgenic mice. J Immunol 185: 211–219 [DOI] [PubMed] [Google Scholar]
- Knox PG, Milner AE, Green NK, Eliopoulos AG, Young LS (2003) Inhibition of metalloproteinase cleavage enhances the cytotoxicity of Fas ligand. J Immunol 170: 677–685 [DOI] [PubMed] [Google Scholar]
- Ligr M, Li Y, Zou X, Daniels G, Melamed J, Peng Y, Wang W, Wang J, Ostrer H, Pagano M, Wang Z, Garabedian MJ, Lee P (2010) Tumor suppressor function of androgen receptor coactivator ARA70alpha in prostate cancer. Am J Pathol 176: 1891–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Li Y, Prabhu V, Young L, Becker KG, Munson PJ, Weng N (2001) Augmentation in expression of activation-induced genes differentiates memory from naive CD4+ T cells and is a molecular mechanism for enhanced cellular response of memory CD4+ T cells. J Immunol 166: 7335–7344 [DOI] [PubMed] [Google Scholar]
- Molinari NA, Ortega-Sanchez IR, Messonnier ML, Thompson WW, Wortley PM, Weintraub E, Bridges CB (2007) The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25: 5086–5096 [DOI] [PubMed] [Google Scholar]
- Nagata S (1999) Fas ligand-induced apoptosis. Ann Rev Genet 33: 29–55 [DOI] [PubMed] [Google Scholar]
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li GM, McCausland M, Kanchan V, Kokko KE, Li S, Elbein R, Mehta AK, Aderem A, Subbarao K, Ahmed R, Pulendran B (2011) Systems biology of vaccination for seasonal influenza in humans. Nat Immunol 12: 786–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell BC, Cheung AF, Simkevich CP, Tam W, Ren X, Mateyak MK, Sedivy JM (2003) A large scale genetic analysis of c-Myc-regulated gene expression patterns. J Biol Chem 278: 12563–12573 [DOI] [PubMed] [Google Scholar]
- Owaki T, Asakawa M, Morishima N, Mizoguchi I, Fukai F, Takeda K, Mizuguchi J, Yoshimoto T (2008) STAT3 is indispensable to IL-27-mediated cell proliferation but not to IL-27-induced Th1 differentiation and suppression of proinflammatory cytokine production. J Immunol 180: 2903–2911 [DOI] [PubMed] [Google Scholar]
- Pan L, Sato S, Frederick JP, Sun XH, Zhuang Y (1999) Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Mol Cell Biol 19: 5969–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelec G, Larbi A (2008) Immunity and ageing in man: Annual Review 2006/2007. Exper Gerontol 43: 34–38 [DOI] [PubMed] [Google Scholar]
- Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, Kennedy K, Wu H, Bennouna S, Oluoch H, Miller J, Vencio RZ, Mulligan M, Aderem A, Ahmed R, Pulendran B (2009) Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol 10: 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvioli S, Bonafe M, Capri M, Monti D, Franceschi C (2001) Mitochondria, aging and longevity--a new perspective. FEBS Lett 492: 9–13 [DOI] [PubMed] [Google Scholar]
- Sasaki S, Sullivan M, Narvaez CF, Holmes TH, Furman D, Zheng NY, Nishtala M, Wrammert J, Smith K, James JA, Dekker CL, Davis MM, Wilson PC, Greenberg HB, He XS (2011) Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies. J Clin Invest 121: 3109–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Suematsu A, Nakashima T, Takemoto-Kimura S, Aoki K, Morishita Y, Asahara H, Ohya K, Yamaguchi A, Takai T, Kodama T, Chatila TA, Bito H, Takayanagi H (2006) Regulation of osteoclast differentiation and function by the CaMK-CREB pathway. Nat Med 12: 1410–1416 [DOI] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N (2003) Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 34: 166–176 [DOI] [PubMed] [Google Scholar]
- Smith KG, Light A, O'Reilly LA, Ang SM, Strasser A, Tarlinton D (2000) bcl-2 transgene expression inhibits apoptosis in the germinal center and reveals differences in the selection of memory B cells and bone marrow antibody-forming cells. J Exp Med 191: 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KG, Nossal GJ, Tarlinton DM (1995) FAS is highly expressed in the germinal center but is not required for regulation of the B-cell response to antigen. Proc Natl Acad Sci USA 92: 11628–11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Cerasoli DM, Dal Porto JM, Shimoda M, Freund R, Fang W, Telander DG, Malvey EN, Mueller DL, Behrens TW, Kelsoe G (1999) Relaxed negative selection in germinal centers and impaired affinity maturation in bcl-xL transgenic mice. J Exp Med 190: 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Ohta H, Takemori T (2001) Fas is required for clonal selection in germinal centers and the subsequent establishment of the memory B cell repertoire. Immunity 14: 181–192 [DOI] [PubMed] [Google Scholar]
- Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, Sasaki S, Imai K, Shibue T, Honda K, Taniguchi T (2003) Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 424: 516–523 [DOI] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K (2003) Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289: 179–186 [DOI] [PubMed] [Google Scholar]
- Thorpe JA, Schwarze SR (2010) IRE1alpha controls cyclin A1 expression and promotes cell proliferation through XBP-1. Cell Stress Chaperones 15: 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuno T, Mejido J, Zhao T, Schmeisser H, Morrow A, Zoon KC (2009) IRF9 is a key factor for eliciting the antiproliferative activity of IFN-alpha. J Immunother 32: 803–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida D, Kawamata H, Omotehara F, Miwa Y, Hino S, Begum NM, Yoshida H, Sato M (2000) Over-expression of TSC-22 (TGF-beta stimulated clone-22) markedly enhances 5-fluorouracil-induced apoptosis in a human salivary gland cancer cell line. Lab Invest 80: 955–963 [DOI] [PubMed] [Google Scholar]
- Vajo Z, Tamas F, Sinka L, Jankovics I (2010) Safety and immunogenicity of a 2009 pandemic influenza A H1N1 vaccine when administered alone or simultaneously with the seasonal influenza vaccine for the 2009-10 influenza season: a multicentre, randomised controlled trial. Lancet 375: 49–55 [DOI] [PubMed] [Google Scholar]
- Villarino A, Hibbert L, Lieberman L, Wilson E, Mak T, Yoshida H, Kastelein RA, Saris C, Hunter CA (2003) The IL-27R (WSX-1) is required to suppress T cell hyperactivity during infection. Immunity 19: 645–655 [DOI] [PubMed] [Google Scholar]
- Webster R, Cox N, Stohr K (2002) WHO Manual on Animal Influenza Diagnosis and Surveillance. World Health OrganizationCDS/CSR/NCS/2002.5 [Google Scholar]
- Weiskopf D, Weinberger B, Grubeck-Loebenstein B (2009) The aging of the immune system. Transpl Int 22: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Xiang Z, Cutler AJ, Brownlie RJ, Fairfax K, Lawlor KE, Severinson E, Walker EU, Manz RA, Tarlinton DM, Smith KG (2007) FcgammaRIIb controls bone marrow plasma cell persistence and apoptosis. Nat Immunol 8: 419–429 [DOI] [PubMed] [Google Scholar]
- Yager EJ, Ahmed M, Lanzer K, Randall TD, Woodland DL, Blackman MA (2008) Age-associated decline in T cell repertoire diversity leads to holes in the repertoire and impaired immunity to influenza virus. J Exp Med 205: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Kawashita N, Kubota-Koketsu R, Inoue Y, Watanabe Y, Ibrahim MS, Ideno S, Yunoki M, Okuno Y, Takagi T, Yasunaga T, Ikuta K (2010) Highly conserved sequences for human neutralization epitope on hemagglutinin of influenza A viruses H3N2, H1N1 and H5N1: Implication for human monoclonal antibody recognition. Biochem Biophys Res Commun 393: 614–618 [DOI] [PubMed] [Google Scholar]
- Yoshimoto T, Morishima N, Mizoguchi I, Shimizu M, Nagai H, Oniki S, Oka M, Nishigori C, Mizuguchi J (2008) Antiproliferative activity of IL-27 on melanoma. J Immunol 180: 6527–6535 [DOI] [PubMed] [Google Scholar]
- Yu C, Minemoto Y, Zhang J, Liu J, Tang F, Bui TN, Xiang J, Lin A (2004) JNK suppresses apoptosis via phosphorylation of the proapoptotic Bcl-2 family protein BAD. Mol Cell 13: 329–340 [DOI] [PubMed] [Google Scholar]
- Zhang CZ, Chen GG, Lai PB (2010) Transcription factor ZBP-89 in cancer growth and apoptosis. Biochim Biophys Acta 1806: 36–41 [DOI] [PubMed] [Google Scholar]
- Zhang J, Xu LG, Han KJ, Wei X, Shu HB (2004) PIASy represses TRIF-induced ISRE and NF-kappaB activation but not apoptosis. FEBS Lett 570: 97–101 [DOI] [PubMed] [Google Scholar]
- Zhang X, Li L, Jung J, Xiang S, Hollmann C, Choi YS (2001) The distinct roles of T cell-derived cytokines and a novel follicular dendritic cell-signaling molecule 8D6 in germinal center-B cell differentiation. J Immunol 167: 49–56 [DOI] [PubMed] [Google Scholar]
- Zhou T, Edwards CK 3rd, Mountz JD (1995) Prevention of age-related T cell apoptosis defect in CD2-fas-transgenic mice. J Exp Med 182: 129–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metadata, exposure history and antibody titer pre- and post-vaccine
Data Availability Statement
Gene modules and their regulatory programs can be accessed at http://www.cs.unc.edu/~vjojic/fluy2/. Probe-level expression data was submitted to the GEO repository under accession number GSE41080.