Abstract
Systemic lupus erythematosus (SLE) is a prototypic multisystem autoimmune disorder where interplay of environmental and genetic risk factors leads to progressive loss of tolerance to nuclear antigens over time, finally culminating in clinical disease. The heterogeneity of clinical manifestations and the disease’s unpredictable course characterized by flares and remissions are very likely a reflection of heterogeneity at the origin of disease, with a final common pathway leading to loss of tolerance to nuclear antigens. Impaired clearance of immune complexes and apoptotic material and production of autoantibodies have long been recognized as major pathogenic events in this disease. Over the past decade the type I interferon cytokine family has been postulated to play a central role in SLE pathogenesis, by promoting feedback loops progressively disrupting peripheral immune tolerance and driving disease activity. The identification of key molecules involved in the pathogenesis of SLE will not only improve our understanding of this complex disease, but also help to identify novel targets for biological intervention.
Keywords: autoantibody, autoantigen, B cells, complement, dendritic cells, genetics, immune complex, interferon, pathogenesis, systemic lupus erythematosus, Toll-like receptor
Introduction
The pathogenesis of systemic lupus erythematosus (SLE) is incompletely understood. Even though the hallmark of the disease is a loss of tolerance to nuclear antigens, clinical manifestations as well as disease severity and course vary from patient to patient. This most likely reflects the heterogeneous genetic background that underlies disease susceptibility. The past few years have witnessed an explosion of SLE genomic studies. Here we summarize recent genetic and transcriptome data that are helping to reconstruct the puzzle of SLE pathogenesis. However, many questions remain to be addressed, including the factors governing disease expression in specific organs, which, with the exception of congenital heart block, remain largely unknown.
SLE has a complex genetic basis
A genetic contribution is important to cause disease even though the concordance rate for SLE is only 25% among monozygotic twins.1 More than 25 genetic risk loci have been identified in recent genome-wide association scans. Despite this impressive progress, it is estimated that less than 10% of the total genomic susceptibility to SLE has been characterized to date.2 The genetic risk for lupus is likely derived from variation in many (perhaps as many as 100) genes, each of modest effect size with odds ratios between 1.15 and 2.0.3
HLA-DRB1, signal transducer and activator of transcription 4 (STAT4) and interferon regulatory factor 5 (IRF5) are the three most frequently observed alleles accounting each for a little more than 1% of the variance in genome-wide association scans.4 Together they point towards an interplay of alterations in the innate and adaptive immune systems: IRF5 is involved in the transcription of type I interferon and pro-inflammatory cytokines triggered by TLR signaling and STAT4 plays a key role in type I and type II IFN signaling. Presentation of epitopes within the grooves of MHC-I or MHC-II defines the choice of targets for the adaptive immune system and thereby explains the towering dominance of MHC in determining genetic susceptibility, not only in SLE but also in many other autoimmune disorders.5
Summarizing current knowledge, genes associated with SLE are involved in the following pathways2–4,6–16 (Figure 1):
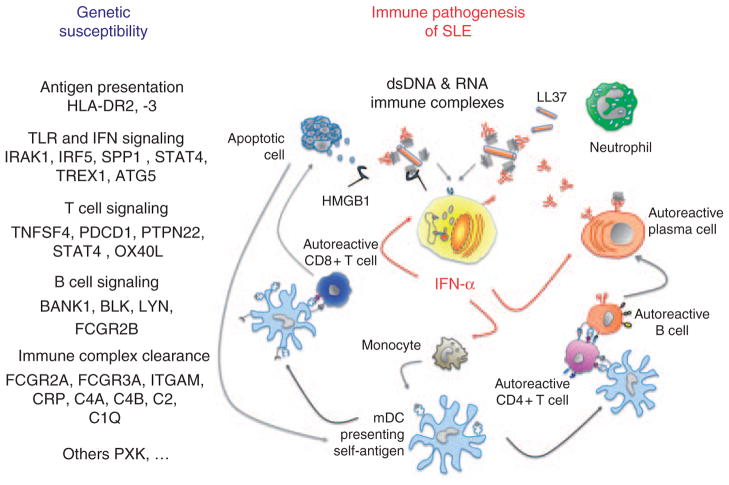
Figure 1.
The IFN-α signature of systemic lupus erythematosus (SLE). Genetic susceptibility to SLE includes genes involved in immune complex clearance, the stimulation of IFN-α production and IFN-α signaling as well as antigen presentation and B- and T-cell signaling contributing to immune pathogenesis of SLE as shown in the right part of this figure. Nucleic acids can act as endogenous triggers of IFN-α production in pDC. Molecules such as LL37 and HMGB1 not only protect self-nucleic acids from extracellular degradation, but are also involved in their uptake through lipid rafts and/or receptors such as RAGE into endosomal TLR7/9 compartments to stimulate IFN-α production. Moreover, immune complexes can bind to FcgRIIa (CD32) on pDC and thereby gain access to endosomes by receptor-mediated endocytosis. IFN-α induces and maintains the generation of mature DCs, which expand and activate rather than delete autoreactive T cells. The latter contribute to tissue damage yielding large numbers of nucleosomes, which can be captured by mature DCs, further amplifying the autoreactive process. Together with IL-6 IFN-α promotes plasmablasts to develop into antibody-secreting plasma cells. Also BLyS/BAFF is induced by IFN-α and contributes to the survival of mature, peripheral B cells. Moreover, IFNα upregulates the expression of IRF7 in pDC, and of TLR7 and IRF7 in mDC and monocytes, thereby increasing the responsiveness to DNA and/or RNA-containing immune complexes with further augmentation of IFN-α synthesis. For a color version of this figure, see online version of this article.
Antigen presentation to the T-cell receptor of CD4+ T cells via HLA-DR (which is expressed primarily on dendritic cells, monocytes and B cells): HLA-DR2, HLA-DR3.
Components of pathways upstream and downstream of type I IFN: (i) components of Toll-like receptor (TLR) signaling pathways (IRAK1, IRF5, IRF7, IRF8, SPP1 and TNFAIP3), (ii) IFN signaling (STAT4), (iii) intracellular DNA degradation (TREX1), (iv) autophagy-related genes (ATG5) which might contribute to IFN production by plasmacytoid dendritic cells.
Signaling molecules activated after engagement of the T-cell receptor (TCR; such as TNFSF4/OX40L, PDCD1, PTPN22, STAT4).
Signaling molecules activated after engagement of the B-cell receptor (BCR; such as BANK1, BLK, LYN, PTPN22).17,18
Molecules involved in the clearance of apoptotic debris and of immune complexes such as FCGR2A/CD32 and FCGR3A/CD16, ITGAM/CD11b, an integrin which functions as complement receptor 3 but is also involved in the extravasation of leukocytes into tissues and in neutrophil phagocytosis and apoptosis; 19 CRP, C4A, C4B, C2 and C1Q, which are important in opsonization.
Other molecules involved in ubiquitination (UBE2L3, TNFAIP3), DNA methylation (MECP2) and other yet undefined pathways such as PXK, XKR6, KIAA1542 or SCUBE1.
Potentially most informative results are to be expected from functional and immunological characterization of the latter group of genes. This will undoubtedly lead to the identification of novel pathogenetic pathways in SLE.
The serologic hallmark of SLE is the production of antinuclear antibodies
Autoantibodies are present many years before the onset of clinical symptoms in SLE. Antinuclear antibodies (ANA), anti-Ro, anti-La and antiphospholipid antibodies appear first (mean 3.4 years), followed by anti-dsDNA antibodies (mean 2 years) and anti-Sm and anti-RNP autoantibodies, which appear around 1 year before diagnosis of SLE and often coincide with the first clinical symptoms.20 Most of these autoantibodies have not been shown to play a pathogenetic role, correlate with disease activity or decrease following high-dose immunosuppressive treatment.
The most common lupus autoantibodies recognize double-stranded DNA (dsDNA) and small nuclear ribonucleoproteins (snRNPs), which are macromolecular complexes of small nuclear RNA (snRNA) and associated proteins. Anti-histone antibodies are frequently found in drug-induced SLE as well as in naturally occurring SLE.
Some of the nuclear autoantigens targeted in SLE have been described clustered in cell-surface blebs during keratinocyte apoptosis.21 These autoantigens can be cleaved by granzyme B, a serine protease present in natural killer and cytotoxic T cells, leading to the generation of unique antigenic fragments not observed during other forms of cell death.22 Other forms of cell death besides apoptosis might contribute to make nuclear antigens accessible to antibodies and cells of the adaptive immune system. In particular, neutrophils release DNA and histones as part of a unique type of cell death characterized by the extrusion of chromatin and proteins from neutrophilic granules into the extracellular space. This novel type of death, which aims at killing bacteria, is known as ‘netosis’ based on the formation of neutrophil extracellular traps (NETs).23,24 In fact, histones have been shown already in the late 1950s to have bactericidal activity when released into the extracellular space.25
Broadening of the antigenic repertoire (‘epitope spreading’) is a characteristic of the SLE immune response. There is evidence supporting that this is an antigen-driven and T-cell-dependent process. Thus, autoreactive B cells require (similarly self-reactive) CD4 T-cell help for autoantibody production. Of note, only little is known about the epitope specificity of autoreactive T cells in SLE.26,27
Impaired clearance of immune complexes in SLE
A characteristic feature of SLE is an impaired clearance and accumulation of autoantigen–autoantibody complexes in tissues such as the kidney glomeruli or the dermo-epidermal junction of the skin in variants of cutaneous lupus erythematosus (‘lupus band’; see the article on ‘Pathogenesis of cutaneous lupus erythematosus’ in this issue). Deficiencies of early components of the classical pathway of complement such as C1q, C2 or C4 are rare, but they confer the strongest genetic susceptibility to an SLE-like disease in humans, with a penetrance rate from 30% (C4 deficiency) to over 90% (C1q deficiency).28–30 Although homozygous mutations are exceedingly rare, nevertheless these ‘experiments of nature’ unequivocally demonstrate the functional consequences of impaired clearance on self-tolerance. Recently, Lood et al. reported that C1q inhibits immune-complex triggered IFN-α production by plasmacytoid dendritic cells (pDC), providing a novel link between complement deficiency and the activation of the type I IFN pathway in SLE as further discussed below.31
The IFN family of cytokines was discovered by its ability to interfere with viral replication,32 although the antiviral potency of individual IFNs varies considerably. IFNs are classified based upon amino acid sequence and recognition of specific receptors into three families.33 Type I IFN comprise IFN-α (which includes 13 subspecies), IFN-β, IFN-ε (expressed in the placenta), IFN-κ (expressed in keratinocytes), and IFN-ω in humans.34,35 Type II IFN consist of a single member, IFN-γ, which has weak antiviral, but potent immunomodulatory functions. Type III IFN, IFN-λ1 (IL-29), IFN-λ2 (IL-28A) and IFN-λ3 (IL-28B), exert antiviral properties.
The IFN-α signature of SLE
Several observations led to the identification of IFN-α as a central player in SLE pathogenesis: IFN-α was found to be elevated in lupus sera,36 most notably during disease flares, and anecdotal reports described the induction of a lupus-like syndrome following treatment with IFN-α for melanoma or hepatitis C.37 The ability of DNA-containing immune complexes from lupus sera to induce IFN-α production by a novel cell type 38–41 later identified as the pDC was described in the late 1990s.42 Furthermore IFN-α regulated gene transcripts were shown to be significantly upregulated in peripheral blood of pediatric and adult SLE patients upon gene expression profiling.43,44 As described above, a genetic association with pathways related to type I IFN transcription and or signaling has been confirmed in SLE (for example, IRF5, STAT4, TNFAIP3 or TREX1).2–4,7,9–13,15,16,45,46 Indeed, an excessive production and/or response to type I IFN explains many of the immune alterations observed in the disease.47,48 Consequently, IFN-α is a logical therapeutic target, a hypothesis that has been tested in a phase I clinical trial as reported recently.49 Moreover, the IFN-α signature is being assessed as a new biomarker for SLE disease activity.49,50
Immature pDC are the main source of type I IFN following endosomal TLR7 or TLR9 ligation
Many cells can produce IFN-α in response to viral infection in small quantities. pDC, previously called ‘naturally interferon-producing cells’, are unique in their capacity to produce vast amounts of IFN-α capable of generating systemic effects.42,51 A single pDC, for example, can synthesize up to one billion IFN-α molecules within 12 hours (or 3–10 pg per cell), which is 200–1000 times more than the amount produced by any other cell type.42,52 This unique ability of pDC can in part be explained by the constitutive expression or TLR7, TLR9 and IRF7.53–57 Upon pDC maturation, IFN-α secretion is switched off, the cell acquires a dendritic morphology and assumes a ‘professional’ antigen-presenting function.52,58
pDC have been described by pathologists as early as the 1970s as an unusual cell with morphological and immunohistochemical features reminiscent of plasma cells, T lymphocytes and monocytes (‘plasmacytoid T cells’ or ‘plasmacytoidmonocytes’). pDC are Lin-BDCA-2+CD123+HLA-DR+ILT7+ and constitute less than 1% of circulating white blood cells.51,52,59 Their frequency is decreased up to 100-fold in the peripheral blood of SLE patients42,60,61 due to increased exodus into inflamed tissue like the kidney and skin62–64. Interestingly, phototesting can reproduce clinically and histologically photosensitive variants of cutaneous lupus erythematosus, including an influx of pDC following UV irradiation65 (see also the review on ‘Photosensitivity, phototesting, and photoprotection in cutaneous lupus erythematosus’ in this issue).
The innate immune system contributes to the loss of tolerance to nucleic acids in SLE
The endosomal localization of TLR7 and TLR9, their inability to recognize endogenous DNA and RNA sequences and the rapid extracellular degradation of free nucleic acids by ubiquitous DNAses and RNAses are assumed to function as physiological protective barriers against ‘accidental’ activation of these potent receptors by self-nucleic acids.
There is experimental evidence that all of these protective mechanisms can be circumvented in SLE: DNAse1 knock-out mice develop a lupuslike disease;66 moreover, DNAse1 deficiency was also observed in some lupus patients.67 Molecules such as LL37, produced by neutrophils or damaged keratinocytes,68 or the ubiquitous HMGB1 not only protect self-nucleic acids from extracellular degradation but are also involved in their uptake through lipid rafts and/or receptors such as RAGE into endosomal TLR7/9 compartments.69,70 Moreover, immune complexes can bind to FcgRIIa (CD32) on pDC and thereby gain access to endosomes by receptor-mediated endocytosis. 71,72 Hypomethylated DNA rich in linear cytosine– guanosine (CpG) sequences and uracil-rich small nuclear RNA (particularly U1 snRNA tightly bound to Sm and other small ribonucloproteins) are infrequent in the human genome; interestingly such unusual sequences are enriched in immune complexes in SLE 73 and are preferentially recognized by endosomal TLRs.74–78
pDCs sense aggregated DNA structures in early endosomes through TLR9. This leads to signaling through a TRAF3-IRAK1-OPN complex, leading to IRF-7 nuclear translocation and induction of type I IFN gene transcription. Linear (monomeric) DNA structures (or their synthetic correlate type B ODN) traffic through early endosomes into more acidic late endolysosomes where TLR7/9 activation recruits a different set of signaling molecules (i.e. NF-kB and IRF5) leading to transcription of inflammatory cytokines (IL-6, TNF-α) and costimulatory molecules (CD80, CD86 and CD40), DC maturation and diminished IFN-α secretion.79 Synthetic compounds specifically targeting TLR9 and TLR7 to prevent immune complex stimulated IFN-α production in pDC promise new therapeutic avenues in the treatment of SLE.80,81 In fact, an ‘old’ drug in the treatment of SLE already targets TLR9: chloroquine directly inhibits CpG-driven activation of TLR9, which might explain one of the actions of this antimalarial at the molecular level in autoimmune diseases.82,83
pDC and type I IFN at the center of lupus pathogenesis
Under steady-state conditions, immature mDC capture antigen and migrate, without maturing, to draining lymph nodes where they present self-peptide MHC complexes in the absence of costimulatory signals to self-reactive T cells, which leads to anergy and deletion.84 Furthermore, immature DC may contribute to peripheral tolerance through the induction and maintenance of regulatory CD4+ T cells. Thus, autoreactive T cells escaping negative selection in the thymus are kept in check by peripheral immune tolerance. IFN-α contributes to disrupting peripheral immune tolerance by promoting mDC maturation.61,85 Moreover, defects in Treg cell activity86 and functional impairment of a novel subset of CD19+CD24hiCD38hi B cells with IL-10/B7 (B7=CD80/CD86) mediated suppressor activity on CD4+ T cells have been recently described as contributing to impaired immune tolerance in SLE.87
IFN-α-induced upregulation of costimulatory molecules such as CD80 and CD86 contributes to the survival, expansion and differentiation of self-reactive T cells. Autoreactive CD4+ T cells provide help to autoreactive CD8+ T cells. Furthermore IFN-α enhances a cytotoxic program of CD8+ T cell maturation characterized by an increased expression of perforin and granzymes.88 These autoreactive CD8+ T cells contribute to tissue damage and to the generation of novel granzyme B dependent autoantigens,22 further fueling immune complex driven production of IFN-α (Figure 1).
Importantly, IFNα upregulates the expression of TLR7 and IRF7 in pDC, mDC and monocytes, thereby increasing the responsiveness to nucleic-acid-containing immune complexes with further augmentation of IFN-α synthesis.89
IFN-α has myelosuppressive effects explaining in part the B-cell and T-cell lymphopenia observed in the peripheral blood of lupus patients. At the same time BLyS/BAFF is induced by IFN-α and contributes to the survival of mature, peripheral B cells. IFN-α promotes the differentiation of activated B cells into plasmablasts and, together with IL-6, permits plasmablasts to develop into antibody-secreting plasma cells.90 Autoreactive B cells generate autoantibodies and thereby provide, through the formation of nucleic-acid-containing immune complexes, a positive feedback loop for enhanced TLR7/9 activation and even more increased IFN-α production. Since B cells express TLR7 and TLR9 similarly to pDC, they too can be targeted by nucleic-acid-containing immune complexes.48,90,91
These effects explain the dramatic upregulation of type I IFN-inducible genes (the so-called ‘IFN signature’) described in the majority of SLE patient’s blood.
Our understanding of the pathogenetic pathways involved in SLE has greatly expanded over the past decade. Defects in the clearance of immune complexes and apoptotic cells, altered B- and T-cell signaling, the production of autoantibodies and the link of all of the above with unabated IFN-α production provide a comprehensive picture of the pathogenic immune alterations taking place in SLE. Targeting B-cell hyperactivity by monoclonal antibodies directed against IFN-inducible proteins such as BAFF/BlyS or its receptor TACI as well as against type I IFN are already being tested in clinical trials in SLE patients. A number of new promising compounds that inhibit TLR7 and TLR9 show promising results in animal studies.80 Despite the incredible progress made in the last few years, we probably have only discovered some islands of fact in a sea of uncertainty.92 We therefore impatiently await the discovery of novel pathways leading to the identification of novel targets for biological intervention and successful treatment of this devastating disease.
Acknowledgments
This work was supported by grants from the National Institutes of Health by R01 AR050770-01, ARO54083-01CORT, NIH ARO55503-01CORT, U19-AI082715-01, the Alliance for Lupus Research (VP), the Mary Kirkland Foundation (VP, GO), the Erwin Schrödinger Research Grant of the Austrian Science Fund (GO) and the Medical Research Fond (MFF) Tirol (GO). We thank Dr Lynn Punaro and Dr Katherine Madson and our patients at the Pediatric Rheumatology Clinic of Texas Scottish Rite Hospital in Dallas for their cooperation in participating in our studies. We thank Dr Jeanine Baisch for helpful discussion of the manuscript.
List of abbreviations
- SLE
Systemic lupus erythematosus
- ANA
Antinuclear antibodies
- IFN
Interferon
- IFN-α
Interferon alpha
- DC
Dendritic cell
- mDC
Myeloid dendritic cell
- pDC
Plasmacytoid dendritic cell
- TLR
Toll-like receptor
- IRF7
Interferon regulatory factor 7
- IRAK1
Interleukin-1 receptor-associated kinase 1
- TNFAIP3
Tumor necrosis factor alpha-induced protein 3
- TREX1
Three prime repair exonuclease 1
- STAT4
Signal transducer and activator of transcription 4
- ATG5
Autophagy protein 5
- TCR
T-cell receptor
- TNFSF4
Tumor necrosis factor (ligand) superfamily member 4 (=OX40L)
- PDCD1
Programmed cell death 1
- PTPN22
Protein tyrosine phosphatase non-receptor type 22
- BCR
B-cell receptor
- BANK1
B-cell scaffold protein with ankyrin repeats 1
- BLK
B lymphoid tyrosine kinase
- LYN
v-yes-1 Yamaguchi sarcoma viral-related oncogene homolog
- FcGR2A
Low-affinity IIa receptor for Fc fragment of IgG (CD32)
- FcGR3A
Low-affinity IIIa receptor for Fc fragment of IgG (CD16)
- ITGAM
Integrin alpha M (complement component 3 receptor 3, CD11b)
- CRP
C-reactive protein
- C4A
Complement component 4A
- C4B
Complement component 4B
- C2
Complement component 2
- C1Q
Complement component 1q
- UBE2L3
Ubiquitin-conjugating enzyme E2L 3
- MECP2
methyl CpG binding protein 2
- PXK
PX domain containing serine/threonine kinase
- XKR6
X Kell blood group precursor-related family member 6
- SCUBE1
Signal peptide-CUB domain-EGF-related
- TRAF3
TNF receptor-associated factor 3
- OPN
Osteopontin
- ODN
Oligodeoxynucleotides
- NFkB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- TNF-α
Tumor necrosis factor alpha
- BAFF/BlyS
B-cell activating factor/B lymphocyte stimulator
- TACI
Transmembrane activator and calcium modulator and cyclophilin ligand interactor
Footnotes
Reprints and permissions: http://www.sagepub.co.uk/journalsPermissions.nav
References
- 1.Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;358:929–939. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 2.Moser KL, Kelly JA, Lessard CJ, Harley JB. Recent insights into the genetic basis of systemic lupus erythematosus. Genes Immun. 2009;10:373–379. doi: 10.1038/gene.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harley IT, Kaufman KM, Langefeld CD, Harley JB, Kelly JA. Genetic susceptibility to SLE: new insights from fine mapping and genome-wide association studies. Nat Rev Genet. 2009;10:285–290. doi: 10.1038/nrg2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gateva V, Sandling JK, Hom G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mathis D, Benoist C. Levees of immunological tolerance. Nat Immunol. 2010;11:3–6. doi: 10.1038/ni.1833. [DOI] [PubMed] [Google Scholar]
- 6.Prokunina L, Castillejo-Lopez C, Oberg F, et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet. 2002;32:666–669. doi: 10.1038/ng1020. [DOI] [PubMed] [Google Scholar]
- 7.Graham RR, Kyogoku C, Sigurdsson S, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci U S A. 2007;104:6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remmers EF, Plenge RM, Lee AT, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harley JB, Alarcon-Riquelme ME, Criswell LA, et al. Genomewide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hom G, Graham RR, Modrek B, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 11.Kozyrev SV, Abelson AK, Wojcik J, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 12.Nath SK, Han S, Kim-Howard X, et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40:152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 13.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham RR, Hom G, Ortmann W, Behrens TW. Review of recent genome-wide association scans in lupus. J Intern Med. 2009;265:680–688. doi: 10.1111/j.1365-2796.2009.02096.x. [DOI] [PubMed] [Google Scholar]
- 15.Jacob CO, Zhu J, Armstrong DL, et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2009;106:6256–6261. doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunninghame Graham DS, Vyse TJ, Fortin PR, et al. Association of LY9 in UK and Canadian SLE families. Genes Immun. 2008;9:93–102. doi: 10.1038/sj.gene.6364453. [DOI] [PubMed] [Google Scholar]
- 17.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol. 2007;179:4704–4710. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 18.Arechiga AF, Habib T, He Y, et al. Cutting edge: the PTPN22 allelic variant associated with autoimmunity impairs B cell signaling. J Immunol. 2009;182:3343–3347. doi: 10.4049/jimmunol.0713370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coxon A, Rieu P, Barkalow FJ, et al. A novel role for the beta 2 integrin CD11b/CD18 in neutrophil apoptosis: a homeostatic mechanism in inflammation. Immunity. 1996;5:653–666. doi: 10.1016/s1074-7613(00)80278-2. [DOI] [PubMed] [Google Scholar]
- 20.Arbuckle MR, McClain MT, Rubertone MV, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349:1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 21.Casciola-Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179:1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casciola-Rosen L, Andrade F, Ulanet D, Wong WB, Rosen A. Cleavage by granzyme B is strongly predictive of autoantigen status: implications for initiation of autoimmunity. J Exp Med. 1999;190:815–826. doi: 10.1084/jem.190.6.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 24.Brinkmann V, Zychlinsky A. Beneficial suicide: why neutrophils die to make NETs. Nat Rev Microbiol. 2007;5:577–582. doi: 10.1038/nrmicro1710. [DOI] [PubMed] [Google Scholar]
- 25.Hirsch JG. Bactericidal action of histone. J Exp Med. 1958;108:925–944. doi: 10.1084/jem.108.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu L, Kaliyaperumal A, Boumpas DT, Datta SK. Major peptide autoepitopes for nucleosome-specific T cells of human lupus. J Clin Invest. 1999;104:345–355. doi: 10.1172/JCI6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaels MA, Kang HK, Kaliyaperumal A, Satyaraj E, Shi Y, Datta SK. A defect in deletion of nucleosome-specific autoimmune T cells in lupus-prone thymus: role of thymic dendritic cells. J Immunol. 2005;175:5857–5865. doi: 10.4049/jimmunol.175.9.5857. [DOI] [PubMed] [Google Scholar]
- 28.Walport MJ. Complement. Second of two parts. N Engl J Med. 2001;344:1140–1144. doi: 10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- 29.Manderson AP, Botto M, Walport MJ. The role of complement in the development of systemic lupus erythematosus. Annu Rev Immunol. 2004;22:431–456. doi: 10.1146/annurev.immunol.22.012703.104549. [DOI] [PubMed] [Google Scholar]
- 30.Harley JB, Kelly JA, Kaufman KM. Unraveling the genetics of systemic lupus erythematosus. Springer Semin Immunopathol. 2006;28:119–130. doi: 10.1007/s00281-006-0040-5. [DOI] [PubMed] [Google Scholar]
- 31.Lood C, Gullstrand B, Truedsson L, et al. C1q inhibits immune complex-induced interferon-alpha production in plasmacytoid dendritic cells: a novel link between C1q deficiency and systemic lupus erythematosus pathogenesis. Arthritis Rheum. 2009;60:3081–3090. doi: 10.1002/art.24852. [DOI] [PubMed] [Google Scholar]
- 32.Vilcek J. Fifty years of interferon research: aiming at a moving target. Immunity. 2006;25:343–348. doi: 10.1016/j.immuni.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 34.Noppert SJ, Fitzgerald KA, Hertzog PJ. The role of type I interferons in TLR responses. Immunol Cell Biol. 2007;85:446–457. doi: 10.1038/sj.icb.7100099. [DOI] [PubMed] [Google Scholar]
- 35.van Boxel-Dezaire AHH, Rani MRS, Stark GR. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Preble OT, Black RJ, Friedman RM, Klippel JH, Vilcek J. Systemic lupus erythematosus: presence in human serum of an unusual acid-labile leukocyte interferon. Science. 1982;216:429–431. doi: 10.1126/science.6176024. [DOI] [PubMed] [Google Scholar]
- 37.Ronnblom LE, Alm GV, Oberg KE. Possible induction of systemic lupus erythematosus by interferon-alpha treatment in a patient with a malignant carcinoid tumour. J Intern Med. 1990;227:207–210. doi: 10.1111/j.1365-2796.1990.tb00144.x. [DOI] [PubMed] [Google Scholar]
- 38.Vallin H, Blomberg S, Alm GV, Cederblad B, Ronnblom L. Patients with systemic lupus erythematosus (SLE) have a circulating inducer of interferon-alpha (IFN-alpha) production acting on leucocytes resembling immature dendritic cells. Clin Exp Immunol. 1999;115:196–202. doi: 10.1046/j.1365-2249.1999.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallin H, Perers A, Alm GV, Ronnblom L. Anti-double-stranded DNA antibodies and immunostimulatory plasmid DNA in combination mimic the endogenous IFN-alpha inducer in systemic lupus erythematosus. J Immunol. 1999;163:6306–6313. [PubMed] [Google Scholar]
- 40.Ronnblom L, Alm GV. A pivotal role for the natural interferon alpha-producing cells (plasmacytoid dendritic cells) in the pathogenesis of lupus. J Exp Med. 2001;194:F59–F63. doi: 10.1084/jem.194.12.f59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovgren T, Eloranta ML, Bave U, Alm GV, Ronnblom L. Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum. 2004;50:1861–1872. doi: 10.1002/art.20254. [DOI] [PubMed] [Google Scholar]
- 42.Siegal FP, Kadowaki N, Shodell M, et al. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 43.Baechler EC, Batliwalla FM, Karypis G, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennett L, Palucka AK, Arce E, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee-Kirsch MA, Gong M, Chowdhury D, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- 46.Feng D, Stone RC, Eloranta ML, et al. Genetic variants and disease-associated factors contribute to enhanced interferon regulatory factor 5 expression in blood cells of patients with systemic lupus erythematosus. Arthritis Rheum. 2010;62:562–573. doi: 10.1002/art.27223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palucka AK, Banchereau J, Blanco P, Pascual V. The interplay of dendritic cell subsets in systemic lupus erythematosus. Immunol Cell Biol. 2002;80:484–488. doi: 10.1046/j.1440-1711.2002.01112.x. [DOI] [PubMed] [Google Scholar]
- 48.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–92. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 49.Yao Y, Richman L, Higgs BW, et al. Neutralization of interferonalpha/beta-inducible genes and downstream effect in a phase I trial of an anti-interferon-alpha monoclonal antibody in systemic lupus erythematosus. Arthritis Rheum. 2009;60:1785–1796. doi: 10.1002/art.24557. [DOI] [PubMed] [Google Scholar]
- 50.Ronnblom L, Alm GV, Eloranta ML. Type I interferon and lupus. Curr Opin Rheumatol. 2009;21:471–477. doi: 10.1097/BOR.0b013e32832e089e. [DOI] [PubMed] [Google Scholar]
- 51.Cella M, Jarrossay D, Facchetti F, et al. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- 52.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 53.Kadowaki N, Ho S, Antonenko S, et al. Subsets of human dendritic cell precursors express different Toll-like receptors and respond to different microbial antigens. J Exp Med. 2001;194:863–869. doi: 10.1084/jem.194.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Izaguirre A, Barnes BJ, Amrute S, et al. Comparative analysis of IRF and IFN-alpha expression in human plasmacytoid and monocyte-derived dendritic cells. J Leukoc Biol. 2003;74:1125–1138. doi: 10.1189/jlb.0603255. [DOI] [PubMed] [Google Scholar]
- 55.Honda K, Ohba Y, Yanai H, et al. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 56.Honda K, Yanai H, Negishi H, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 57.Ronnblom L, Pascual V. The innate immune system in SLE: type I interferons and dendritic cells. Lupus. 2008;17:394–399. doi: 10.1177/0961203308090020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grouard G, Rissoan MC, Filgueira L, Durand I, Banchereau J, Liu YJ. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J Exp Med. 1997;185:1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dzionek A, Sohma Y, Nagafune J, et al. BDCA-2, a novel plasmacytoid dendritic cell-specific type II C-type lectin, mediates antigen capture and is a potent inhibitor of interferon alpha/beta induction. J Exp Med. 2001;194:1823–1834. doi: 10.1084/jem.194.12.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cederblad B, Blomberg S, Vallin H, Perers A, Alm GV, Ronnblom L. Patients with systemic lupus erythematosus have reduced numbers of circulating natural interferon-alpha-producing cells. J Autoimmun. 1998;11:465–470. doi: 10.1006/jaut.1998.0215. [DOI] [PubMed] [Google Scholar]
- 61.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294:1540–1543. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 62.Blomberg S, Eloranta ML, Cederblad B, Nordlin K, Alm GV, Ronnblom L. Presence of cutaneous interferon-alpha producing cells in patients with systemic lupus erythematosus. Lupus. 2001;10:484–490. doi: 10.1191/096120301678416042. [DOI] [PubMed] [Google Scholar]
- 63.Farkas L, Beiske K, Lund-Johansen F, Brandtzaeg P, Jahnsen FL. Plasmacytoid dendritic cells (natural interferon-alpha/beta-producing cells) accumulate in cutaneous lupus erythematosus lesions. Am J Pathol. 2001;159:237–243. doi: 10.1016/s0002-9440(10)61689-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tucci M, Quatraro C, Lombardi L, Pellegrino C, Dammacco F, Silvestris F. Glomerular accumulation of plasmacytoid dendritic cells in active lupus nephritis: role of interleukin-18. Arthritis Rheum. 2008;58:251–262. doi: 10.1002/art.23186. [DOI] [PubMed] [Google Scholar]
- 65.Obermoser G, Schwingshackl P, Weber F, et al. Recruitment of plasmacytoid dendritic cells in ultraviolet irradiation-induced lupus erythematosus tumidus. Br J Dermatol. 2009;160:197–200. doi: 10.1111/j.1365-2133.2008.08873.x. [DOI] [PubMed] [Google Scholar]
- 66.Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Moroy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 67.Botto M, Dell’Agnola C, Bygrave AE, et al. Homozygous C1q deficiency causes glomerulonephritis associated with multiple apoptotic bodies. Nat Genet. 1998;19:56–59. doi: 10.1038/ng0598-56. [DOI] [PubMed] [Google Scholar]
- 68.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang Y-h, Homey B. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007:449. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 69.Tian J, Avalos AM, Mao SY, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 70.Latz E, Schoenemeyer A, Visintin A, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 71.Bave U, Magnusson M, Eloranta ML, Perers A, Alm GV, Ronnblom L. Fc gamma RIIa is expressed on natural IFN-alpha-producing cells (plasmacytoid dendritic cells) and is required for the IFN-alpha production induced by apoptotic cells combined with lupus IgG. J Immunol. 2003;171:3296–3302. doi: 10.4049/jimmunol.171.6.3296. [DOI] [PubMed] [Google Scholar]
- 72.Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Invest. 2005;115:407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sano H, Morimoto C. DNA isolated from DNA/anti-DNA antibody immune complexes in systemic lupus erythematosus is rich in guanine-cytosine content. J Immunol. 1982;128:1341–1345. [PubMed] [Google Scholar]
- 74.Barrat FJ, Meeker T, Gregorio J, et al. Nucleic acids of mammalian origin can act as endogenous ligands for Toll-like receptors and may promote systemic lupus erythematosus. J Exp Med. 2005;202:1131–1139. doi: 10.1084/jem.20050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vollmer J, Tluk S, Schmitz C, et al. Immune stimulation mediated by autoantigen binding sites within small nuclear RNAs involves Toll-like receptors 7 and 8. J Exp Med. 2005;202:1575–1585. doi: 10.1084/jem.20051696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nat Rev Immunol. 2006;6:823–835. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Savarese E, Chae OW, Trowitzsch S, et al. U1 small nuclear ribonucleoprotein immune complexes induce type I interferon in plasmacytoid dendritic cells through TLR7. Blood. 2006;107:3229–3234. doi: 10.1182/blood-2005-07-2650. [DOI] [PubMed] [Google Scholar]
- 78.Krieg AM, Vollmer J. Toll-like receptors 7, 8, and 9: linking innate immunity to autoimmunity. Immunol Rev. 2007;220:251–269. doi: 10.1111/j.1600-065X.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 79.Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- 80.Barrat FJ, Coffman RL. Development of TLR inhibitors for the treatment of autoimmune diseases. Immunol Rev. 2008;223:271–283. doi: 10.1111/j.1600-065X.2008.00630.x. [DOI] [PubMed] [Google Scholar]
- 81.Vollmer J, Krieg AM. Immunotherapeutic applications of CpG oligodeoxynucleotide TLR9 agonists. Adv Drug Deliv Rev. 2009;61:195–204. doi: 10.1016/j.addr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 82.Rutz M, Metzger J, Gellert T, et al. Toll-like receptor 9 binds single-stranded CpG-DNA in a sequence- and pH-dependent manner. Eur J Immunol. 2004;34:2541–2550. doi: 10.1002/eji.200425218. [DOI] [PubMed] [Google Scholar]
- 83.Lafyatis R, York M, Marshak-Rothstein A. Antimalarial agents: closing the gate on Toll-like receptors? Arthritis Rheum. 2006;54:3068–3070. doi: 10.1002/art.22157. [DOI] [PubMed] [Google Scholar]
- 84.Banchereau J, Briere F, Caux C, et al. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 85.Santini SM, Lapenta C, Logozzi M, et al. Type I interferon as a powerful adjuvant for monocyte-derived dendritic cell development and activity in vitro and in Hu-PBL-SCID mice. J Exp Med. 2000;191:1777–1788. doi: 10.1084/jem.191.10.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ehrenstein MR, Evans JG, Singh A, et al. Compromised function of regulatory T cells in rheumatoid arthritis and reversal by anti-TNFalpha therapy. J Exp Med. 2004;200:277–285. doi: 10.1084/jem.20040165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blair PA, Norena LY, Flores-Borja F, et al. CD19(+)CD24(hi)CD38(hi) B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic Lupus Erythematosus patients. Immunity. 2010;32:129–140. doi: 10.1016/j.immuni.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 88.Blanco P, Pitard V, Viallard JF, Taupin JL, Pellegrin JL, Moreau JF. Increase in activated CD8+T lymphocytes expressing perforin and granzyme B correlates with disease activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2005;52:201–211. doi: 10.1002/art.20745. [DOI] [PubMed] [Google Scholar]
- 89.Ganguly D, Chamilos G, Lande R, et al. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J Exp Med. 2009;206:1983–1994. doi: 10.1084/jem.20090480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jego G, Palucka AK, Blanck JP, Chalouni C, Pascual V, Banchereau J. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity. 2003;19:225–234. doi: 10.1016/s1074-7613(03)00208-5. [DOI] [PubMed] [Google Scholar]
- 91.Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 92.Baccala R, Hoebe K, Kono DH, Beutler B, Theofilopoulos AN. TLR-dependent and TLR-independent pathways of type I interferon induction in systemic autoimmunity. Nat Med. 2007;13:543–551. doi: 10.1038/nm1590. [DOI] [PubMed] [Google Scholar]