Abstract
The generation of recombinant single-chain antibodies from either non-immune or immune phage display antibody libraries is an effective means to obtain high affinity antibodies against a specific target. Non-immune libraries contain a wide variety of antibodies but these are often low affinity. Immune libraries contain a high frequency of high affinity antibodies, but are typically limited to a single antigen. Due to the VH and VL recombination that occurs during antibody library construction, we hypothesized that an immune antibody library produced against one member of a protein family would contain antibodies specific for other members of the same protein family. Here, we tested this hypothesis by mining an existing anti-human Toll-like receptor-2 (hTLR2) antibody library for antibodies specific for other members of the TLR family. This procedure, referred to as homolog mining, proved to be effective. Using a cell-based system to pan and screen the anti-TLR2 library, we identified single chain antibodies specific for three of the four hTLR2 homologs we targeted. The antibodies identified, anti-murine TLR2, anti-hTLR5, and anti-hTLR6, bind specifically to their target, with no cross-reactivity to hTLR2 or other TLRs tested. These results demonstrate that combinatorial re-assortment of VH and VL fragments from multiple sources during Ab library construction increases Ab repertoire complexity, allowing antibody libraries produced by immunization with one antigen to be used to obtain antibodies specific to related antigens. The principle of homolog mining may be extended to other protein families and will facilitate and accelerate antibody production processes.
Keywords: Toll-like receptor, recombinant antibody, scFv, phage-displayed antibody library, cell surface antigen, flow cytometry
1. Introduction
The production of single chain variable fragment (scFv) antibodies using phage display (McCafferty et al., 1990; Winter et al., 1994; Marks and Marks, 1996) has several advantages over traditional hybridoma technologies (Kohler and Milstein, 1975). First, the number of phage antibodies that can be generated is much larger than the number of hybridomas that can be produced in the same period of time, or from the same number of immunized animals (Carmen and Jermutus, 2002). Second, large numbers of phage antibodies can be screened rapidly, in some cases using high-throughput methods (Winter et al., 1994). Third, during phage library construction, the heavy chain variable regions (VH) and light chain variable regions (VL) of individual B cells are randomly reassorted to create unique antibodies that were not present in the immune repertoire of the donor animal. In this manner, phage display can yield antibodies specific for epitopes that might not have generated monoclonal antibodies due to tolerance as well as antibodies against antigens that are too toxic to use as immunogens.
ScFvs are usually generated using one of two general strategies. The first involves screening non-immune phage libraries. Non-immune libraries are typically highly complex (109–1011 clones), consisting of scFvs formed from the VH and VL fragments of a large number of naïve donor animals or humans (Sheets et al., 1998; Huie et al., 2001; Amersdorfer et al., 2002; Carmen and Jermutus, 2002). The advantage of this strategy is that a single highly complex library can be used to screen for many different targets. The disadvantages of this strategy are that the frequency of scFvs specific for a given Ag are low, thereby necessitating a large amount of screening, and that the affinity of the scFvs identified are often only moderate, making it necessary to employ affinity maturation through mutagenesis to achieve the required affinity (Kontermann and Dubel, 2001; Amersdorfer et al., 2002) The alternative strategy is to screen immune libraries, which are generated from the B cells of immunized animals or humans after an infection. Immune libraries are typically smaller (105–106 clones) than non-immune libraries but usually contain a much higher frequency of scFvs that bind with high affinity to the Ag used for immunization. For this reason, immune libraries typically yield a greater number of candidate scFvs than non-immune libraries. The disadvantage of using immune libraries is that a new library must be used for each target Ag. This involves both a dedicated series of immunized animals or patient samples and the effort required for library construction itself.
The utility of immune phage display antibody libraries would be improved if they could be used to obtain antibodies against multiple distinct target antigens. We hypothesized that, due to the reassortment of VH and VL regions, an immune library generated against one antigen would contain scFvs that recognize members of the same protein family at a reasonable frequency and thereby eliminate the need to generate immune libraries for each family member. Here, we tested this hypothesis by mining an existing anti-human Toll-like receptor-2 (hTLR2) library for scFvs that are specific for other members of the Toll-like receptor (TLR) family. TLRs are important mediators of innate immune response, but investigation of these receptors has been hampered by a lack of antibodies capable of binding to cell surface TLRs. Anti-TLR antibody generation has proven problematic because TLRs can be difficult to express as recombinant proteins and because antibodies raised against TLR proteins often fail to recognize these proteins in their native form expressed on the cell surface (Lipes et al., 2008). In previous work we addressed these problems by establishing an entirely cell-based procedure to produce scFvs that bind to native proteins on the surface of cells (Lipes et al., 2008). This allowed the generation of numerous scFvs that bind cell-surface hTLR2. In the current study, we sought to extend the use of our existing anti-TLR2 scFv library by using our cell-based system to select scFvs specific for other members of the TLR family from this library. This strategy, which we refer to as homolog mining, proved to be feasible, as we were able to identify scFvs specific for mTLR2, hTLR5, and hTLR6 that did not cross-react with the original immunogen, hTLR2. These results demonstrate that a single immune library can be mined for recombinant Abs against multiple homologous cell surface proteins using a procedure that efficiently selects for scFvs that bind those proteins in their native form. This strategy should extend the utility of existing immune phage display libraries and provide a marked increase in the efficiency with which Abs against families of complex cell surface proteins can be generated.
2. Materials and Methods
2.1 TLR expression constructs and cell lines
mTLR2-HA-pUNO and hTLR2-HA-pUNO plasmids were purchased from InvivoGen (San Diego, CA). The 300.19 mouse tumor cell line (Kearney et al., 1979) (kindly provided by Dr. T. Tedder, Duke University, Durham, NC) was transfected with hTLR2-HA-pUNO by electroporation and used for immunizations. For library selection and flow cytometry assays, HEK293 cells (ATCC, Manassas, VA) were transfected with hTLR2-HA-pUNO or mTLR2-HA-pUNO using Superfect (Quiagen, Valencia, CA). hTLR1-HA-pUNO, hTLR5-HA-pUNO, and hTLR6-HA-pUNO transfected HEK cells were purchased from InvivoGen. The term TLR-HEK used in the following sections refers to all mTLR2, hTLR1, hTLR5, and hTLR6-HEKs, unless otherwise noted. Expression of TLRs was verified by probing Western blots of cell extracts with anti-HA antibody. The YFP-HEK cells for TLR-negative controls were kindly provided by Dr. R. Lefkowitz (Duke University).
2.2 scFv library panning by cell-based selection
Construction of the anti-hTLR2 immune phage library was described previously (Lipes et al., 2008). Panning of this library was performed using the biopanning and rapid analysis of selected interactive ligand (BRASIL) approach (Giordano et al., 2001) with slight modifications. All solutions and incubations were at 4 °C unless otherwise noted. HEK293 and TLR-HEK cells were harvested with Versene (Invitrogen, Carlsbad, CA), washed in Dulbecco’s PBS and resuspended at 107 cells/ml in DMEM (Invitrogen)/1% BSA Fraction V (Sigma, St. Louis, MO). An aliquot of anti-hTLR2 scFv phage library containing 5×109 M13 phage particles was added to 150 ul of HEK293 cells suspension and rotated for two hours. This solution was layered on top of 200 ul of a mixture of dibutyl phthalate:cyclohexane (15:1) (Sigma) and spun at 10,000g in a microfuge for 10 minutes. The aqueous phase containing pre-cleared phage was used to resuspend a cell pellet containing 1.5×106 TLR-HEK cells. This suspension was rotated for 1.5 hours. The cells were washed three times in 200 ul DMEM/1% BSA, then resuspended in 200 ul DMEM/1% BSA and layered over 200 ul of dibutyl phthalate:cyclohexane (15:1). After centrifugation (10000g × 10 min), the tube was flash frozen in liquid nitrogen. The cell pellet with bound phage in the tube tip was removed with a tube cutter (GeneMate). The cell pellet was resuspended in 10 ml of log-phase TG-1 cells. Following a 30 minute incubation at 37°C, the cells were concentrated by centrifugation and plated on two 15 cm 2xYT/AG plates. The plates were incubated for 16 hours at 30°C. The resulting colonies were scraped into 2xYT and used to make a glycerol frozen stock
2.3 Phage Rescue
M13 phage particles for selections and screening were obtained by rescue with M13K07 phage (New England Biolabs, Ipswich, MA). To prepare concentrated phage, 12 ml 2xYT/AG (2xYT supplemented with 2% glucose and 0.1 mg/ml ampicillin) was inoculated with pooled or individual TG-1 clones and grown at 37 °C until OD600 reached 0.7. 2×1011 M13K07 helper phage were added and allowed to infect for 30 minutes at 37°C then the culture was shaken at 200 rpm for 30 minutes at 37°C. Bacteria were pelleted at 3500g for 10 minutes and the medium replaced with 50 ml of 2xYT supplemented with ampicillin and kanamycin. The culture was shaken at 37°C for 30 minutes, then at 30°C for 16 hours. The culture was centrifuged as above, and the supernatant subjected to sequential precipitations in 0.2 volumes of ice-cold 20% PEG-8000/2.5 M NaCl to concentrate the phage particles. Phage particles were pelleted at 10,000g for 10 minutes. Following the last precipitation, the phage were resuspended in 1 ml PBS and stored at 4°C. Phage titers were determined by infections of TG-1 cells with serially diluted phage.
Individual phage clones were rescued for flow cytometry screens in sterile 2 ml/well 96-well plates (Continental Lab Products, San Diego, CA). Briefly, 400 ul of 2xYT/AG in each well was inoculated with a phage clone and shaken at 200 rpm for 16 hours at 30°C. Aliquots from the wells of this master plate were used to inoculate a rescue plate containing 400 ul of 2xYT/AG supplemented with 2×1010 phage/ml of M13K07 helper phage. The rescue plate was shaken at 37°C for 4 hours. After centrifugation (3000g, 10 minutes), bacteria pellets were resuspended in 400 ul of 2xYT/AK and shaken for 16 hours at 30°C. The plate was centrifuged again as above to pellet bacteria and the supernatant containing phage particles was transferred to a fresh plate for screening assays.
2.4 Flow cytometric screening and clone validation
Procedures for phage clone screening and phage and purified scFv binding assays were performed as previously described (Lipes et al., 2008). Briefly, TLR-HEK and YFP-HEK were mixed in equal numbers, plated in 96-well U-bottom plates at 5 ×105 cells/well, stained with phage or scFv preparations as primary reagents, washed, stained with secondary and tertiary reagents, and subjected to flow cytometric analysis using a BD LSRII flow cytometer. Data were analyzed using Flowjo software (TrecStar Inc, Ashland, OR). To determine TLR-specific binding, TLR-HEK and YFP-HEK cell populations were gated separately based on YFP fluorescence in the FITC channel, and the mean fluorescence intensity (MFI) of allophycocyanin (APC) was determined for each population. TLR-specific staining was calculated as the TLR-HEK MFI minus the YFP-HEK MFI of APC. Staining of cells with purified biotinylated scFvs was performed on mTLR2-HEK, J774A.1 (Ralph and Nakoinz, 1977b) cells, and mouse lung cells (see below). The staining procedure was as above except the staining buffer contained 5% normal mouse serum and 5% normal rat serum. 1 ug/ml biotinylated scFvs were used as the primary staining reagent, and 0.2 ug/ml allophycocyanin-conjugated streptavidin (BD Biosciences, San Jose, CA) was used as the secondary staining reagent.
2.5 DNA fingerprinting
ScFv inserts obtained by PCR from infected bacteria (Gussow and Clackson, 1989) were subjected to restriction fragment length polymorphism or “DNA fingerprint” analysis. PCR was performed using 2ul of TG-1 culture, 200 uM dNTPs, 200 uM pCANTAB 5E primers (scFvGWfwd: GCGGCCCAGCCGGCC; scFvGWrev: CTGGAACCGCGTG), 3 units EasyA DNA polymerase (Stratagene, La Jolla, CA), and manufacturer’s buffer in 50 ul. After amplification (95°C for 2 min, 30 cycles of 95°C × 30 sec, 58°C for 30 sec, 72°C for 1 min; and 72°C for 10 minutes), aliquots of the product were digested with Sau3A I or BstN I (New England Biolabs) then fractionated on 2% agarose TAE gels.
2.6 Expression of soluble scFvs in Drosophila
ScFv inserts were amplified from pCANTAB 5E (GE Life Sciences, Buckinghamshire, UK) using above PCR conditions and ligated with topoisomerase into the Gateway entry vector pCR8Topo (Invitrogen). The Drosophila expression vector pMTBiP/V5His (Invitrogen) was converted to a Gateway destination vector by replacing the multiple cloning sequence with recombination sites for the bacteriophage enzyme LRII (Invitrogen) and a BirA Avitag site (Avidity, Denver, CO). After recombining scFv inserts into pMTBiP, the resulting plasmids and pCoBlast (Invitrogen) were co-transfected into Schneider 2 (S2) cells using Cellfectin (Invitrogen) and selected in 25 mg/ml blasticidin to obtain stable tranfectants. ScFv expression was induced with 0.75 mM CuSO4 in Sf900-II serum-free medium (Invitrogen) for 36 to 48 hours at 28°C. ScFvs were purified from culture supernatant by ion metal affinity chromatography (Talon resin, Clontech, Mountain View, CA), and subjected to SDS-PAGE with silver staining and BCA assay (Pierce, Rockford, IL) to assess purity and quantity. To make biotinylated-scFvs, purified scFvs were subject to in vitro BirA biotinylation (Avidity) as described in manufacture’s instructions.
2.7 Primary mouse lung cells
8–12 week-old C57BL/6 mice received 12.5 ug LPS intranasally and lung parenchymal cells were harvested after 24 hours as described previously (Lin et al., 2008). Briefly, lungs were perfused with 3 ml HBSS, minced and digested with 1 mg/ml of collagenase for 40 min at 37°C. Cells were dissociated by passing through a 70-um mesh strainer. Dendritic cells (DC) and macrophages were enriched by passing through a 17% Metrizamide gradient at room temperature. Red blood cells were lysed using AKC buffer (0.15 M NH4Cl, 10mM KHCO3, 0.1 mM EDTA, pH7.2–7.4). The resulting single cell suspensions were subjected to staining and flow cytometry analysis to identify neutrophils, DC, macrophages, and monocytes as described (Lin et al., 2008).
3. Results
3.1 Library enrichment and antibody clone screening against murine TLR2
To determine the feasibility of obtaining scFvs against hTLR2 homologs from an anti-hTLR2 library, we used the library, selection, and screening procedures we have previously described (Lipes et al., 2008). In brief, multiple BALB/c and C57BL/6 mice were immunized with hTLR2-transfected 300.19 cells and relative anti-hTLR2 IgG sera titers determined in a flow cytometric assay. High titer animals were used to construct an anti-hTLR2 M13 phage-scFv library, which had a theoretical complexity of 3×106. To enrich for clones specific for TLRs other than hTLR2, we employed a cell-based selection procedure, BRASIL, which relies on differential centrifugation to obtain Ag-specific phage clones (Giordano et al., 2001). In this procedure, the scFv phage library is first pre-cleared over parental HEK293 cells, subjected to differential centrifugation, and then the remaining phage in the supernatant are incubated with target TLR-HEK cells. Another differential centrifugation through an organic phase removes loosely associated non-specific phage. The phage remaining bound to TLR-HEK cells are then used to generate enriched phage stocks for iterative rounds of selections. Once enriched, the phage library is arrayed as individual clones in 96 well plates. The raw phage preparations, in plates, are screened for their ability to bind target (TLR-HEK) but not control (YFP-HEK) cells in a high throughput flow cytometric assay. The resulting positive clones are subjected to restriction fragment length polymorphism or “DNA fingerprint” analysis to eliminate duplicates, prepared as purified phage, and tested again by flow cytometry to confirm their specific binding to TLR-HEK cells. The scFv inserts of confirmed clones are subcloned into a Drosophila expression vector, expressed as soluble scFvs in Schneider 2 (S2) cells, purified, biotinylated, and validated for their ability to specifically bind TLR-HEK cells in a flow cytometric assay.
To test homolog mining, we first attempted to identify clones specific for mTLR2, the receptor most closely related to hTLR2. The hTLR2 scFv library was subjected to three iterative rounds of BRASIL selection using mTLR2-HEK cells for positive selection. 43 (51%) of the 84 clones screened in a flow cytometric assay were positive for mTLR2 binding (Table I, Fig. 1A). DNA fingerprinting of 32 clones revealed 18 (56%) to be unique (Table I). Nine of these unique clones were used to generate purified phage stocks and tested for their ability to bind mTLR2. Five (56%) of these nine anti-mTLR2 phage clones proved to be positive in this confirmatory test (Table I, Fig. 1B), and were subcloned and expressed as purified soluble scFvs in S2 cells. All five clones retained their capacity to specifically bind to mTLR2-HEK cells when expressed as soluble scFvs (Table I, Fig. 1B). Importantly, none of these anti-mTLR2 clones demonstrated binding to human TLR2 (Fig. 3 and data not shown).
Table 1.
Summary of numbers of clones and positive rates for screening/verification at each step of anti-TLR scFv generation.
| Screened by | Initial Screen - Raw Phage | Fingerprint | Purified phage Binding to cells | scFv binding to cells | |
|---|---|---|---|---|---|
|
| |||||
| # of clones screened | # of clones positive | # of unique clones | # of clones positive | # of clones positive | |
| hTLR2 HEK | 168 | 78 (46%) | 9/23 (39%) | 8/9 (89%) | 5/5 (100%) |
|
| |||||
| mTLR2-HEK | 84 | 43 (51%) | 18/32 (56%) | 5/9 (56%) | 5/5 (100%) |
|
| |||||
| hTLR5-HEK R3 | 186 | 39 (21%) | 18/39 (46%) | 2/17 (12%) | 1/2 (50%) |
|
|
|||||
| R4* | 92 | 22 (24%) | 2/19 (11%) | 1/2 (50%) | 1/2 (50%) |
|
| |||||
| hTLR6-HEK | 186 | 11 (6%) | 6/8 (75%) | 1/6 (17%) | 1/1 (100%) |
|
| |||||
| hTLR1-HEK | 94 | 0 (0%) | N/A | N/A | N/A |
Numbers in fractions indicate “number of positive clones/number of clones tested” and percentages in parentheses indicate positive rates for each test.
Results after performing a 4th round of selection (R4) for hTLR5 versus the standard 3 rounds (R3) of selection.
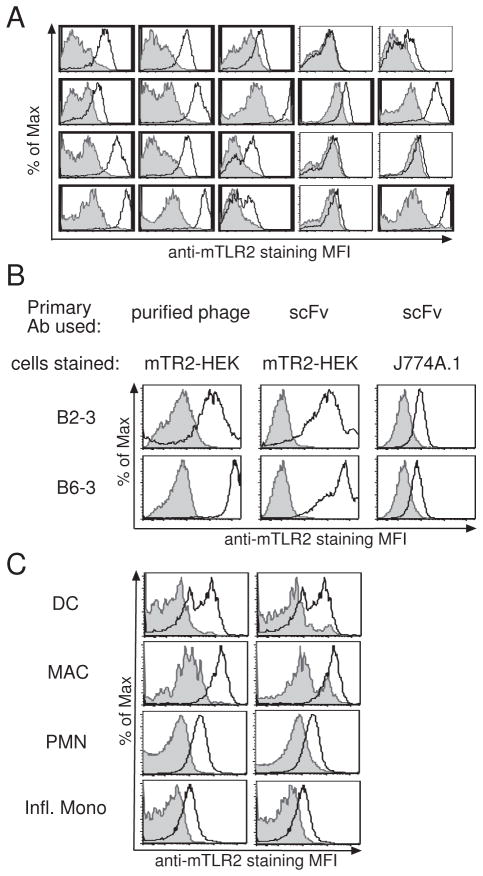
Figure 1. Identification and characterization of anti-mTLR2 antibody clones.
(A) Screening anti-mTLR2 antibody clones. Raw phage preparations of individual clone were used to stain mTLR2-HEK (white) or YFP-HEK (gray) cells. 20 representative clones are shown here. Positive clones are indicated in bold. (B) Validation of anti-mTLR2 binding. Purified phage (left) or corresponding scFvs (center) were used as primary Ab to stain on mTLR2-HEK cells (white) or YFP-HEK (gray). Right panel shows J774A.1 cells stained with anti-mTLR2 scFvs (white) or with anti-Tie2 scFv (gray). (C) mTLR2-scFv bind to primary mouse lung cells. Purified anti-mTLR2 scFv was used as primary Ab to stain mouse lung cells (white). In the left panel, anti-Tie2 antibody was used as a negative control antibody (gray). In the right panel, cells harvested from a mTLR2 konck-out mouse were used as negative control cells (gray). Lungs were harvested from intranasally LPS-treated mouse and myeloid cells populations were identified as described by Lin et al.
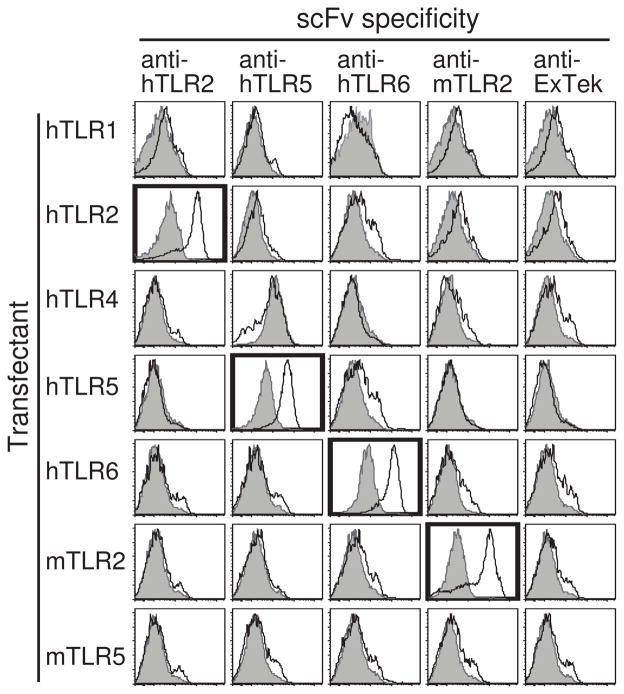
Figure 3. Anti-TLR antibody clones specifically bind their target Ag but not other TLRs.
scFvs against hTLR2 (clone B11), hTLR5 (clone 3B5), hTLR6 (clone 3F7), mTLR2 (clone B2–3), or Tie2 extra-cellular domain (ExTek, clone 1D6) were used as primary antibodies to stain hTLR1-, hTLR2-, hTLR4-, hTLR5-, hTLR6-, mTLR2- or mTLR5-HEK (white), with YFP-HEK as negative control cells (gray). All scFv clones obtained to date show the same specificity to their target TLR Ag and the result of one representative clone is shown here for each group. Positive stainings are outlined in bold.
To determine whether the anti-mTLR2 scFvs identified are able to detect endogenous mTLR2, these clones were tested for their ability to stain a murine monocyte/macrophage cell line, J774A.1 (Ralph and Nakoinz, 1977a). Despite the low level of mTLR2 expression in this cell line, all scFvs were able to detect mTLR2 on the surface of these cells (Fig. 1B). One anti-mTLR2 scFv (D2–3) was also tested for its ability to stain primary lung cells. As shown in figure 1C, clone D2–3 detected expression of mTLR2 on inflammatory monocytes, neutrophils, macrophages, and dendritic cells obtained from the lungs of LPS-treated mice. No such staining was seen on cells obtained from TLR2-deficient mice or when staining was performed with an irrelevant scFv (anti-Tie2 clone ExTek 1D6). These findings demonstrate that it is possible to obtain anti-mTLR2 antibodies that have sufficient affinity to detect endogenous mTLR2 from a library raised against hTLR2.
3.2 Homolog mining for anti-hTLR1, hTLR5, and hTLR6 scFvs
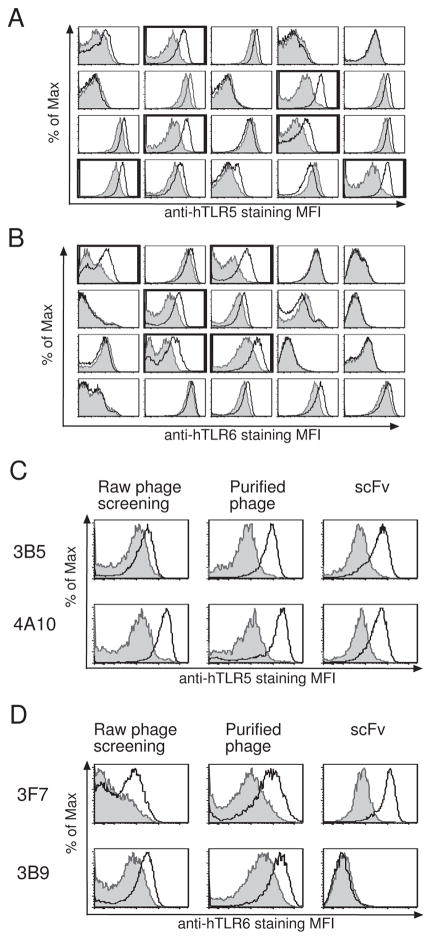
Having demonstrated that homolog mining works in principle, we sought to test this procedure with other, less homologous, members of the TLR family. Separate cell-based selections of the hTLR2 library were performed as above for hTLR1, hTLR5 and hTLR6. After 3 rounds of selection, arrays of the enriched clones were screened in a flow cytometric assay. These initial screens yielded 39 (21%)of 186 positive clones for hTLR5 and 11 (6%) of 186 positive clones for hTLR6 (Table 1, Fig. 2A and 2B). No positive clones were obtained for hTLR1 from a screen of 94 clones (Table 1). After elimination of duplicate clones by DNA fingerprinting (Table 1), the activity of clones prepared as purified phage was validated for binding to the selected TLR. When tested by flow cytometry, 2 (12%) of 17 anti-hTLR5 phage clones and 1 (17%) of 6 anti-TLR6 clones bound their respective target (Fig. 2C, 2D and Table I). Without antibodies available to use as a positive controls in screening hTLR5 and hTLR6, it was more difficult to distinguish positive clones from background signals (Fig. 2A, 2B and data not shown) than it was for previous TLR2 measurements that included a positive control. This resulted in a higher rate of false positives for hTLR5 and hTLR6. When prepared as purified scFvs in Drosophila cells, 1 out of 2 (50%) anti-TLR5 scFvs bound to hTLR5-HEK cells, while 1 out of 1 (100%) anti-hTLR6 scFvs bound to hTLR6-HEK cells (Fig. 2C, 2D and Table I). In an attempt to obtain more anti-hTLR5 clones, we performed a fourth round of BRASIL selection for hTLR5. This resulted in similar frequency of positive clones in the initial screening step (22 out of 92, or 24%), but a much lower frequency of unique clones (2 out of 19, or 11%) than seen after three rounds of selection (Table I, hTLR5-HEK R4).
Figure 2. Identification of anti-hTLR5 or anti-hTLR6 antibody clones.
(A&B) Screening anti-hTLR5 (A) or anti-hTLR6 (B) antibody clones. Individual clones of raw phage preparation were used as primary Ab to stain hTLR5- (A, white), hTLR6- (B, white), or YFP- (A & B, gray) HEK cells. 20 representative clones were shown here. Positive clones are indicated in bold. (C&D) Flow cytometry staining results for two representative hTLR5 (C) or hTLR6 (D) antibody clones. Raw phage preparations (left), purified phage (center), or purified scFvs (right) were used as primary antibody to stain hTLR5- (C, white), hTLR6- (D, white) or YFP- (gray) HEK cells.
3.3 Specificity of TLR scFv clones
A potential pitfall of homolog mining is that it may select for scFvs that bind to protein domains conserved among multiple receptors and thereby only identify clones that cross-react with multiple TLRs. To ensure that the scFvs we identified are specific to individual TLRs, we examined the binding of purified anti-TLR scFvs mined from the hTLR2 library to all of the TLRs we have expressed. The anti-Tie2 scFv, ExTek clone 1D6, was again used as negative control. As shown in figure 3, all of the scFv clones we identified bound only to their target and displayed no cross-reactivity to other members of the TLR family. In the case of TLR2, scFvs demonstrated no cross-species reactivity. The anti-TLR scFvs isolated from the hTLR2 library thus exhibit exquisite specificity for their target TLR. This result suggests that homolog mining selects for scFvs that recognize epitopes that are not fully conserved among TLRs.
3.4 Origin of anti-homolog scFvs
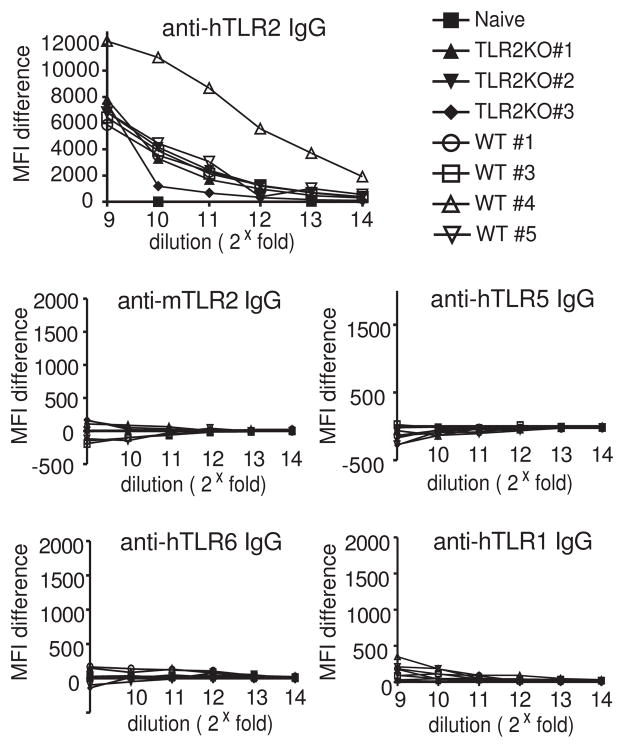
As shown above, our scFv phage display library, which was raised against hTLR2, contains clones specific for TLRs other than hTLR2. It is not clear if such clones represent antibody molecules formed in vivo after immunization or if they were created during library construction in the course of VH-VL re-assortment. To address this question, we examined the reactivity of the sera of the individual hTLR2-immunized mice used to construct the phage library. Using a flow cytometric assay we have previously described (Lipes et al., 2008), serial dilutions of each serum sample were tested for binding to hTLR1, hTLR2, mTLR2, hTLR5, and hTLR6.
As we have shown previously (Lipes et al., 2008), sera from responsive hTLR2-immunized mice contain detectable anti-TLR2 antibodies at dilutions up to 212 (Fig. 4). In contrast, the reactivity of these sera against hTLR1, mTLR2, hTLR5 and hTLR6 is no greater than that of unimmunized mice (Fig. 4). Antibodies against these TLRs are either not present in the original antibody repertoire or represent an extremely small fraction of it. This finding strongly suggests that the scFvs we identified are the products of combinatorial re-assortment of heavy and light chain variable fragments.
Figure 4. Sera from hTLR2-immunized mice does not contain detectable levels of IgG against hTLR1, hTLR5, hTLR6 or mTLR2.
Mice were immunized with hTLR2 by a cell-based immunization method and sera were harvested 7 days after the 5th boost. Mice sera were used as primary staining reagents in flow cytometry to test their binding on hTLR2-, mTLR2-, hTLR5-, hTLR6- or hTLR1-HEK, respectively. Ag-specific IgG titer was detected by secondary staining of APC-conjugated goat-anti-mouse IgG antibody. Serum from naïve mice were used as negative control.
4. Discussion
The screening of immune scFv phage display libraries is an effective means to generate large numbers of high affinity single-chain antibodies against a single antigen. The major impediment to the routine use of this strategy has been the need to construct a new library for each individual antigenic target. Here, we demonstrate that a novel strategy, homolog mining, can be used to significantly extend the utility of immune scFv libraries. By homolog mining an existing anti-hTLR2 library, we were able to obtain scFvs specific for three of the four hTLR2 homologs we targeted. These results demonstrate that, although not foolproof, homolog mining has the potential to markedly reduce the number of libraries required to generate antibodies against a family of proteins. It appears that homolog mining is widely applicable. Preliminary results in our lab indicate that an anti-Tie2 scFv library can be mined for antibodies against other receptor tyrosine kinases (Lipes and Kenan, manuscript in preparation).
Our findings demonstrate that a significant number of anti-homolog scFvs are present in our immune library. These were most likely generated via the combinatorial re-assortment of anti-hTLR2, and perhaps background, VH and VL fragments during library construction. Consistent with this view, we were unable to detect anti-homolog binding activity in the sera of the mice used for library construction. We have no way to assess the extent to which reassortment increased the diversity and complexity of our library over that from the cells from which it was derived. However, previous studies suggest that this increase can be significant. In one elegant study, transgenic mice were engineered to express a single immunoglobulin mu heavy chain derived from a vesicular stomatitis virus Indiana serotype-neutralizing (VSV-Ind) antibody (Senn et al., 2003). This heavy chain could not undergo rearrangements and was expressed in combination with endogenous kappa or lambda chains. The mice expressed a B cell repertoire biased heavily toward VSV-Ind, with 30 to 60 percent of the peripheral B cells showing VSV-Ind specificity. Despite this bias, the mice were able to mount successful B cell responses when immunized with the VSV-New Jersey serotype virus as well as other unrelated antigens such as bacterial porins. These studies showed combinatorial L chain variability alone is capable of generating a complex antibody repertoire. In another study, induction of further VH and VL re-assortment with Cre recombinase in a naive scFv library increased its calculated theoretical complexity from 107 to 1011 (Sblattero and Bradbury, 2000; Cen et al., 2006). When this library was used in selections for 18 recombinant targets, an average of 6 specific antibodies were obtained per target.
By comparison, even though the number of antibodies contained in our anti-hTLR2 scFv library (3×106) is five orders of magnitude lower than the Cre-reassorted library and three to four orders of magnitude lower than the number contained in a highly complex non-immune library (Sheets et al., 1998; Carmen and Jermutus, 2002), we were able to obtain multiple specific phage clones for most targets by screening a single 96-well plate. This finding suggests that immunization with hTLR2 provides numerous affinity matured VH and VL regions directed at TLR epitopes that, when re-assorted, can bind TLRs other than the original immunogen. It would be reasonable to expect that scFvs generated by such re-assortment and selected against a homolog would bind to both the homolog and the original immunogen. However, all of the Abs generated from these selections bound only to their target TLR without cross-reactivity to the library immunogen hTLR2 or any other TLRs tested (Fig. 3). antigens. One possible explantation for this is that the entirely cell-based techniques for selection and screening employed here allowed the receptors to be fully post-translationally modified and to adopt their native, membrane bound structures, emphasizing subtle structural differences between homologs. Purified recombinant proteins often lack such post-translational modifications, do not fold into their native structures, and may have normally internal epitopes exposed. Homologous domains may therefore adopt more similar structures in these conditions, thereby increasing the potential for antibody cross-reactivity. In selections we have previously performed using recombinant hTLR2 protein, we saw appreciable cross-reactivity with recombinant hTLR4 protein (data not shown), suggesting that cell-based methods offer improved antibody specificity. The impressive level of specificity exhibited by our anti-TLR antibodies establishes the feasibility of using existing immune libraries for selections with related antigens, and suggests that cell-based selections may, in general, yield fewer cross-reactive antibodies than selections with recombinant proteins.
There are several practical aspects of homolog mining and scFv generation that should be emphasized. First, we have found that the ability to generate scFvs that bind to either the immunogen or its homologs is highly dependent on the initial response to immunization. In our hands, libraries generated from mice that display only low-titer responses to the immunogen have failed to generate functional scFvs. Second, the hTLR2 scFv library used in these studies was constructed by pooling material from six mice of two different strains. We believe that the strategy of incorporating multiple immune responses from multiple mouse strains into the library captures a more diverse antibody repertoire than would be obtained from a single animal or mouse strain. A similar approach has been used to harvest immune responses from patients with autoimmune diseases (de Wildt et al., 1996) and from convalescent serum (Marks and Winter, 1992; Amersdorfer et al., 2002) to make human immune libraries.
Another practical concern is determining the threshold at which anti-homolog clones are considered positive in initial screens and, thereby, the sensitivity and specificity of this assay. In instances where a control antibody is not available, setting this threshold correctly can be problematic. In addition, the concentration of phage in the raw phage preparations used for screening can vary markedly between wells, leading to large differences in signal that are independent of the actual binding affinity. The relatively high level of background staining seen in our anti-TLR5 and anti-TLR6 screens was likely due to a low level of expression of these TLRs on HEK cells, and perhaps a lower affinity of the scFvs specific for these TLRs. Thus, the high rate of false positives seen in these screens is not a general feature for this technique, but will depend on scFv affinities, target expression levels, and the criteria determined by the user to select positive clones.
In order to be certain our TLR positive clones were emerging as the result of successful enrichment through our cell-based selections, we screened the original library without enrichment as well as pools of phage resulting from only two rounds of enrichment. No phage clones binding to any TLRs were seen in either instance (data not shown). A fourth round of selection performed for some of the receptors did not yield a more enriched population, but rather a much less diverse pool of positive clones (Table 1, hTLR5-HEK R4 and data not shown) as well as a distinct population of WT-HEK binding clones (data not shown). Thus, three rounds of enrichment were required and optimal to obtain positive clones. Taken together, these results indicate the anti-mTLR2, -hTLR5, and -hTLR6 antibody clones generated were not random results of large scale screening of the anti-hTLR2 library.
The exception to our successful screening results was the hTLR1 selection, in which we did not discover any anti-hTLR1 antibodies among the 94 clones screened. A more exhaustive screen may have revealed some positive clones but the more likely explanation is that a low level of hTLR1 expression in the cells used for panning may have not permitted sufficient enrichment during iterative selection. Western blot measurements showed that expression of hTLR1 in HEK cells was lower than that of the other TLR HEK cell lines utilized (data not shown).
While these studies illustrate a valuable new application for immune antibody libraries, highly complex non-immune antibody libraries remain the best option to obtain antibodies to the widest variety of targets, particularly antigens that are non-immunogenic due to tolerance or agents that are too toxic to use for immunizations (Nowakowski et al., 2002). However, construction of non-immune libraries is difficult, requiring extensive reagent optimization to obtain vast amounts of variable region fragments and necessitating hundreds of electroporations. Thus, investigators without access to a non-immune library may wish to instead attempt selections using an immune library against an antigen related to their target of interest.
Acknowledgments
We thank John Whitesides, Patti McDermott, and Danielle King of Duke Human Vaccine Institute Flow Cytometry facility for their excellent technical assistance. This work was supported by NIH grant U19-AI056572.
Abbreviation used
- Ab
antibody
- Ag
antigen
- BSA
bovine serum albumin
- IgG
immunoglobulin G
- MFI
mean florescent intensity
- scFv
single-chain variable fragment
- hTLR
human toll-like receptor
- mTLR
murine toll-like receptor
- YFP
yellow fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amersdorfer P, Wong C, Smith T, Chen S, Deshpande S, Sheridan R, Marks JD. Genetic and immunological comparison of anti-botulinum type A antibodies from immune and non-immune human phage libraries. Vaccine. 2002;20:1640–1648. doi: 10.1016/s0264-410x(01)00482-0. [DOI] [PubMed] [Google Scholar]
- Carmen S, Jermutus L. Concepts in antibody phage display. Brief Funct Genomic Proteomic. 2002;1:189–203. doi: 10.1093/bfgp/1.2.189. [DOI] [PubMed] [Google Scholar]
- Cen X, Bi Q, Zhu S. Construction of a large phage display antibody library by in vitro package and in vivo recombination. Appl Microbiol Biotechnol. 2006;71:767–72. doi: 10.1007/s00253-006-0334-5. [DOI] [PubMed] [Google Scholar]
- de Wildt RM, Finnern R, Ouwehand WH, Griffiths AD, van Venrooij WJ, Hoet RM. Characterization of human variable domain antibody fragments against the U1 RNA-associated A protein, selected from a synthetic and patient-derived combinatorial V gene library. Eur J Immunol. 1996;26:629–39. doi: 10.1002/eji.1830260319. [DOI] [PubMed] [Google Scholar]
- Giordano RJ, Cardo-Vila M, Lahdenranta J, Pasqualini R, Arap W. Biopanning and rapid analysis of selective interactive ligands. Nat Med. 2001;7:1249–53. doi: 10.1038/nm1101-1249. [DOI] [PubMed] [Google Scholar]
- Gussow D, Clackson T. Direct clone characterization from plaques and colonies by the polymerase chain reaction. Nucleic Acids Research. 1989;17:4000–4003. doi: 10.1093/nar/17.10.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huie MA, Cheung MC, Muench MO, Becerril B, Kan YW, Marks JD. Antibodies to human fetal erythroid cells from a nonimmune phage antibody library. Proc Natl Acad Sci U S A. 2001;98:2682–7. doi: 10.1073/pnas.051631798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney JF, Radbruch A, Liesegang B, Rajewsky K. A New Mouse Myeloma Cell Line that Has Lost Immunoglobulin Expression but Permits the Construction of Antibody-Secreting Hybrid Cell Lines. J Immunol. 1979;123:1548–1550. [PubMed] [Google Scholar]
- Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Kontermann R, Dubel S. Antibody Engineering. Springer; New York: 2001. [Google Scholar]
- Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. CCR2+ Monocyte-Derived Dendritic Cells and Exudate Macrophages Produce Influenza-Induced Pulmonary Immune Pathology and Mortality. J Immunol. 2008;180:2562–2572. doi: 10.4049/jimmunol.180.4.2562. [DOI] [PubMed] [Google Scholar]
- Lipes BD, Chen YH, Ma H, Staats HF, Kenan DJ, Gunn MD. An Entirely Cell-Based System to Generate Single-Chain Antibodies against Cell Surface Receptors. Journal of Molecular Biology. 2008;379:261–272. doi: 10.1016/j.jmb.2008.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks C, Marks JD. Phage libraries--a new route to clinically useful antibodies. N Engl J Med. 1996;335:730–3. doi: 10.1056/NEJM199609053351008. [DOI] [PubMed] [Google Scholar]
- Marks JD, Winter G. An artificial immune system for making antibodies. Behring Inst Mitt. 1992:6–12. [PubMed] [Google Scholar]
- McCafferty DP, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: filamentous phage displaying antibody variable fragments. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- Nowakowski A, Wang C, Powers DB, Amersdorfer P, Smith TJ, Montgomery VA, Sheridan R, Blake R, Smith LA, Marks JD. Potent neutralization of botulinum neurotoxin by recombinant oligoclonal antibody. Proc Natl Acad Sci U S A. 2002;99:11346–11350. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph P, Nakoinz I. Antibody-Dependent Killing of Erythrocyte and Tumor Targets by Macrophage-Related Cell Lines: Enhancement by PPD and LPS. J Immunol. 1977a;119:950–954. [PubMed] [Google Scholar]
- Ralph P, Nakoinz I. Direct Toxic Effects of Immunopotentiators on Monocytic, Myelomonocytic, and Histiocytic or Macrophage Tumor Cells in Culture. Cancer Res. 1977b;37:546–550. [PubMed] [Google Scholar]
- Sblattero D, Bradbury A. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat Biotech. 2000;18:75–80. doi: 10.1038/71958. [DOI] [PubMed] [Google Scholar]
- Senn BM, Lopez-Macias C, Kalinke U, Lamarre A, Isibasi A, Zinkernagel RM, Hengartner H. Combinatorial immunoglobulin light chain variability creates sufficient B cell diversity to mount protective antibody responses against pathogen infections. Eur J Immunol. 2003;33:950–61. doi: 10.1002/eji.200323340. [DOI] [PubMed] [Google Scholar]
- Sheets MD, Amersdorfer P, Finnern R, Sargent P, Lindqvist E, Schier R, Hemingsen G, Wong C, Gerhart JC, Marks JD. Efficient construction of a large nonimmune phage antibody library: The production of high-affinity human single-chain antibodies to protein antigens. Proc Natl Acad Sci U S A. 1998;95:6157–6162. doi: 10.1073/pnas.95.11.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making Antibodies by Phage Display Technology. Annual Review of Immunology. 1994;12:433. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]