Abstract
Autosomal DNA polymorphisms can provide new information and understanding of both the origins of and relationships among modern Native American populations. At the same time that autosomal markers can be highly informative, they are also susceptible to ascertainment biases in the selection of the markers to use. Identifying markers that can be used for ancestry inference among Native American populations can be considered separate from identifying markers to further the quest for history. In the current study we are using data on nine Native American populations to compare the results based on a large haplotype-based dataset with relatively small independent sets of SNPs. We are interested in what types of limited datasets an individual laboratory might be able to collect are best for addressing two different questions of interest. First, how well can we differentiate the Native American populations and/or infer ancestry by assigning an individual to her population(s) of origin? Second, how well can we infer the historical/evolutionary relationships among Native American populations and their Eurasian origins. We conclude that only a large comprehensive dataset involving multiple autosomal markers on multiple populations will be able to answer both questions; different small sets of markers are able to answer only one or the other of these questions. Using our largest dataset we see a general increasing distance from Old World populations from North to South in the New World except for an unexplained close relationship between our Maya and Quechua samples.
Keywords: PCA, structure, Native Americans, ancestry, SNPs, haplotypes
Anthropologists have had a long-standing interest in the origins of Native Americans and the relationships among modern Native American populations. Studies have been based on morphology (e.g., Powell and Neves, 1999), on blood group and protein polymorphisms (e.g., Cavalli-Sforza et al., 1994; Estrada-Mena et al., 2010), on linguistics (Hunley et al., 2007; Ruhlen, 1998), as well as on DNA polymorphisms (a) on mitochondrial DNA, both contemporary (Fagundes et al, 2008; Tamm et al., 2007) and ancient (Gilbert et al., 2008), (b) on the non-recombining region of the Y chromosome (Mahli et al., 2008, 2010), and (c) on the autosomes (Schroeder et al., 2009; Wang et al., 2007). Our recent studies of autosomal DNA polymorphisms provide new information and understanding of the evolutionary origins and relationships of Native American populations. At the same time they are highly informative regarding the gaps in our knowledge in these areas and the difficulties in obtaining additional historical insights.
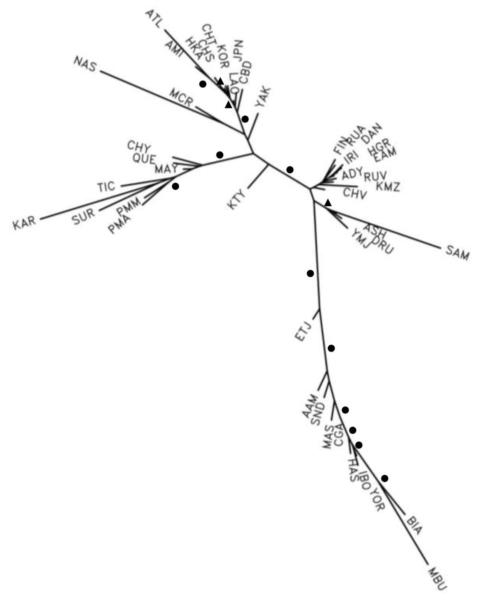
It is clear that with very large datasets, outcomes can be strongly supported statistically. For example, in Fig. 1 the bootstrap values for many of the various branches of the tree are greater than 90%; most of the major branches separating groups of populations have bootstrap values of 100%. Thus, many inferences can be made with confidence about the relationships and relative timing of historical divergences of populations, especially assuming the origin of modern humans in Africa.
Fig. 1.
The best-fitting least squares tree based on 506 haplotyped loci in 45 populations using abbreviations in Table 1. The haplotyped loci encompass over 3000 independent alleles involving 2556 separate SNPs. The circles indicate those branches that were found to be present in 100% of 1000 bootstrap analyses. Note, the segment lengths should be proportional to t/2Ne; recent very small effective population sizes are reflected in the long branches to Mbuti pygmies, Samaritans, Atayal, Nasioi, and Karitiana.
Unfortunately, very few Native American populations have been studied for as many SNPs as on the HGDP-CEPH populations (Li et al., 2008) with five Native American populations. More comprehensive studies of Native American populations are becoming possible as more populations are studied for large numbers of SNPs (e.g., Yang et al., 2010). Even then only small sets of data are likely to be available for many of Native American populations. However, because it is not possible for all labs to type their population samples for a vast number of polymorphisms, it would be useful to have knowledge of what kind of datasets are useful for different questions. In the current study we are comparing the results with a large haplotype-based dataset of 506 haplotype loci with three different and relatively small independent sets of SNPs and haplotypes. We use three different statistical methods to address two different questions on the limited datasets an individual laboratory might be able to collect: (1) How well can we differentiate the Native American populations and/or infer ancestry by assigning an individual to her population(s) of origin? and (2) How well can we infer the Eurasian origins of, and ancestral relations among, Native Americans populations? We have genotyped individuals from a global set of 48 populations, including nine Native American populations, for three sets of markers of the size an individual small lab could assemble. To evaluate the results, we compare them to a much larger dataset of haplotypes we use as a reference. The first dataset consists of 168 SNPs identified in two previous studies for use in ancestry inference, admixture estimation, and sample matching in case-control studies, especially genome wide association studies (GWAS). The second dataset consists of the same number of SNPs selected essentially at random from the several thousand SNPs typed for reasons other than high global allele frequency variation on the same populations. The third dataset consists of 168 multiallelic haplotyped loci selected for their Native American Informativeness (In) from among the 506 haplotyped loci. Results using the full set of 506 haplotyped loci constitute the reference. The analyses of these datasets allow some conclusions with respect to Native American populations but at least as important they reveal different types of datasets are best at answering different questions.
METHODS
Samples and SNPs
Samples
The samples consist of DNA from a total of 2404 individuals, 396 of whom are Native Americans. Table 1 provides the name and sample size for each of the 48 population samples, nine of which are Native Americans (for some of the analyses in this study, only eight of these populations were available). All samples were collected with informed consent under protocols approved by the IRB at Yale and other relevant IRBs. More complete descriptions of all of the population samples are in ALFRED (http://alfred.med.yale.edu) associated with the allele frequencies. We note that for five of the Native American populations we report on here, data exist as part of the HGDP-CEPH dataset both for STRPs (Rosenberg et al., 2002; Zhivotovsky et al., 2003) and for SNPs (Li et al., 2008; Pemberton et al., 2008). However, in those cases we have SNP data on additional individuals in those populations and have used the larger dataset.
Table 1.
The populations, number of individuals sampled in each population, and the three-letter abbreviation used in the figures.
| Population Name |
N | Abbreviation | Population Name |
N | Abbreviation |
|---|---|---|---|---|---|
| Biaka | 68 | BIA | Komi Zyriani | 46 | KMZ |
| Mbuti | 37 | MBU | Khanty | 49 | KTY |
| Yoruba | 77 | YOR | Keralite | 30 | KER |
| Ibo | 48 | IBO | Yakut | 50 | YAK |
| Hausa | 38 | HAS | Nasioi | 23 | NAS |
| Chagga | 44 | CGA | Micronesian | 33 | MCR |
| Masaai | 20 | MAS | Laotian | 118 | LAO |
| Sandawe | 40 | SND | Cambodian | 23 | CBD |
| Zaramo | 36 | ZRM | Chinese, SF | 56 | CHS |
| Afr. American | 86 | AAM | Chinese, T | 49 | CHT |
| Ethiopian Jews | 32 | ETJ | Hakka | 41 | HKA |
| Yemenite Jews | 40 | YMJ | Japanese | 48 | JPN |
| Druze | 97 | DRU | Koreans | 54 | KOR |
| Samaritans | 39 | SAM | Ami | 38 | AMI |
| Ashkenazi Jews | 78 | ASH | Atayal | 40 | ATL |
| Adygei | 53 | ADY | Cheyenne | 54 | CHY |
| Chuvash | 42 | CHV | Pima, Arizona | 50 | PMA |
| Hungarian | 89 | HGR | Pima, Mexico | 53 | PMM |
| Russian, Archangel | 33 | RUA | Maya | 48 | MAY |
| Russian, Vologda | 47 | RUV | Guihiba | 11 | GHB |
| Finns | 34 | FIN | Quechua | 22 | QUE |
| Danes | 49 | DAN | Ticuna | 64 | TIC |
| Irish | 112 | IRI | R. Surui | 42 | SUR |
| Eur. American | 71 | EAM | Karitiana | 52 | KAR |
Polymorphisms
The “506 haplotype” dataset is the largest comprehensively genotyped on all populations; it consists of 2556 SNPs organized into 506 multiallelic haplotyped loci. Each locus is largely or completely independent at the population level, i.e., there is little or no linkage disequilibrium (LD) between loci. These loci were derived from densely typed regions of the genome being studied for different reasons. Each locus was selected by breaking larger regions at gaps in linkage disequilibrium that separated different sets of SNPs with high intermarker LD. The global pattern of LD across each larger region was visualized using HAPLOT (Gu et al., 2005). The number of alleles (haplotypes) in each of the 506 loci varied from 3 to 15 for a total of 3052 independent alleles. The first set of small selected markers consists of 168 SNPs: the128 reported by Kosoy et al. (2009) and the 40 SNPs selected by Nievergelt (in prep.). These two sets were independently selected by the Informativeness (In) statistic (N A Rosenberg et al., 2003, as implemented in INFOCALC; Rosenberg, 2005), using different sets of populations and SNPs. The SNPs reported by Kosoy et al. (2009) were selected specifically to distinguish among three sets of populations--African, European, and Native American populations--based on SNPs typed in their lab and contained in the Illumina 300K genome-wide array. Nievergelt’s 40 SNPs were selected to maximize distinction among four continental regions using the HGDP populations and genotypes in the Li et al (2008) dataset. For comparison we developed two additional datasets based on the same number of loci. One consists of 168 randomly selected (i.e., without regard to In or Fst) SNPs from among over 3000 SNPs typed on these same populations; the other dataset consists of 168 haplotyped loci selected for In among Native Americans from the set of 506 haplotyped loci described above and used as the basis for Fig. 1. Data on those loci were not available for one of the Native American populations (Guihiba), but are otherwise comparable to the 168 SNP datasets: the same individuals are in all datasets. All three sets of 168 sites and sites within haplotypes were genotyped using TaqMan® SNP Genotyping Assays. All datasets are available from the authors.
Statistical Methods
The many statistical genetic methodologies available for studying populations have different assumptions and differences in the way they graphically summarize the results. A combination of these methods should allow a more comprehensive picture of which methods and datasets are best for inferring the origins of New World populations (including at least relative times, demographies, and population similarities) as well as which are best for inferring ancestry of an individual.
Principal Components Analysis (PCA)
Of the methods used in this report, PCA is the method with the fewest genetic assumptions about populations. PCA analyses used the GenAlEx 6.4 software (http://www.anu.edu.au/BoZo/GenAlEx/) and XLSTAT (version 2009.4.07) to determine the major factors accounting for the variance among populations and individuals using the three datasets: (1) 168 “random” SNPs, (2) 168 “informativeness” SNPs, and (3) 168 “informativeness” haplotyped loci. PCA for individuals was based on the individual multisite genotypes; PCA for populations was based on pairwise genetic distance matrices.
Genetic Distance
The tau distances (Kidd and Cavalli-Sforza 1974) were calculated for each dataset and used for PCA analyses and for the tree analyses except that the bootstrap analyses used the Reynolds distance (Reynolds et al., 1983), which is virtually identical numerically.
Phylogenetic Tree
The commonly used neighbor joining method assumes an additive tree and generates a tree that is a good approximate least squares solution for the input data, but the tree is not necessarily the best solution considering all possible tree structures nor is it an exact solution for the tree structure. Under the standard assumptions of genetic drift, the tau genetic distance matrix should be additive and yield a tree structure in which the branch lengths represent additive components in units of t/2Ne. Although it is impossible to examine every possible tree, it is possible to derive an exact least squares solution for a particular tree structure since each structure corresponds to a different set of linear equations. Using arbitrarily specified, random, and/or neighbor joining tree structures to start, we have used a search algorithm shown to improve the fit of an additive tree structure to the genetic distances (Kidd and Sgaramella-Zonta, 1971). The trees illustrated are these exact least squares solutions for the “best” of the many trees examined for each dataset.
Bootstraps
The bootstrap values for each tree were generated by software programs that are part of the Phylogeny Inference Package (PHYLIP) software (Felsenstein 1989; Felsenstein, 2005; PHYLIP version 3.61), using the gene frequencies for each population and SEQBOOT, which uses the GENDIST, NEIGHBOR, and CONSENSE programs. The largest values are indicated on the corresponding branches of the “best” least squares trees in the figures.
Structure
structure (version 2.3.3; Pritchard et al., 2000; Falush et al., 2007) was also used to evaluate and to distinguish among individuals in these populations and to examine heterogeneity within populations. The burn-in was set at 20,000 iterations, followed by 10,000 MCMC iterations, with a model of correlated allele frequencies specified. Ten independent replicates at each “K” level were evaluated using CLUMPP (Jakobsson and Rosenberg, 2007). The solution with the highest likelihood among the ten solutions at each K value was plotted using DISTRUCT 1.1 (Rosenberg, 2004). The matrix of pairwise similarities of the G values of replicate runs was used to identify the runs with similar and different overall patterns. When focusing on Native American populations we included four populations as outgroups based on the global analyses: Danes, Khanty, Yakut, and Taiwanese Han. The Danes and Taiwanese Han were chosen because they were the most homogeneous in the European and East Asian clusters, respectively, in a global analysis. The Khanty and Yakut were chosen as the only Siberian populations available; they are also clearly distinct from the European and East Asian groups. Because structure patterns are highly dependent on the set of populations analyzed, these outgroups allow the possibility of finding similarities with Old World populations and/or significant Old World ancestry in individuals in the Native American populations.
RESULTS
Global population relationships based on our largest random set of the most informative markers—506 haplotyped loci based on 2556 single nucleotide polymorphisms (SNPs)—are shown in Fig. 1 as a comparison for our studies using SNPs and selected haplotypes to focus more specifically on identifying markers that can be used for understanding genetic/evolutionary relationships among Native American populations and/or for inferring ancestry for an individual among Native American populations. The tree in Fig. 1 is similar to the one in Tishkoff and Kidd (2004), but is based on much more genetic information.
PCA
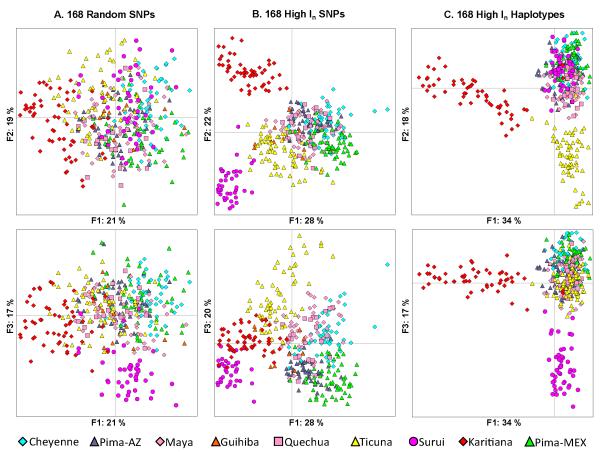
Fig. 2 presents the PCA results for the individuals in the Native American populations based on the first three factors for each of the three datasets. The random SNPs (Fig. 2A) yield highly overlapping sets of individuals but at the fringes one can see Karitiana on the left on factor 1 and Surui at the bottom on factor 3. The high In SNPs (Fig. 2B) provide clearer distinctions between Karitiana and Surui on factors 1 and 2 and Ticuna on factor 3. The high In multiallelic haplotyped loci (Fig. 2C) provide the tightest clusters and the clearest distinctions with Karitiana separated on factor 1, Surui and Ticuna separated on both factors 2 and 3. For all three sets of data the other populations show some distinctions, but with considerable overlap of individuals. The first three principal components explain 57% of the variance for the random SNPs but, interestingly, 69% of the variance for both the In SNPs and haplotyped loci. We also note that in all three instances, the second and third principal components are nearly equal in the per cent of overall variance explained. As expected, the PCA analyses of the genetic distances between pairs of populations yielded plots roughly corresponding to the centroids of the population clusters in Fig. 2 (data not shown), providing no support for any other population distinctions. In the presence of Eurasian outgroups, these Native American individuals are clearly part of one distinct group, but the proportional differentiation among the Native Americans is much smaller than between Eurasian and Native American individuals.
Fig. 2.
The first three factors of PCA analyses of Native American populations for three datasets. (A) The first pair of figures presents factor 1 vs factor 2 and factor 1 vs factor 3 of 168 SNPs chosen at random with regard to Informativeness or Fst. (B) The second pair of figures presents the same views for PCA of 168 SNPs selected specifically for their global Informativeness. (C) The third pair of figures presents these views for PCA of 168 haplotyped loci selected for their Informativeness in Native American populations from the 506 haplotyped loci used to produce the tree in Fig. 1.
Trees
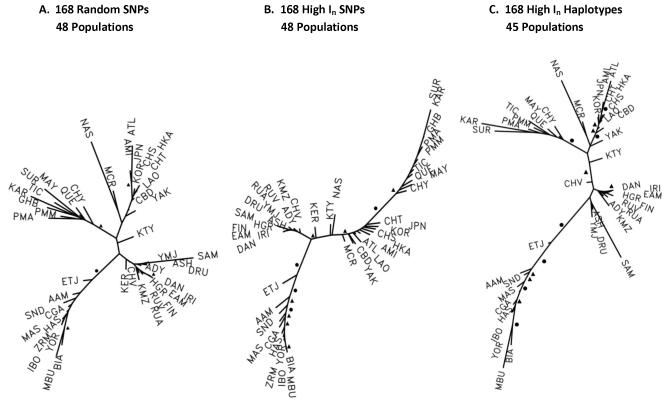
Fig. 3 presents the three “best” trees for the three datasets. Because the pairwise distances from each Native American population to all other populations also provide information on relationships among the Native Americans, all populations are included in the analyses, though we are focusing here on the relationships of the Native American populations. For each dataset at least several dozen different tree structures were evaluated. For each dataset searches starting from several quite different tree structures, including the neighbor joining tree, converged to a set of similar “better” trees of which the best is illustrated. In addition to comparisons among these three datasets, we note the similarities and differences with the tree in Fig. 1 based on the much larger dataset. Whether a tree can accurately represent true phylogenetic relationships can be debated, but a tree can be considered a graphical representation of complex population relationships, under the stated assumptions, that cannot be otherwise represented in two or three dimensions.
Fig. 3.
The “best” additive genetic trees from the pairwise tau genetic distances of the populations with the bootstrap values of 90 to 99% indicated by filled triangles adjacent to the relevant branch and bootstrap values of 100% indicated by filled circles. The trees in Figs 3A, 3B, and 3C are based on the same datasets underlying the PCA analyses in Fig. 2, respectively.
The tree based on 168 random SNPs (Fig. 3A) has very few high bootstrap values with only two of the main branches reaching 90%: the branch separating African populations from all others and the branch separating the Native Americans from all others. Three pairs of similar populations are also on branches reaching 90% bootstraps. This tree is very similar in its major structure to the tree in Fig. 1: four main branches, an African, a Native American, an East Asian and Pacific, and a mostly European and South West Asian branch. The Khanty in Western Siberia are distinct in both. In the Native American branch the Cheyenne are most proximal to the rest of the tree, the two Pima groups, Maya, and Quechua branch off more distally, and then the three Amazonian groups and the Colombians are most distal. Note that Guihiba (Colombians), the Keralites, and Zaramo have data for SNPs but not for the majority of haplotypes used in Fig. 1 and Fig. 3C. Fig. 3B based on 168 high In SNPs shows a very different pattern with respect to East Asia, the Pacific and Native Americans. The pattern within Native Americans is not dramatically different from that shown in Fig. 1, but it is more “stretched”. However, the Native Americans, though separated by 100% bootstraps, appear to be related to the Far East Asians as compared to having a distinct lineage of Central Asian origin. With >90% bootstraps, the Cheyenne are placed closer to Old World populations. In the tree based on the 168 high In haplotypes (Fig. 3C) Native Americans show a pattern very similar to the patterns in Fig. 1 and Fig. 3A.
Structure
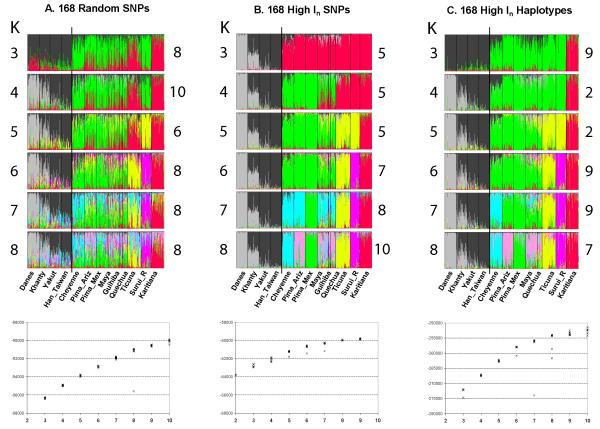
Global structure analyses of the 506 and168 haplotyped loci and the 168 “informativeness” SNPs always showed the Native American populations as a distinct group from K=4 through K=9 (data not shown); therefore, analyses shown here focus on the Native American populations but with representative outgroups as described. Fig. 4 presents the structure analyses of Native American populations based on the three smaller datasets for K=3-8. The lower graphs of Figs. 4A, 4B, and 4C are plots of the likelihoods for all 10 replicates at each K value. The out-groups are distinct from K=2 for all datasets. For the random SNPs (Fig. 4A), though individual populations begin to show some obvious differences at various levels of K, the strongest visual impression is the high variability among individuals within each population. Among the Native Americans at K=5 & 6 one can distinguish the three Amazonian populations (yellow, bright pink, and red) but even that distinction becomes much more blurred at higher values of K. It would be difficult to draw a strong conclusion about distinctions at any K value among the other six populations. Though the likelihoods at higher values of K up to K=12 continue to increase (not shown), albeit somewhat more slowly, the emerging patterns never become visually clearer than at K=8. This visual impression and the gradual plateauing of the likelihood values suggest that K=8 is the conservative stopping point for the structure analyses.
Fig. 4.
Structure analyses and likelihood plots of three datasets of Native American populations. (A), (B), and (C) are analyses of the same populations underlying Figs. 2 and 3, but now including four “outgroup” populations. Each color in the figure represents how individuals are assigned to each of the specified K clusters. The lower graphs in the figure present the ln likelihoods for the replicate runs of each analysis at each K value.
In strong contrast, the high In dataset of SNPs (Fig. 4B) shows the Native Americans as a distinct group from the Eurasian outgroup populations with a very clear differentiation among those Eurasian groups virtually unchanged from K=3 through K=8. At K=6 through K=8, one sees clearly and consistently the three Amazonian populations being cleanly differentiated (yellow, bright pink, and red). At K=7 the Mexican Pima emerge as distinct, at K=8 the Cheyenne become distinct, but the Arizona Pima, Maya, Quechua, and Guihiba are all very similar though collectively distinct from the remainder. K=8 is the point at which likelihoods begin to plateau with increasing K values.
For the 168 haplotype dataset (Fig. 4c) there is clearly much more data reflected in the much smaller ln likelihoods compared to the other datasets. The patterns at K=5 and K=6 are largely the same as in Fig. 4b; however, the outgroups, especially the Khanty, do not show as clear a pattern. At K=7 the additional clustering distinguishes the Cheyenne but leaves the Pima groups, Maya, and Quechua largely indistinguishable. Finally, at K=8 the patterns in Figs. 4b and 4c converge though it is obvious that some individuals show differences. We also note that at K=8 the likelihoods have begun to plateau.
DISCUSSION
It is reasonably clear that if one is willing/able to genotype a set of populations for many thousands of polymorphisms (as illustrated in Fig. 1), it is possible to answer two questions: (1) How are those populations related? (2) How distinguishable is each from the others? If, however, one is limited in the resources available for genotyping vast numbers of polymorphisms for many samples, it would be useful to know how to select those markers that are most likely to be most informative for the question asked. The object would be to minimize the genotyping effort and expense while retaining the capacity to answer the questions posed when new populations are studied. We have shown that modest numbers of markers selected using different criteria cannot answer both of the two questions considered. The random SNPs appear to represent evolution, as well as can be expected with little information per SNP, but in PCA and structure analyses they provide little hope of accurately assigning an individual to her population(s) of origin among Native Americans. Thus, random SNPs appear better at answering the historical/evolutionary relationships. In contrast these high In SNPs appear much better at discriminating among populations, but give what we believe to be an incorrect evolutionary/historical relationship to Old World populations.
This observed difference fits our logical expectation. Each randomly selected marker should be an independent realization of genetic drift. As such, ancestry should be reflected by the consensus of many such realizations. In contrast, markers selected because they have very different frequencies among populations must represent either a tail of the random distribution, or differential selection pressures in different populations. Thus, markers selected to distinguish populations will distort the true ancestral relationships.
Haplotype data should have the least bias because the SNPs used were selected only because of the loci nearby with no prior knowledge of any evolutionary pattern or the amount of allele frequency variation world-wide. While the individual SNPs can be expected to show a bias for highest heterozygosity in Europeans, given historical ascertainment, conversion of nearby SNPs into haplotypes mitigates that bias. Assuming that the data underlying Fig. 1 (2,556 SNPs assembled into 506 multiallelic loci with 3,052 independent alleles) provides a standard against which we can compare three much smaller datasets, we find that datasets selected in different ways do provide different answers to the two questions. The more central Asian origin of Native Americans is supported by many different analyses of autosomal as well as Y and mtDNA and is generally accepted as conclusive relative to an origin from the far East Asians, such as Chinese and Japanese. The relationships among the Native Americans are not well agreed upon other than a general North to South pattern in agreement with the probable migration patterns.
We find that PCA provides little useful information on Native Americans other than confirming that the three Amazonian populations (Ticuna, Surui, and Karitiana) are highly differentiated from other populations and among themselves. This is consistent with recent high levels of genetic drift attributable to the small population sizes for the Surui and Karitiana and probably for our sample of the Ticuna.
In all the tree and structure analyses, including for the data underlying Fig. 1, the Maya and Quechua present as very similar with the Quechua appearing more closely related to the North American populations than to the other South American populations. This has recently been seen in an independent set of autosomal data (Yang et al., 2010). The extent to which this similarity results from pre-Columbian contact between Meso-American and Western Andean populations or West Coast migration into South is unclear, and the present analyses do not allow clarification. It is clearly a question for further study. However, we see no evidence that the similarity results from more recent “shared” admixture with Old World populations.
Most significant to this paper we note that our large dataset on 506 haplotyped loci shows the divergence of Native American populations from Eurasian populations is distinctly not from East Asian populations, but rather would be closer to Central Asia. Lack of comparable data on multiple Central Asian populations for current analyses limits considerably our ability to infer the location(s) of the ancestral gene pool, though the East Asian populations studied are clearly excluded as a significant part of that ancestral gene pool. Another implication of the resulting tree (Fig. 1) is that the Old World gene pool underlying Native American populations diverged considerably before the present day Far East Asian populations (as sampled in this analysis) diverged from one another.
CONCLUSIONS
We conclude that a single dataset consisting of a small number of markers cannot answer both of the two questions: phylogenetic origins and ancestry differentiation. Random SNPs and haplotyped loci are the best markers for determining the evolutionary relationships on a global level among populations when genetic distances are used. However, small numbers of such markers do not work well for distinguishing among a small number of populations. Specifically for Native American populations, we conclude that the SNPs selected for high global In are good for distinguishing among the Native American populations but give an incorrect ancestral relationship to Eurasian populations. Conversely, a relatively small set of randomly selected SNPs can give a reasonable estimate of the broad pattern of modern human historical differentiation. The data that appear to be best for both questions consist of haplotyped loci, but whether or not this is a trait of haplotyped loci or simply a question of amount of information is not clear. Using all datasets we see three specific patterns among the Native American populations. The Maya and Quechua samples are close and it is not explainable by common European admixture. The Ticuna, Surui, and Karitiana are the most distant and outlying of the Native Americans. The Cheyenne are the most similar to the Eurasians, but we cannot exclude European admixture. Until more populations are studied for a single large set of markers, researchers need to be concerned about the criteria for selecting SNPs when interpreting the genetics of Native American populations.
ACKNOWLEDGEMENTS
We thank FL Black, D Goldman, J Kennedy, W Knowler, L Shultz, K Weiss, and H Groot for their help over the past 20+ years in assembling the samples of Native American populations. We extend special thanks to the many hundreds of individuals who volunteered to give blood samples.
This research was supported in part by USPHS grants AA009379 and GM057672 (to KKK & JRK).
REFERENCES
- Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton University Press; Princeton, New Jersey: 1994. p. 518. [Google Scholar]
- Estrada-Mena B, Estrada FJ, Ulloa-Arvizu R, Guido M, Méndez R, Coral R, Canto T, Granados J, Rubí-Castellanos R, Rangel-Villalobos H, García-Carrancá A. Blood group O alleles in Native Americans: implications in the peopling of the Americas. Am J Phys Anthropol. 2010;142:85–94. doi: 10.1002/ajpa.21204. [DOI] [PubMed] [Google Scholar]
- Fagundes NJ, Kanitz R, Bonatto SL. A reevaluation of the Native American mtDNA genome diversity and its bearing on the models of early colonization of Beringia. PLoS One. 2008;3:e3157. doi: 10.1371/journal.pone.0003157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure: Extensions to linked loci and correlates allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Using the quantitative genetic threshold model for inferences between and within species. Philos Trans R Soc Lond B Biol Sci. 2005;360:1427–34. doi: 10.1098/rstb.2005.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Mathematics vs. Evolution: Mathematical Evolutionary Theory. Science. 1989;9246:941–2. doi: 10.1126/science.246.4932.941. [DOI] [PubMed] [Google Scholar]
- Gilbert MT, Kvisild T, Grønnow B, Andersen PK, Metspalu E, Reidla M, Tamm e, Axelsson E, Götherström A, Campos PF, Rasmussen M, Metspalu M, Higham TF, Schwenninger JL, Nathan R, De Hoog CJ, Koch A, Møller LN, Andreasen C, Meldgaard M, Villems R, Bendixen C, Willersley E. Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science. 2008;320:1787–9. doi: 10.1126/science.1159750. [DOI] [PubMed] [Google Scholar]
- Gu S, Pakstis AJ, Kidd KK. HAPLOT: a graphical comparison of haplotype blocks, tagSNP sets and SNP variation for multiple populations. Bioinformatics. 2005;21:3938–3939. doi: 10.1093/bioinformatics/bti649. [DOI] [PubMed] [Google Scholar]
- Hunley KL, Cabana GS, Merriwether DA, Long JC. A formal test of linguistic and genetic coevolution in native Central and South America. Am J Phys Anthropol. 2007;132:622–31. doi: 10.1002/ajpa.20542. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–6. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kidd KK, Cavalli-Sforza LL. The role of genetic drift in the differentiation of Icelandic and Norwegian cattle. Evolution. 1974;28:381–395. doi: 10.1111/j.1558-5646.1974.tb00759.x. [DOI] [PubMed] [Google Scholar]
- Kidd KK, Sgaramella-Zonta LA. Phylogenetic Analysis: Concepts and methods. Am J Hum Genet. 1971;23:235–252. [PMC free article] [PubMed] [Google Scholar]
- Kosoy R, Nassir R, Tian C, White PA, Butler LLM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, De La Vega FM, Seldin MF. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Human Mutation. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Malhi RS, Cybulski JS, Tito RY, Johnson J, Harry H, Dan C. Brief communication: mitochondrial haplotype C4c confirmed as a founding genome in the Americas. Am J Phys Anthropol. 2010;141:494–7. doi: 10.1002/ajpa.21238. [DOI] [PubMed] [Google Scholar]
- Malhi RS, Gonzalez-Oliver A, Schroeder KB, Kemp BM, Greenberg JA, Dobrowski SZ, Smith DG, Resendez A, Karafet T, Hammer M, Zegura S, Brovko T. Distribution of Y chromosomes among native North Americans: a study of Athapaskan population history. Am J Phys Anthropol. 2008;137:412–24. doi: 10.1002/ajpa.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ, Jakobsson M, Conrad DF, Coop G, Wall JD, Pritchard JK, Patel PI, Rosenberg NA. Using population mixtures to optimize the utility of genomic databases: linkage disequilibrium and association study design in India. Ann Hum Genet. 2008;72(Pt 4):535–46. doi: 10.1111/j.1469-1809.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JF, Neves WA. Craniofacial morphology of the first Americans: Pattern and process in the peopling of the New World. Am J Phys Anthropol. Suppl. 1999;29:153–88. doi: 10.1002/(sici)1096-8644(1999)110:29+<153::aid-ajpa6>3.3.co;2-c. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: Basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Algorithms for Selecting Informative Marker Panels for Population Assignment. J Comp. Biol. 2005;12:1183–1201. doi: 10.1089/cmb.2005.12.1183. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73:1402–22. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. (Program Note) DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Sci. 2002;298:2361–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Ruhlen M. The origin of the Na-Dene. Proc Natl Acad Sci U S A. 1998;95:13994. doi: 10.1073/pnas.95.23.13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KB, Jakobsson M, Crawford MH, Schurr TG, Boca SM, Conrad DF, Tito RY, Osipova LP, Tarskaia LA, Zhadanov SI, Wall JD, Pritchard JK, Malhi RS, Smith DG, Rosenberg NA. Haplotypic background of a private allele at high frequency in the Americas. Mol Biol Evol. 2009;26:995–1016. doi: 10.1093/molbev/msp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm E, Kivisild T, Reidla M, Metspalu M, Smith DG, Mulligan CJ, Bravi CM, Rickards O, Martinez-Labarga C, Khusnutdinova EK, Fedorova SA, Golubenko MV, Stepanov VA, Gubina MA, Zhadanov SI, Ossipova LP, Damba L, Voevoda MI, Dipierri JE, Villems R, Malhi RS. Beringian standstill and spread of Native American founders. PLoS One. 2007 Sep 5;:e829. doi: 10.1371/journal.pone.0000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Kidd KK. Implications of biogeography of human populations for ‘race’ and medicine. Nat Genet. 2004;36(11 Suppl):S21–7. doi: 10.1038/ng1438. Review. [DOI] [PubMed] [Google Scholar]
- Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, Rojas W, Parra MV, Molina JA, Gallo C, Mazzotti G, Poletti G, Hill K, Hurtado AM, Labuda D, Klitz W, Barrantes R, Bortolini MC, Salzano FM, Petzl-Erler ML, Tsuneto LT, Llop E, Rothhammer F, Excoffier L, Feldman MW, Rosenberg NA, Ruiz-Linares A. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang NN, Mazières S, Bravi C, Ray N, Wang S, Burley MW, Bedoya G, Rojas W, Parra MV, Molina JA, Gallo C, Poletti G, Hill K, Hurtado AM, Petzl-Erler ML, Tsuneto LT, Klitz W, Barrantes R, Llop E, Rothhammer F, Labuda D, Salzano FM, Bortolini MC, Excoffier L, Dugoujon JM, Ruiz-Linares A. Contrasting patterns of nuclear and mtDNA diversity in Native American populations. Ann Hum Genet. 2010;74:525–38. doi: 10.1111/j.1469-1809.2010.00608.x. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genome-wide microsatellite markers. Am J Hum Genet. 2003;72:1171–1186. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]