Abstract
To date the genus Corynebacterium comprises 88 species. More than half of these are connected to human and animal infections, with the most prominent member of the pathogenic species being Corynebacterium diphtheriae, which is also the type species of the genus. Corynebacterium species are characterized by a complex cell wall architecture: the plasma membrane of these bacteria is followed by a peptidoglycan layer, which itself is covalently linked to a polymer of arabinogalactan. Bound to this, an outer layer of mycolic acids is found which is functionally equivalent to the outer membrane of Gram-negative bacteria. As final layer, free polysaccharides, glycolipids, and proteins are found. The composition of the different substructures of the corynebacterial cell envelope and their influence on pathogenicity are discussed in this paper.
1. The Genus Corynebacterium
The genus Corynebacterium belongs to the class of Actinobacteria (high G+C Gram-positive bacteria) and comprises a collection of morphologically similar, irregular- or club-shaped nonsporulating (mico)aerobic microorganisms [1, 2]. To date, 88 species were taxonomically classified [3]. More than half of these, that is, 53 species, are occasional or rare causes of infections, with the most prominent member of the pathogenic species being Corynebacterium diphtheriae, which is also the type species of the whole genus. Several pathogenic species are considered to be part of the human skin flora, for example, Corynebacterium amycolatum or Corynebacterium jeikeium, others are considered as zoonotic agents, for example, Corynebacterium pseudotuberculosis, Corynebacterium ulcerans, or Corynebacterium xerosis [3]. Biotechnologically important species used for the industrial production of nucleotides and amino acids are Corynebacterium ammoniagenes, Corynebacterium efficiens, and Corynebacterium glutamicum. C. glutamicum especially is dominating the field of white (bacterial) biotechnology with a production of two million tons of L-glutamate and 1.8 million tons of L-lysine per year [4] and an increasing application as platform organism for the industrial production of various metabolites [5].
Based on their medical importance as etiological agent of diphtheria and their enormous biotechnological potential, C. diphtheriae and C. glutamicum, respectively, have been the best investigated Corynebacterium species for many years. In fact, these were also the first species for which genome sequences became available [6–8]. With the development of high throughput sequencing techniques, to date numerous genome sequences are available including those of Corynebacterium aurimucosum [9], Corynebacterium bovis [10], various C. diphtheriae strains [11–13], C. jeikeium [14], Corynebacterium kroppenstedtii [15], several C. pseudotuberculosis strains [16–24], Corynebacterium resistens [25], two C. ulcerans strains [26], Corynebacterium urealyticum [27], and Corynebacterium variabile [28]. This wealth of genome information now provides a basis for the characterization of corynebacterial cell envelope components and corresponding genes both on the whole cell level by global analysis techniques (genomics, transcriptomics, and proteomics) and on a molecular level by specific approaches.
2. General Cell Envelope Architecture
Almost all Corynebacterium species are characterized by a complex cell wall architecture: the plasma membrane of these bacteria is covered by a peptidoglycan layer, which itself is covalently linked to arabinogalactan, an additional heteropolysaccharide meshwork. Bound to this, an outer layer of mycolic acids is found which is functionally equivalent to the outer membrane of Gram-negative bacteria. As top layer, outer surface material composed of free polysaccharides, glycolipids, and proteins (including S-layer proteins, pili, and other surface proteins) is found (reviewed in [29, 30], see also Figure 1).
Figure 1.
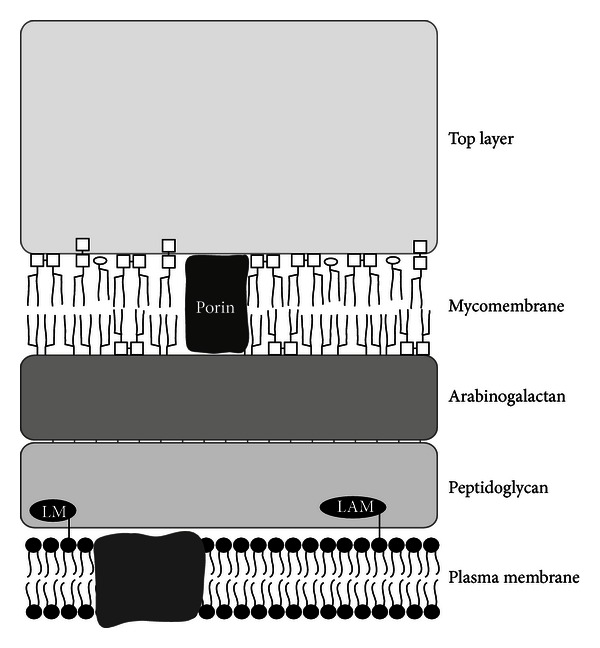
The corynebacterial cell envelope. A schematic representation of the general envelope structure of corynebacteria is shown. The different layers are found in almost all species, and distribution of single components like lipomannan (LM) and lipoarabinomannan (LAM) might differ (for details, see text). Porin proteins might also be present in top layer, S-layer, and pili proteins not shown (figure courtesy of S. Morbach, Friedrich-Alexander-Universität Erlangen-Nürnberg).
The corynebacterial cell envelope has been investigated by different optical techniques. Electron microscopy of thin sections after freeze substitution revealed a layered cell envelope organization, comprising a plasma membrane, a thick electron-dense layer, an electron-transparent layer, and a thin outer layer [31–33]. This picture resembles the typical mycobacterial cell envelope appearance. The electron-dense layer is traditionally interpreted as peptidoglycan, the electron-transparent layer as mycolic acid layer. When comparing the thickness determined from the electron microscopic pictures of corynebacteria and mycobacteria, the question arose, why their electron-transparent layer has a similar thickness despite the fact that mycobacterial mycolic acids have about a threefold length compared to corynebacterial ones [33]. A solution of this problem was indicated by cryoelectron tomography studies. These revealed a typical outer membrane with a bilayer structure in Mycobacterium smegmatis, Mycobacterium bovis, and C. glutamicum. The thickness of mycobacterial outer layers was smaller than expected and, based on this observation, an alternative model for mycolic acid distribution was proposed, implicating a folding of mycolic acids [34, 35].
The general structure and composition of the corynebacterial cell envelope were earlier reviewed by [29, 30] in respect to biochemical and genetic properties especially in C. glutamicum. The aim of this paper will be the presentation of cell envelope properties with a broader focus on different corynebacteria and the importance of cell envelope components for pathogen host interaction.
3. Cytoplasmic Membrane and Fatty Acid Synthesis
3.1. Plasma Membrane Composition
The cytoplasmic or plasma membrane is the main diffusion barrier of cells and separates cytoplasm and environment. As in other bacteria, the corynebacterial plasma membrane is mainly composed of phospholipids, assembled into a lipid bilayer, which additionally contains other polar lipids besides a great variety of proteins crucial for transport processes and bioenergetics of the cell.
The main phospholipid found in C. glutamicum is phosphatidylglycerol, followed by diphosphatidylglycerol, phosphatidylinositol, and minor amounts of phosphatidylinositol dimannosides (PIM2) [33, 36–38]. Fatty acids dominating in the plasma membrane are the saturated palmitic acid (16:0) and the desaturated decenoic acid (18:1) [39–42]. However, as in other bacteria, fatty acid composition might change significantly depending on environmental conditions such as low or high temperature [43] or the carbon source available [42].
3.2. Fatty Acid Synthesis
Fatty acids are synthesized by successive cycles of multistep reactions [44]. Two distinct types of these fatty acid synthases (FASs) are distinguished based on their general composition: the FAS-II type, characteristically found in bacteria, contains the minimum seven functional domains necessary for fatty acid synthesis organized in one polypeptide, whereas the FAS-I type, typically found in eukaryotes, is comprised of a large multifunctional protein complex. Interestingly and as an exception from the rule FAS-I proteins are found in members of the Corynebacterineae. C. glutamicum, and C. efficiens contain even two FAS-I-type complexes, FAS-IA and FAS-IB, which were functionally characterized in detail for C. glutamicum [42]. FAS-IA is essential in C. glutamicum, while FAS-IB is not; however, FAS-IB-devoid mutant strains exhibit an altered pattern of fatty and mycolic acids, showing that FAS-IB is active and necessary to generate the typical wild-type fatty acid profile [42]. The regulatory mechanism allowing adaptation of FAS activity to environmental stimuli (see above) is unknown.
C. glutamicum does not only have genes coding for two FAS-I but also genes coding for FAS-II [42], which are involved in elongation of mycolic acid chains in mycobacteria. Since C. glutamicum does not contain elongated mycolic acids and FAS-II is absent in other corynebacteria such as C. diphtheriae, it was speculated that these proteins might play a minor physiological role or even not be functional in C. glutamicum [42].
3.3. Lipomannan and Lipoarabinomannan
As in other pro- and eukaryotes, the corynebacterial lipid bilayer of the cytoplasmic membrane is not symmetric. In corynebacteria, the proposed reason for asymmetry is an insertion of glycoconjugates in the outer sheet of the cytoplasmic membrane [29]. Lipomannan (LM) and lipoarabinomannan (LAM) derivatives were found in different Corynebacterium species [33]. They might be inserted into the plasma membrane via covalently bound palmitic acid or decenoic acid molecules, besides their appearance in the outer surface material of corynebacteria. Distribution of LM- and LAM-like substances seems to be species-specific: in C. glutamicum LM-like molecules are dominating, in C. xerosis and C. amycolatum LAM-like substances were preferentially found, while a C. diphtheriae strain showed an almost equal distribution of LM and LAM derivatives [33]. An excellent review dealing with the synthesis of PIM, LM, and LAM derivatives was published recently [45].
3.4. Lipoarabinomannan as a Virulence Factor
A 10 kDa lipoarabinomannan polymer was identified in C. diphtheriae and designated CdiLAM. The basic structure of CdiLAM shows similarity to mycobacterial LAM; however, in contrast to lipoarabinomannans in other Actinobacteria, CdiLAM presents an unusual substitution at position 4 of α 1→6 mannan backbone by α-D-Araf. CdLAM might be part of the plasma membrane but is also located at the surface of C. diphtheriae (see the following) and facilitates binding to epithelial cells [46].
The role of lipoarabinomannan with respect to initiation of immune responses was addressed in C. glutamicum recently [47]. Characterization of a C. glutamicum strain devoid of α(1→2) arabinofuranosyltransferase AftE, revealed that AftE is involved in the synthesis of arabinans of LAM. Absence of AftE leads to a hypermannosylated variant of LAM, designated hLM. Both, LAM and hLM were able to modulate the initiation of immune response by interacting with TLR2. As shown by a number of in vitro assays, arabinose branching of lipoarabinomannan impacts T-helper-cell differentiation and LAM as well as hLM activate dendritic cells via TLR2. Interestingly, alterations of lipoarabinomannan seem to be discriminated by TLR2 and signal pathway induction by hLM was shown to be broader. In accordance with this observation, hLM was shown to be a stronger inducer of immune responses in mice.
4. The Cell Wall Heteropolysaccharide Meshwork
In contrast to, for example, Escherichia coli or Bacillus subtilis, the cell wall skeleton of corynebacteria and related taxa is not exclusively composed of peptidoglycan, but, in addition, a layer of arabinogalactan is covalently bound to the peptidoglycan, which itself is linked to mycolic acids (for review see [29, 33, 48]).
4.1. Peptidoglycan
As in other bacteria, the glycan part of the murein sacculus is composed of alternating β-1,4-linked N-acetylglucosamine and N-acetyl muramic acid units, which form the glycan part of the macromolecule. Cross-linking between different glycan polymers occurs via peptide side chains attached to the carboxyl group of muramic acid via peptide bonds. Corynebacterial peptidoglycan is directly cross-linked, as shown for C. bovis, Corynebacterium pseudodiphthericum, C. pseudotuberculosis, C. striatum, C. ulcerans, and C. xerosis [49]. Interpeptide bridges as found in other Gram-positives are absent. In summary, the peptidoglycan of Corynebacterium in sensu stricto is of the A1γ type [49]. In C. diphtheriae, the major peptide units found are the tetrapeptide L-Ala-D-Glu-meso-DAP-D-Ala and the tripeptide L-Ala-D-Glu-meso-DAP [50]. Interestingly, only a portion of the peptide side chains are cross-linked via D-Ala-meso-DAP bridges, while the others are supposed to be connected by DAP-DAP bridges [29].
Due to the similar peptidoglycan structure, as well as the homology and synteny of genes involved in cell wall synthesis, peptidoglycan synthesis is assumed to be similar to that in E. coli [34] (for a topical review on E. coli cell wall synthesis, see [51]) and can be separated into three distinct parts. First, the building blocks of peptidoglycan have to be synthesized in the cytoplasm. For this purpose, UDP-N-acetylglucosamine is synthesized and partially converted to UDP-N-acetylmuramic acid by the murA and murB gene products [30, 52]. Next, UDP-N-acetylmuramyl-pentapeptide is formed, a process in which the murE, murF, murD, and murC gene products are involved.
The second step of building block synthesis is located at the cytoplasmic membrane and involves transfer of a phospho-N-acetylmuramyl-pentapeptide to polyprenol phosphate catalyzed by the mraY gene product and resulting in lipid I. As in many other bacteria, undecaprenol (C55) might be the polyprenol used, since at least in C. glutamicum this compound is used for polyprenyl monophosphomannose synthesis [53]. Next, N-acetylglucosamine is transferred from UDP-N-acetylglucosamine to lipid I. This is catalyzed by murG and yields lipid II or N-acetylglucosamine-β-(1,4)-N-acetylmuramyl(pentapeptide)-pyrophosphoryl-polyprenol. The generated lipid intermediate mediates the transport of the hydrophilic disaccharide pentapeptide precursor from the cytoplasm across the hydrophobic plasma membrane to the peptidoglycan layer.
As third step, disaccharide pentapeptide precursors are integrated into the growing peptidoglycan meshwork by transglycosylation and transpeptidation reactions catalyzed by penicillin binding proteins [54]. Typically, several copies of these can be found in different Corynebacterium species such as C. diphtheriae [52] and C. glutamicum [55] (for review, see [30]). For C. glutamicum five out of nine putative penicillin binding proteins were shown to be functional in peptidoglycan synthesis [55].
A strict coordination of the described steps of cell wall synthesis is crucial for survival of bacteria, since otherwise the cells would be prone to disruption due to the high internal turgor pressure. The regulation of this process and cell division has been reviewed for C. glutamicum [54] and other Actinobacteria [56] recently.
4.2. Linker Unit
As shown in mycobacteria, arabinogalactan is covalently bound to the murein sacculus via a polysaccharide linker unit making up phosphodiester bonds to about 10% of the muramic acid residues of the peptidoglycan [57]. The mycobacterial linker unit consists of galactose, rhamnose, and N-acetylglucosamine linked as Galf-(1→4)-Rhap-(1→3)-GlcNAc via a 1-O-phosphoryl bond of GlcNAc to the 6-OH position of muramic acid [58]. Studies of C. diphtheriae revealed a very similar linkage profile of arabinogalactan to that of Mycobacterium tuberculosis [30, 33].
4.3. Arabinogalactan
Arabinogalactan is a heteropolysaccharide consisting of D-arabinose and D-galactose present in their furanose form (Araf, Galf). In C. glutamicum alternating β(1→5) and β(1→6)-linked Galf residues form a linear chain of up to 30 sugar moieties, which is bound to peptidoglycan by the linker unit. The galactan chain is decorated with arabinan, branched from the C5 of β(1→6) linked Galf. In C. glutamicum three branched arabinan domains are linked per galactan domain at the 8th, 10th, and 12th Galf residues. Enzymes involved in arabinogalactan synthesis are well characterized in C. glutamicum [59–62] and have been reviewed recently [30].
Whereas C. glutamicum arabinogalactan consists exclusively of arabinose and galactose, C. diphtheriae arabinogalactan contains significant amounts of mannose, and C. amycolatum and C. xerosis arabinogalactans are characterized by additional glucose content [33]. In any case, arabinogalactan provides a covalent connection not only to peptidoglycan but also to the outer membrane layer.
5. Mycolic Acids and Trehalosyl Mycolates
5.1. Composition and Biosynthesis
A second permeability barrier equivalent to the outer membrane of Gram-negative bacteria is a key feature of the CMN group (Corynebacterium, Mycobacterium, and Nocardia) of Actinobacteria. The functionality of this barrier is critically influenced by its mycolic acid content [63]. The inner half of the corynebacterial mycolic acid layer is mainly formed by mycolic acids esterified to the 5-OH group of the penultimate β(1→2)-linked or ultimate Araf residue of arabinogalactan, while in the outer sheet, trehalose and glycerol esterified mycolic acids are predominating. Additionally, minor amounts of free mycolic acids are found. A recent biochemical disclosure of the outer membrane of C. glutamicum showed that the lipids composing the mycomembrane consist almost exclusively of mycolic acids derivatives, whereas only minor amounts, if any, of phospholipids and lipomannans were detected [64].
In corynebacteria, fatty acids with around 30 carbon atoms (corynomycolates), in nocardia with about 50 carbon atoms (nocardomycolates), and in mycobacteria with about 70 to ninety carbon atoms (eumycolates) are found [29]. In contrast to the linear fatty acids of the phospholipids, mycolic acids are α-branched β-hydroxy fatty acids, requiring carboxylation and condensation of two fatty acids for their synthesis [65]. The enzymes involved in these steps were identified by mutation analyses in C. glutamicum, and two carboxylases were identified to be essential for mycolic (and fatty) acid synthesis, AccD2 and AccD3 [66]. These are conserved in Corynebacterineae and provide the crucial carboxylated intermediate for condensation of merochain and α-branch [67]. Further proteins involved in mycolic acid synthesis in C. glutamicum and related organisms are AccD1, involved in malonyl-CoA synthesis, Pks, a ketoacyl synthase involved in fatty acid elongation, and FadD, a fatty acid acyl-AMP ligase ([65, 67]; for a recent review see [30]).
Three pathways for trehalose synthesis were described in C. glutamicum: the OtsA-OtsB pathway synthesizing trehalose from UDP-glucose and glucose-6-phosphate, the TreY-TreZ pathway using malto-oligosaccharides or α-1,4-glucans as substrate, and the TreS pathway using maltose as an educt for trehalose synthesis [68, 69]. It is suggested that the transfer of mycolic acid moieties to trehalose occurs outside the cytoplasm [70], since in absence of internally synthesized trehalose, either trehalose or externally added glucose, maltose and maltotriose, can be used as substrate for mycolic acid modification and result in the corresponding di- and monocorynomycolates [70].
The production of arabinogalactan-linked mycolates, trehalosylmonocorynomycolates, and trehalosyldicorynomycolates indicates the presence of mycolyltransferases. In fact, proteins similar to the mycobacterial antigen 85 showing mycoloyltransferase activity, also designated fibronectin-binding protein, were identified in corynebacteria. The first member was the PS1 protein from C. glutamicum [71]. Later it was shown that six of these proteins are present in this species, five in C. efficiens, and four in C. diphtheriae [29, 72, 73]. The enzymes are fully redundant in C. glutamicum in respect to mycoloyl moiety transfer to trehalose and partially redundant with respect to transfer of arabinogalactan [29, 72, 74]. The proteins associated with mycolic acid transport across the plasma membrane were characterized in M. smegmatis and C. glutamicum recently [75]. In C. glutamicum, four mmpL genes encode large membrane proteins associated with mycolate metabolism and transport, which have partially redundant function [75].
Besides the plasma membrane lipids, the outer membrane fatty acid composition also has to be adapted to different temperatures, a crucial process for function of the mycolic acid layer as a diffusion barrier [76]. In fact, one stress-induced protein, designated ElrF, was identified, which is conserved in Corynebacterineae and plays a role in the regulation of outer membrane lipid composition in response to heat stress [77].
5.2. Corynomycolates and Pathogenicity
Almost all Corynebacterium and Mycobacterium species are characterized by a complex cell wall architecture comprising an outer layer of mycolic acids, which is functionally equivalent to the outer membrane of Gram-negative bacteria, not only in respect to its physiological role as a permeability barrier, but also as important component of host pathogen interaction. It has long been known that constituents of the mycolic acid layer may be immune stimulatory and effecting macrophage function. These effects are best investigated in M. tuberculosis [78], where trehalose dimycolate, also designated as cord factor, inhibits fusion events inside the host macrophage and at the same time contributes to macrophage activation. However, limited data for corynebacteria are available. Investigations of C. pseudotuberculosis (formerly Corynebacterium ovis) indicate a lethal effect of outer membrane lipids on caprine and murine macrophages. Lipid extracts of C. pseudotuberculosis had negative effects on glycolytic activity, viability, and membrane integrity [79]. When macrophages were infected with C. pseudotuberculosis, uptake of the bacteria and lysosome fusion was functional, but bacteria survived internalization. However, the macrophages were destroyed [80]. For C. glutamicum priming and activation of murine macrophages by trehalose dimycolates were reported [81].
6. Top Layer
Cell surface molecules extracted from different corynebacteria consist of over 90% carbohydrates and of a minor portion (less than 10%) of proteins. Analyses of surface saccharides of C. diphtheriae strains indicated the presence of sugars such as N-acetylglucosamine, N-acetylgalactosamine, galactose, mannose, and sialic acid [82]. In C. amycolatum and C. xerosis, a neutral glucan consisting mainly of glucose with an apparent mass of 110 kDa was found, in addition to arabinomannans consisting of arabinose and mannose in a 1 : 1 ratio of 13 and 1.7 kDa [29, 33]. Additionally, the above-mentioned lipoarabinomannan and lipomannan were detected in the outer layer, besides trehalose dicorynomycolate, trehalose monocorynomycolate, and phospholipids. Interestingly, the same lipid composition was found for whole bacteria, indicating that all classes of lipid molecules were exposed on the corynebacterial cell surface [33], in contrast to the mycobacterial situation [83]. Also, a more distinct composition of plasma membrane and mycomembrane was reported for C. glutamicum [64].
While Puech and coworkers could only detect a few bands on coomassie-stained SDS polyacrylamide gels for different corynebacteria, proteome analyses supported the idea that a plethora of proteins is exposed on the surface of corynebacteria. Corresponding studies carried out for C. diphtheriae [84], C. efficiens [85], C. glutamicum [64, 85, 86], C. jeikeium [87], and C. pseudotuberculosis [88] revealed a significant number of proteins for every single species. Often, these proteins are uncharacterized; some clearly have functions for nutrient uptake and growth, while others are involved in host pathogen interactions.
6.1. Corynebacterial Porins
Since the permeability barrier of the mycolic acid layer most likely hinders nutrient uptake by plasma membrane transporters, the outer membrane has to be selectively permeabilized to allow growth. For this purpose, porin proteins are inserted into the membrane. In general these are functionally equivalent to Gram-negative porins but resemble a completely different structure, multimers of α-helical subunits instead of the Gram-negative trimeric β-barrels.
Corynebacterial porins are best investigated in C. glutamicum, where several different channel-forming proteins were identified, namely, PorA, PorB, PorC, and PorH [89–92]. Homologs of these were found in Corynebacterium callunae, C. efficiens [93], and C. diphtheria [94]. Despite the fact that C. amycolatum does not have corynemycolic acids and contains only small amounts of extractable lipids [33, 95, 96], a channel-forming protein was also isolated from this pathogenic species [97]. Reconstitution experiments with purified PorA and PorH from C. glutamicum and homology studies indicated that the major cell wall channels of C. callunae, C. diphtheriae, C. efficiens, and C. glutamicum are formed of two porins, one of the PorA and one of the PorH type [98]. Furthermore, for C. glutamicum PorA and PorH, an O-mycolation was shown, an extremely unusual modification [99].
Comparative studies of C. glutamicum wild type and a PorA-lacking mutant strain revealed a drastically decreased susceptibility of the mutant towards ampicillin, kanamycin, streptomycin, tetracycline, and gentamicin [92]. Corresponding studies with pathogenic corynebacteria are missing; however, based on the structural and functional similarities depicted above, differences in the porin repertoire might be one reason for different antibiotic susceptibility observed for the different species.
6.2. S-Layer Proteins
In some Corynebacterium strains, the cell surface is covered with a crystalline surface layer composed of single protein species, which are anchored in the corynebacterial outer membrane [100]. In C. glutamicum, the S-layer protein PS2 has an apparent molecular weight of 63 kDa and is anchored in the mycomembrane by a C-terminal hydrophobic domain [101]. Electron and atomic force microscopy applications revealed a highly ordered hexagonal S-layer [31, 101–104] and a certain degree of variability in different C. glutamicum isolates [103]. The PS2 encoding cspB gene is located on a genomic island [103], a situation often found for virulence genes; however, no distinct phenotype was found for S-layer mutant strains.
6.3. Sialidase
Sialidases, also designated as neuraminidases, are glycosyl hydrolases that catalyze the removal of terminal sialic acid residues from a variety of glycoconjugates of the host surface [105]. The sugar is subsequently metabolized or used to decorate the surface of the pathogen. In fact, C. diphtheriae exposes sialic acids on its outer surface [82]. Sialidase activity was first identified in a crude preparation of diphtheria toxin [106], and sialidase production and composition of cell surface carbohydrates of C. diphtheriae seem to be directly depending on iron concentration in the medium [107–109]. A putative exosialidase, designated NanH, was identified in C. diphtheriae. Biochemical studies revealed trans-sialylation activity; however, it is still unclear if NanH is involved in sialic acids decoration or not [107].
6.4. Pili
Proteinaceous protrusions such as fimbriae and pili are pivotal players for the attachment of bacteria to abiotic and biotic surfaces. In corynebacteria, pili were first reported in Corynebacterium renale [110]. Later detailed molecular biological analyses were carried out in C. diphtheriae (for review, see [111]). C. diphtheriae type strain NCTC13129 produces three distinct pilus structures, SpaA-, SpaD-, and SpaH-type pili, which are polymerized by specific class C sortases [112] and covalently linked to cell surface by sortase F [113]. Each type of pilus is composed of the name-giving shaft proteins, SpaA, SpaD, and SpaH, and minor pili subunits, that is, SpaB, SpaC, SpaE, SpaF, SpaG, and SpaI. For SpaB and SpaC, a function in cell line specificity of pathogen attachment was shown [114]. Ott and coworkers observed that different C. diphtheriae isolates are characterized by a different pili repertoire [115] which is characterized by different biophysical properties [116]. Interestingly, pili formation and adhesion rate are not strictly coupled processes and bald strains are also able to attach to host cells [115], indicating the presence of adhesion factors besides pili.
Besides in C. diphtheriae, pilus gene clusters were found in several pathogenic corynebacteria including Corynebacterium accolens, C. amycolatum, C. aurimucosum, Corynebacterium glucuronolyticum, C. jeikeium, Corynebacterium pseudogenitalium, C. striatum, Corynebacterium tuberculostearicum, and C. urealyticum, [111].
6.5. 67-72p Hemagglutinin
Besides pili, C. diphtheriae exhibits nonfimbrial surface proteins, 67-72p [117]. Recent studies indicated that 67-72p is encoded by a single gene, DIP0733 [118]. The corresponding gene product, DIP0733, enables C. diphtheriae to recognize and bind specifically to human erythrocytes, a process-designated hemagglutination. In vitro experiments with protein-coated latex beads indicated that DIP0733 contributes to invasion and induction of apoptosis in HEp-2 cells [118].
6.6. NlpC/P60 Proteins
Pathogen factors responsible for adhesion are at least partially characterized; however, the molecular background of invasion remains unclear. Based on a comprehensive analysis of proteins secreted by C. diphtheriae [84], Ott and coworker [119] started to characterize the surface-associated protein DIP1281, annotated as invasion associated protein. DIP1281 I is a member of the NlpC/P60 family, a large superfamily of several diverse groups of proteins [120], including putative proteases and probably invasion-associated proteins. They are found in bacteria, bacteriophages, RNA viruses, and eukaryotes and various members are highly conserved among nonpathogenic and pathogenic corynebacteria such as C. diphtheriae, C. efficiens, C. glutamicum, and C. jeikeium. DIP1281 mutant cells completely lacked the ability to adhere to host cells and consequently to invade these. Based on proteome, fluorescence and atomic force microscopy, it was concluded that DIP1281 is a pleotropic effector of the C. diphtheriae outer surface rather than a specific virulence factor [119], an idea that was supported by results obtained for corresponding mutants of the nonpathogenic C. glutamicum R strain [121]. Nevertheless, proteins of this family seem to play a major role in corynebacterial cell surface organization and cell separation. Furthermore, the results obtained with C. diphtheriae mutant strains [119] support the idea that the corynebacterial cell envelope components are important determinants of host pathogen interactions.
7. Concluding Remarks
Corynebacteria show a fascinating complex cell wall synthesis and architecture (see also Figure 1). Enormous progress was made in the characterization of fatty acids, sugar moieties, and cell wall polysaccharides; however, the protein content of the top envelope layer has only been characterized partly, despite its probable importance for intercellular communication and host pathogen interaction, as targets for therapy or its importance for biotechnological production. Furthermore, porin and S-layer proteins may have interesting biotechnological functions in respect to surface display of proteins and setup of nanostructures [100, 122].
References
- 1.Ventura M, Canchaya C, Tauch A, et al. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiology and Molecular Biology Reviews. 2007;71(3):495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhi XY, Li WJ, Stackebrandt E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. International Journal of Systematic and Evolutionary Microbiology. 2009;59(3):589–608. doi: 10.1099/ijs.0.65780-0. [DOI] [PubMed] [Google Scholar]
- 3.Bernard K. The genus Corynebacterium and other medically relevant coryneform-like bacteria. Journal of Clinical Microbiology. 2012;50(10):3152–3158. doi: 10.1128/JCM.00796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels—Corynebacterium glutamicum as versatile cell factory. Current Opinion in Biotechnology. 2012;23(4):631–640. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Vertes AA, Inui M, Yukawa H. Postgenomic approaches to using corynebacteria as biocatalysts. Annual Review of Microbiology. 2012;66:521–550. doi: 10.1146/annurev-micro-010312-105506. [DOI] [PubMed] [Google Scholar]
- 6.Cerdeño-Tárraga AM, Efstratiou A, Dover LG, et al. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Research. 2003;31(22):6516–6523. doi: 10.1093/nar/gkg874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikeda M, Nakagawa S. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Applied Microbiology and Biotechnology. 2003;62(2-3):99–109. doi: 10.1007/s00253-003-1328-1. [DOI] [PubMed] [Google Scholar]
- 8.Kalinowski J, Bathe B, Bartels D, et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. Journal of Biotechnology. 2003;104(1–3):5–25. doi: 10.1016/s0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- 9.Trost E, Götker S, Schneider J, et al. Complete genome sequence and lifestyle of black-pigmented Corynebacterium aurimucosum ATCC 700975 (formerly C. nigricans CN-1) isolated from a vaginal swab of a woman with spontaneous abortion. BMC Genomics. 2010;11(1, article 91) doi: 10.1186/1471-2164-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schröder J, Glaub A, Schneider J, Trost E, Tauch A. Draft genome sequence of Corynebacterium bovis DSM, 20582, which causes clinical mastitis in dairy cows. Journal of Bacteriology. 2012;194(16, article 4437) doi: 10.1128/JB.00839-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trost E, Blom J, Soares Sde C, et al. Pangenomic study of Corynebacterium diphtheriae that provides insights into the genomic diversity of pathogenic isolates from cases of classical diphtheria. Endocarditis, and Pneumonia, Journal of Bacteriology. 2012;194(12):3199–3215. doi: 10.1128/JB.00183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sangal V, Tucker NP, Burkovski A, Hoskisson PA. Draft genome sequence of Corynebacterium diphtheriae biovar intermedius NCTC, 5011. Journal of Bacteriology. 2012;194(17, article 4738) doi: 10.1128/JB.00939-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sangal V, Tucker NP, Burkovski A, Hoskisson PA. The draft genome sequence of Corynebacterium diphtheriae bv. mitis NCTC, 3529 reveals significant diversity between the primary disease-causing biovars. Journal of Bacteriology. 2012;194(12, article 3269) doi: 10.1128/JB.00503-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tauch A, Kaiser O, Hain T, et al. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. Journal of Bacteriology. 2005;187(13):4671–4682. doi: 10.1128/JB.187.13.4671-4682.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tauch A, Schneider J, Szczepanowski R, et al. Ultrafast pyrosequencing of Corynebacterium kroppenstedtii DSM44385 revealed insights into the physiology of a lipophilic Corynebacterium that lacks mycolic acids. Journal of Biotechnology. 2008;136(1-2):22–30. doi: 10.1016/j.jbiotec.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Cerdeira LT, Pinto AC, Schneider MP, et al. Whole-genome sequence of Corynebacterium pseudotuberculosis PAT10 strain isolated from sheep in Patagonia, Argentina. Journal of Bacteriology. 2011;193(22):6420–6421. doi: 10.1128/JB.06044-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerdeira LT, Schneider MP, Pinto AC, et al. Complete genome sequence of Corynebacterium pseudotuberculosis strain CIP 52. 97, isolated from a horse in Kenya. Journal of Bacteriology. 2011;193(24):7025–7026. doi: 10.1128/JB.06293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopes T, Silva A, Thiago R, et al. Complete genome sequence of Corynebacterium pseudotuberculosis strain Cp267, isolated from a llama. Journal of Bacteriology. 2012;194(13):3567–3568. doi: 10.1128/JB.00461-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pethick FE, Lainson AF, Yaga R, et al. Complete genome sequences of Corynebacterium pseudotuberculosis strains 3/99-5 and 42/02-A, isolated from sheep in scotland and Australia, respectively. Journal of Bacteriology. 2012;194(17):4736–4737. doi: 10.1128/JB.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pethick FE, Lainson AF, Yaga R, et al. Complete genome sequence of Corynebacterium pseudotuberculosis strain 1/06-A, isolated from a Horse in North America. Journal of Bacteriology. 2012;194(16):4476–4476. doi: 10.1128/JB.00922-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos RTJ, Silva A, Carneiro AR, et al. Genome sequence of the Corynebacterium pseudotuberculosis Cp316 strain, isolated from the abscess of a Californian horse. Journal of Bacteriology. 2012;194(23):6620–6621. doi: 10.1128/JB.01616-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva A, Ramos RTJ, Carneiro AR, et al. Complete genome sequence of Corynebacterium pseudotuberculosis Cp31, isolated from an Egyptian buffalo. Journal of Bacteriology. 2012;194(23):6663–6664. doi: 10.1128/JB.01782-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silva A, Schneider MPC, Cerdeira L, et al. Complete genome sequence of Corynebacterium pseudotuberculosis I19, a strain isolated from a cow in Israel with bovine mastitis. Journal of Bacteriology. 2011;193(1):323–324. doi: 10.1128/JB.01211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trost E, Ott L, Schneider J, et al. The complete genome sequence of Corynebacterium pseudotuberculosis FRC41 isolated from a 12-year-old girl with necrotizing lymphadenitis reveals insights into gene-regulatory networks contributing to virulence. BMC Genomics. 2010;11(1, article 728) doi: 10.1186/1471-2164-11-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schröder J, Maus I, Meyer K, et al. Complete genome sequence, lifestyle, and multi-drug resistance of the human pathogen Corynebacterium resistens DSM 45100 isolated from blood samples of a leukemia patient. BMC Genomics. 2012;13(1, article 141) doi: 10.1186/1471-2164-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trost E, Al-Dilaimi A, Papavasiliou P, et al. Comparative analysis of two complete Corynebacterium ulcerans genomes and detection of candidate virulence factors. BMC Genomics. 2011;12, article 383 doi: 10.1186/1471-2164-12-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tauch A, Trost E, Tilker A, et al. The lifestyle of Corynebacterium urealyticum derived from its complete genome sequence established by pyrosequencing. Journal of Biotechnology. 2008;136(1-2):11–21. doi: 10.1016/j.jbiotec.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Schröder J, Maus I, Trost E, Tauch A. Complete genome sequence of Corynebacterium variabile DSM 44702 isolated from the surface of smear-ripened cheeses and insights into cheese ripening and flavor generation. BMC Genomics. 2011;12, article 545 doi: 10.1186/1471-2164-12-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daffé M. The cell envelope of corynebacteria. In: Eggeling L, Bott M, editors. Handbook of Corynebacterium glutamicum. Boca Raton, Fla, USA: Taylor & Francis; 2005. pp. 121–148. [Google Scholar]
- 30.Eggeling L, Gurdyal SB, Alderwick L. Structure and synthesis of the cell wall. In: Burkovski A, editor. Corynebacteria. Norfolk, UK: Caister Academic Press; 2008. pp. 267–294. [Google Scholar]
- 31.Chami M, Bayan N, Dedieu JC, Leblon G, Shechter E, Gulik-Krzywicki T. Organization of the outer layers of the cell envelope of Corynebacterium glutamicum: a combined freeze-etch electron microscopy and biochemical study. Biology of the Cell. 1995;83(2-3):219–229. doi: 10.1016/0248-4900(96)81311-6. [DOI] [PubMed] [Google Scholar]
- 32.Marienfeld S, Uhlemann EM, Schmid R, Krämer R, Burkovski A. Ultrastructure of the Corynebacterium glutamicum cell wall. Antonie van Leeuwenhoek. 1997;72(4):291–297. doi: 10.1023/a:1000578811089. [DOI] [PubMed] [Google Scholar]
- 33.Puech V, Chami M, Lemassu A, et al. Structure of the cell envelope of corynebacteria: importance of the non-covalently bound lipids in the formation of the cell wall permeability barrier and fracture plane. Microbiology. 2001;147(5):1365–1382. doi: 10.1099/00221287-147-5-1365. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(10):3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niederweis M, Danilchanka O, Huff J, Hoffmann C, Engelhardt H. Mycobacterial outer membranes: in search of proteins. Trends in Microbiology. 2010;18(3):109–116. doi: 10.1016/j.tim.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoischen C, Kramer R. Membrane alteration is necessary but not sufficient for effective glutamate secretion in Corynebacterium glutamicum . Journal of Bacteriology. 1990;172(6):3409–3416. doi: 10.1128/jb.172.6.3409-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nampoothiri K, Hoischen C, Bathe B, et al. Expression of genes of lipid synthesis and altered lipid composition modulates L-glutamate efflux of Corynebacterium glutamicum . Applied Microbiology and Biotechnology. 2002;58(1):89–96. doi: 10.1007/s00253-001-0861-z. [DOI] [PubMed] [Google Scholar]
- 38.Shibukawa M, Kurima M, Oruchi S. L-Glutamic acid fermentation with molasses. XII. Relationship between the kind of phospholipids and their fatty acid composition in the mechanism of extracellular accumulation of L-glutamate. Agricultural and Biological Chemistry. 1970;34(8):1136–1141. [Google Scholar]
- 39.Brennan PJ, Lehane DP. The phospholipids of corynebacteria. Lipids. 1971;6(6):401–409. doi: 10.1007/BF02531377. [DOI] [PubMed] [Google Scholar]
- 40.Collins MD, Goodfellow M, Minnikin DE. Fatty acid composition of some mycolic acid-containing coryneform bacteria. Journal of General Microbiology. 1982;128(11):2503–2509. doi: 10.1099/00221287-128-11-2503. [DOI] [PubMed] [Google Scholar]
- 41.Minnikin DE, Goodfellow M, Collins MD. Lipid composition in the classification an identification of coryneform and relaxed taxa. In: Bousfield IJ, Galley G, editors. Coryneform Bacteria. London, UK: Academic Press; 1978. p. p. 85. [Google Scholar]
- 42.Radmacher E, Alderwick LJ, Besra GS, et al. Two functional FAS-I type fatty acid synthases in Corynebacterium glutamicum . Microbiology. 2005;151(7):2421–2427. doi: 10.1099/mic.0.28012-0. [DOI] [PubMed] [Google Scholar]
- 43.Özcan N, Ejsing CS, Shevchenko A, Lipski A, Morbach S, Krämer R. Osmolality, temperature, and membrane lipid composition modulate the activity of betaine transporter BetP in Corynebacterium glutamicum . Journal of Bacteriology. 2007;189(20):7485–7496. doi: 10.1128/JB.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rock CO, Cronan JE. Escherichia coli as a model for the regulation of dissociable (type II) fatty acid biosynthesis. Biochimica et Biophysica Acta. 1996;1302(1):1–16. doi: 10.1016/0005-2760(96)00056-2. [DOI] [PubMed] [Google Scholar]
- 45.Mishra AK, Krumbach K, Rittmann D, et al. Lipoarabinomannan biosynthesis in Corynebacterineae: the interplay of two α(1→2)-mannopyranosyltransferases MptC and MptD in mannan branching. Molecular Microbiology. 2011;80(5):1241–1259. doi: 10.1111/j.1365-2958.2011.07640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreira LO, Mattos-Guaraldi AL, Andrade AFB. Novel lipoarabinomannan-like lipoglycan (CdiLAM) contributes to the adherence of Corynebacterium diphtheriae to epithelial cells. Archives of Microbiology. 2008;190(5):521–530. doi: 10.1007/s00203-008-0398-y. [DOI] [PubMed] [Google Scholar]
- 47.Mishra AK, Krumbach K, Rittmann D, et al. Deletion of manC in Corynebacterium glutamicum results in a phospho-myo-inositol mannoside- and lipoglycan-deficient mutant. Microbiology. 2012;158(7):1908–1917. doi: 10.1099/mic.0.057653-0. [DOI] [PubMed] [Google Scholar]
- 48.Sutcliffe IC. Macroamphiphilic cell envelope components of Rhodococcus equi and closely related bacteria. Veterinary Microbiology. 1997;56(3-4):287–299. doi: 10.1016/s0378-1135(97)00097-7. [DOI] [PubMed] [Google Scholar]
- 49.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriological Reviews. 1972;36(4):407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kato K, Strominger JL, Kotani S. Structure of the cell wall of Corynebacterium diphtheriae. I. Mechanism of hydrolysis by the L-3 enzyme and the structure of the peptide. Biochemistry. 1968;7(8):2762–2773. doi: 10.1021/bi00848a010. [DOI] [PubMed] [Google Scholar]
- 51.Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiology and Molecular Biology Reviews. 2005;69(4):585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dover LG, Cerdeño-Tárraga AM, Pallen MJ, Parkhill J, Besra GS. Comparative cell wall core biosynthesis in the mycolated pathogens, Mycobacterium tuberculosis and Corynebacterium diphtheriae . FEMS Microbiology Reviews. 2004;28(2):225–250. doi: 10.1016/j.femsre.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 53.Gibson KJC, Eggeling L, Maughan WN, et al. Disruption of Cg-Ppm1, a polyprenyl monophosphomannose synthase, and the generation of lipoglycan-less mutants in Corynebacterium glutamicum . The Journal of Biological Chemistry. 2003;278(42):40842–40850. doi: 10.1074/jbc.M307988200. [DOI] [PubMed] [Google Scholar]
- 54.Letek M, Fiuza M, Ordóñez E, et al. Cell growth and cell division in the rod-shaped actinomycete Corynebacterium glutamicum . Antonie van Leeuwenhoek. 2008;94(1):99–109. doi: 10.1007/s10482-008-9224-4. [DOI] [PubMed] [Google Scholar]
- 55.Valbuena N, Letek M, Ordóñez E, et al. Characterization of HMW-PBPs from the rod-shaped actinomycete Corynebacterium glutamicum: peptidoglycan synthesis in cells lacking actin-like cytoskeletal structures. Molecular Microbiology. 2007;66(3):643–657. doi: 10.1111/j.1365-2958.2007.05943.x. [DOI] [PubMed] [Google Scholar]
- 56.Letek M, Fiuza M, Villadangos AF, Mateos LM, Gil JA. Cytoskeletal proteins of Actinobacteria . International Journal of Cell Biology. 2012;2012:10 pages. doi: 10.1155/2012/905832.905832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lederer E, Adam A, Ciorbaru R, Petit JF, Wietzerbin J. Cell walls of mycobacteria and related organisms; chemistry and immunostimulant properties. Molecular and Cellular Biochemistry. 1975;7(2):87–104. doi: 10.1007/BF01792076. [DOI] [PubMed] [Google Scholar]
- 58.McNeil M, Daffe M, Brennan PJ. Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. The Journal of Biological Chemistry. 1990;265(30):18200–18206. [PubMed] [Google Scholar]
- 59.Alderwick LJ, Dover LG, Seidel M, et al. Arabinan-deficient mutants of Corynebacterium glutamicum and the consequent flux in decaprenylmonophosphoryl-D-arabinose metabolism. Glycobiology. 2006;16(11):1073–1081. doi: 10.1093/glycob/cwl030. [DOI] [PubMed] [Google Scholar]
- 60.Alderwick LJ, Radmacher E, Seidel M, et al. Deletion of Cg-emb in Corynebacterianeae leads to a novel truncated cell wall arabinogalactan, whereas inactivation of Cg-ubiA results in an Arabinan-deficient mutant with a cell wall galactan core. The Journal of Biological Chemistry. 2005;280(37):32362–32371. doi: 10.1074/jbc.M506339200. [DOI] [PubMed] [Google Scholar]
- 61.Seidel M, Alderwick LJ, Birch HL, Sahm H, Eggeling L, Besra GS. Identification of a novel arabinofuranosyltransferase AftB involved in a terminal step of cell wall arabinan biosynthesis in Corynebacterianeae, such as Corynebacterium glutamicum and Mycobacterium tuberculosis . The Journal of Biological Chemistry. 2007;282(20):14729–14740. doi: 10.1074/jbc.M700271200. [DOI] [PubMed] [Google Scholar]
- 62.Seidel M, Alderwick LJ, Sahm H, Besra GS, Eggeling L. Topology and mutational analysis of the single Emb arabinofuranosyltransferase of Corynebacterium glutamicum as a model of Emb proteins of Mycobacterium tuberculosis . Glycobiology. 2007;17(2):210–219. doi: 10.1093/glycob/cwl066. [DOI] [PubMed] [Google Scholar]
- 63.Gebhardt H, Meniche X, Tropis M, Krämer R, Daffé M, Morbach S. The key role of the mycolic acid content in the functionality of the cell wall permeability barrier in Corynebacterineae . Microbiology. 2007;153(5):1424–1434. doi: 10.1099/mic.0.2006/003541-0. [DOI] [PubMed] [Google Scholar]
- 64.Marchand CH, Salmeron C, Raad RB, et al. Biochemical disclosure of the mycolate outer membrane of Corynebacterium glutamicum . Journal of Bacteriology. 2012;194(3):587–597. doi: 10.1128/JB.06138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Portevin D, de Sousa-D’Auria C, Houssin C, et al. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(1):314–319. doi: 10.1073/pnas.0305439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gande R, Dover LG, Krumbach K, et al. The two carboxylases of Corynebacterium glutamicum essential for fatty acid and mycolic acid synthesis. Journal of Bacteriology. 2007;189(14):5257–5264. doi: 10.1128/JB.00254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Portevin D, de Sousa-D’Auria C, Montrozier H, et al. The acyl-AMP ligase FadD32 and AccD4-containing acyl-CoA carboxylase are required for the synthesis of mycolic acids and essential for mycobacterial growth: identification of the carboxylation product and determination of the acyl-CoA carboxylase components. The Journal of Biological Chemistry. 2005;280(10):8862–8874. doi: 10.1074/jbc.M408578200. [DOI] [PubMed] [Google Scholar]
- 68.Tzvetkov M, Klopprogge C, Zelder O, Liebl W. Genetic dissection of trehalose biosynthesis in Corynebacterium glutamicum: inactivation of trehalose production leads to impaired growth and an altered cell wall lipid composition. Microbiology. 2003;149(7):1659–1673. doi: 10.1099/mic.0.26205-0. [DOI] [PubMed] [Google Scholar]
- 69.Wolf A, Krämer R, Morbach S. Three pathways for trehalose metabolism in Corynebacterium glutamicum ATCC13032 and their significance in response to osmotic stress. Molecular Microbiology. 2003;49(4):1119–1134. doi: 10.1046/j.1365-2958.2003.03625.x. [DOI] [PubMed] [Google Scholar]
- 70.Tropis M, Meniche X, Wolf A, et al. The crucial role of trehalose and structurally related oligosaccharides in the biosynthesis and transfer of mycolic acids in Corynebacterineae . The Journal of Biological Chemistry. 2005;280(28):26573–26585. doi: 10.1074/jbc.M502104200. [DOI] [PubMed] [Google Scholar]
- 71.Joliff G, Mathieu L, Hahn V, et al. Cloning and nucleotide sequence of the csp1 gene encoding PS1, one of the two major secreted proteins of Corynebacterium glutamicum: the deduced N-terminal region of PS1 is similar to the Mycobacterium antigen 85 complex. Molecular Microbiology. 1992;6(16):2349–2362. doi: 10.1111/j.1365-2958.1992.tb01410.x. [DOI] [PubMed] [Google Scholar]
- 72.Brand S, Niehaus K, Pühler A, Kalinowski J. Identification and functional analysis of six mycolyltransferase genes of Corynebacterium glutamicum ATCC 13032: the genes cop1, cmt1, and cmt2 can replace each other in the synthesis of trehalose dicorynomycolate, a component of the mycolic acid layer of the cell envelope. Archives of Microbiology. 2003;180(1):33–44. doi: 10.1007/s00203-003-0556-1. [DOI] [PubMed] [Google Scholar]
- 73.de Sousa-D’Auria C, Kacem R, Puech V, et al. New insights into the biogenesis of the cell envelope of corynebacteria: identification and functional characterization of five new mycoloyltransferase genes in Corynebacterium glutamicum . FEMS Microbiology Letters. 2003;224(1):35–44. doi: 10.1016/S0378-1097(03)00396-3. [DOI] [PubMed] [Google Scholar]
- 74.Kacem R, de Sousa-D’Auria C, Tropis M, et al. Importance of mycoloyltransferases on the physiology of Corynebacterium glutamicum . Microbiology. 2004;150(1):73–84. doi: 10.1099/mic.0.26583-0. [DOI] [PubMed] [Google Scholar]
- 75.Varela C, Rittmann D, Singh A, et al. MmpL genes are associated with mycolic acid metabolism in mycobacteria and corynebacteria. Chemistry and Biology. 2012;19(4):498–506. doi: 10.1016/j.chembiol.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Y, Shi F, Tao G, Wang X. Purification and structure analysis of mycolic acids in Corynebacterium glutamicum . Journal of Microbiology. 2012;50(2):235–240. doi: 10.1007/s12275-012-1459-0. [DOI] [PubMed] [Google Scholar]
- 77.Meniche X, Labarre C, de Sousa-D’Auria C, et al. Identification of a stress-induced factor of Corynebacterineae that is involved in the regulation of the outer membrane lipid composition. Journal of Bacteriology. 2009;191(23):7323–7332. doi: 10.1128/JB.01042-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Indrigo J, Hunter RL, Jr., Actor JK. Cord factor trehalose 6,6′-dimycolate (TDM) mediates trafficking events during mycobacterial infection of murine macrophages. Microbiology. 2003;149(8):2049–2059. doi: 10.1099/mic.0.26226-0. [DOI] [PubMed] [Google Scholar]
- 79.Hard GC. Comparative toxic effect of the surface lipid of Corynebacterium ovis on peritoneal macrophages. Infection and Immunity. 1975;12(6):1439–1449. doi: 10.1128/iai.12.6.1439-1449.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tashjian JJ, Campbell SG. Interaction between caprine macrophages and Corynebacterium pseudotuberculosis: an electron microscopic study. American Journal of Veterinary Research. 1983;44(4):690–693. [PubMed] [Google Scholar]
- 81.Chami M, Andréau K, Lemassu A, et al. Priming and activation of mouse macrophages by trehalose 6,6′-dicorynomycolate vesicles from Corynebacterium glutamicum . FEMS Immunology and Medical Microbiology. 2002;32(2):141–147. doi: 10.1111/j.1574-695X.2002.tb00546.x. [DOI] [PubMed] [Google Scholar]
- 82.Mattos-Guaraldi AL, Cappelli EA, Previato JO, Formiga LCD, Andrade AFB. Characterization of surface saccharides in two Corynebacterium diphtheriae strains. FEMS Microbiology Letters. 1999;170(1):159–166. doi: 10.1111/j.1574-6968.1999.tb13369.x. [DOI] [PubMed] [Google Scholar]
- 83.Ortalo-Magné A, Lemassu A, Lanéelle MA, et al. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. Journal of Bacteriology. 1996;178(2):456–461. doi: 10.1128/jb.178.2.456-461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hansmeier N, Chao TC, Kalinowski J, Pühler A, Tauch A. Mapping and comprehensive analysis of the extracellular and cell surface proteome of the human pathogen Corynebacterium diphtheriae . Proteomics. 2006;6(8):2465–2476. doi: 10.1002/pmic.200500360. [DOI] [PubMed] [Google Scholar]
- 85.Hansmeier N, Chao TC, Pühler A, Tauch A, Kalinowski J. The cytosolic, cell surface and extracellular proteomes of the biotechnologically important soil bacterium Corynebacterium efficiens YS-314 in comparison to those of Corynebacterium glutamicum ATCC 13032. Proteomics. 2006;6(1):233–250. doi: 10.1002/pmic.200500144. [DOI] [PubMed] [Google Scholar]
- 86.Hermann T, Finkemeier M, Pfefferle W, Wersch G, Krämer R, Burkovski A. Two-dimensional electrophoretic analysis of Corynebacterium glutamicum membrane fraction and surface proteins. Electrophoresis. 2000;21(3):654–659. doi: 10.1002/(SICI)1522-2683(20000201)21:3<654::AID-ELPS654>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 87.Hansmeier N, Chao TC, Daschkey S, et al. A comprehensive proteome map of the lipid-requiring nosocomial pathogen Corynebacterium jeikeium K411. Proteomics. 2007;7(7):1076–1096. doi: 10.1002/pmic.200600833. [DOI] [PubMed] [Google Scholar]
- 88.Pacheco LG, Slade SE, Seyffert N, et al. A combined approach for comparative exoproteome analysis of Corynebacterium pseudotuberculosis . BMC Microbiology. 2011;11, article 12 doi: 10.1186/1471-2180-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lichtinger T, Rieß FG, Burkovski A, et al. The low-molecular-mass subunit of the cell wall channel of the gram-positive Corynebacterium glutamicum: immunological localization, cloning and sequencing of its gene porA. European Journal of Biochemistry. 2001;268(2):462–469. doi: 10.1046/j.1432-1033.2001.01913.x. [DOI] [PubMed] [Google Scholar]
- 90.Hünten P, Costa-Riu N, Palm D, Lottspeich F, Benz R. Identification and characterization of PorH, a new cell wall channel of Corynebacterium glutamicum . Biochimica et Biophysica Acta - Biomembranes. 2005;1715(1):25–36. doi: 10.1016/j.bbamem.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 91.Costa-Riu N, Maier E, Burkovski A, Krämer R, Lottspeich F, Benz R. Identification of an anion-specific channel in the cell wall of the Gram-positive bacterium Corynebacterium glutamicum . Molecular Microbiology. 2003;50(4):1295–1308. doi: 10.1046/j.1365-2958.2003.03754.x. [DOI] [PubMed] [Google Scholar]
- 92.Costa-Riu N, Burkovski A, Krämer R, Benz R. PorA represents the major cell wall channel of the gram-positive bacterium Corynebacterium glutamicum . Journal of Bacteriology. 2003;185(16):4779–4786. doi: 10.1128/JB.185.16.4779-4786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hünten P, Schiffler B, Lottspeich F, Benz R. PorH, a new channel-forming protein present in the cell wall of Corynebacterium efficiens and Corynebacterium callunae . Microbiology. 2005;151(7):2429–2438. doi: 10.1099/mic.0.27903-0. [DOI] [PubMed] [Google Scholar]
- 94.Schiffler B, Barth E, Daffé M, Benz R. Corynebacterium diphtheriae: identification and characterization of a channel-forming protein in the cell wall. Journal of Bacteriology. 2007;189(21):7709–7719. doi: 10.1128/JB.00864-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Collins MD, Burton RA, Jones D. Corynebacterium amycolatum sp. nov. a new mycolic acid-less Corynebacterium species from human skin. FEMS Microbiology Letters. 1988;49(3):349–352. [Google Scholar]
- 96.Barreau C, Bimet F, Kiredjian M, Rouillon N, Bizet C. Comparative chemotaxonomic studies of mycolic acid-free coryneform bacteria of human origin. Journal of Clinical Microbiology. 1993;31(8):2085–2090. doi: 10.1128/jcm.31.8.2085-2090.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dörner U, Schiffler B, Lanéelle MA, Daffé M, Benz R. Identification of a cell-wall channel in the corynemycolic acid-free Gram-positive bacterium Corynebacterium amycolatum . International Microbiology. 2009;12(1):29–38. [PubMed] [Google Scholar]
- 98.Barth E, Barceló MA, Kläckta C, Benz R. Reconstitution experiments and gene deletions reveal the existence of two-component major cell wall channels in the genus Corynebacterium . Journal of Bacteriology. 2010;192(3):786–800. doi: 10.1128/JB.01142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huc E, Meniche X, Benz R, et al. O-mycoloylated proteins from Corynebacterium: an unprecedented post-translational modification in bacteria. The Journal of Biological Chemistry. 2010;285(29):21908–21912. doi: 10.1074/jbc.C110.133033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bayan N, Houssin C, Chami M, Leblon G. Mycomembrane and S-layer: two important structures of Corynebacterium glutamicum cell envelope with promising biotechnology applications. Journal of Biotechnology. 2003;104(1–3):55–67. doi: 10.1016/s0168-1656(03)00163-9. [DOI] [PubMed] [Google Scholar]
- 101.Chami M, Bayan N, Peyret JL, Gulik-Krzywicki T, Leblon G, Shechter E. The S-layer protein of Corynebacterium glutamicum is anchored to the cell wall by its C-terminal hydrophobic domain. Molecular Microbiology. 1997;23(3):483–492. doi: 10.1046/j.1365-2958.1997.d01-1868.x. [DOI] [PubMed] [Google Scholar]
- 102.Scheuring S, Stahlberg H, Chami M, Houssin C, Rigaud JL, Engel A. Charting and unzipping the surface layer of Corynebacterium glutamicum with the atomic force microscope. Molecular Microbiology. 2002;44(3):675–684. doi: 10.1046/j.1365-2958.2002.02864.x. [DOI] [PubMed] [Google Scholar]
- 103.Hansmeier N, Bartels FW, Ros R, et al. Classification of hyper-variable Corynebacterium glutamicum surface-layer proteins by sequence analyses and atomic force microscopy. Journal of Biotechnology. 2004;112(1-2):177–193. doi: 10.1016/j.jbiotec.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 104.Dupres V, Alsteens D, Pauwels K, Dufrêne YF. In vivo imaging of S-layer nanoarrays on Corynebacterium glutamicum . Langmuir. 2009;25(17):9653–9655. doi: 10.1021/la902238q. [DOI] [PubMed] [Google Scholar]
- 105.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiology and Molecular Biology Reviews. 2004;68(1):132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Blumberg BS, Warren I. The effect of sialidase on transferrins and other serum proteins. Biochimica et Biophysica Acta. 1961;50(1):90–101. [Google Scholar]
- 107.Kim S, Oh DB, Kwon O, Kang HA. Identification and functional characterization of the NanH extracellular sialidase from Corynebacterium diphtheriae . Journal of Biochemistry. 2010;147(4):523–533. doi: 10.1093/jb/mvp198. [DOI] [PubMed] [Google Scholar]
- 108.de Oliveira Moreira L, Andrade AFB, Vale MDD, et al. Effects of iron limitation on adherence and cell surface carbohydrates of Corynebacterium diphtheriae strains. Applied and Environmental Microbiology. 2003;69(10):5907–5913. doi: 10.1128/AEM.69.10.5907-5913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Warren L, Spearing CW. Sialidase (Neuraminidase) of Corynebacterium diphtheriae . Journal of Bacteriology. 1963;86:950–955. doi: 10.1128/jb.86.5.950-955.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yanagawa R, Otsuki K, Tokui T. Electron microscopy of fine structure of Corynebacterium renale with special reference to pili. Japanese Journal of Veterinary Research. 1968;16(1):31–37. [PubMed] [Google Scholar]
- 111.Rogers EA, Das A, Ton-That H. Adhesion by pathogenic corynebacteria. Advances in Experimental Medicine and Biology. 2011;715:91–103. doi: 10.1007/978-94-007-0940-9_6. [DOI] [PubMed] [Google Scholar]
- 112.Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends in Microbiology. 2004;12(5):228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 113.Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Research in Microbiology. 2005;156(3):289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 114.Mandlik A, Swierczynski A, Das A, Ton-That H. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Molecular Microbiology. 2007;64(1):111–124. doi: 10.1111/j.1365-2958.2007.05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ott L, Höller M, Rheinlaender J, Schäffer TE, Hensel M, Burkovski A. Strain-specific differences in pili formation and the interaction of Corynebacterium diphtheriae with host cells. BMC Microbiology. 2010;10, article 257 doi: 10.1186/1471-2180-10-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rheinlaender J, Gräbner A, Ott L, Burkovski A, Schaffer TE. Contour and persistence length of Corynebacterium diphtheriae pili by atomic force microscopy. European Biophysics Journal. 2012;41(6):561–570. doi: 10.1007/s00249-012-0818-4. [DOI] [PubMed] [Google Scholar]
- 117.Colombo AV, Hirata R, Jr., de Souza CMR, et al. Corynebacterium diphtheriae surface proteins as adhesins to human erythrocytes. FEMS Microbiology Letters. 2001;197(2):235–239. doi: 10.1111/j.1574-6968.2001.tb10609.x. [DOI] [PubMed] [Google Scholar]
- 118.Sabbadini PS, Assis MC, Trost E, et al. Corynebacterium diphtheriae 67-72p hemagglutinin, characterized as the protein DIP0733, contributes to invasion and induction of apoptosis in HEp-2 cells. Microbial Pathogenesis. 2012;52(3):165–176. doi: 10.1016/j.micpath.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 119.Ott L, Höller M, Gerlach RG, et al. Corynebacterium diphtheriae invasion-associated protein (DIP1281) is involved in cell surface organization, adhesion and internalization in epithelial cells. BMC Microbiology. 2010;10, article 2 doi: 10.1186/1471-2180-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biology. 2003;4(2, article R11) doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tsuge Y, Ogino H, Teramoto H, Inui M, Yukawa H. Deletion of cgR_1596 and cgR_2070, encoding NlpC/P60 proteins, causes a defect in cell separation in Corynebacterium glutamicum R. Journal of Bacteriology. 2008;190(24):8204–8214. doi: 10.1128/JB.00752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tateno T, Hatada K, Tanaka T, Fukuda H, Kondo A. Development of novel cell surface display in Corynebacterium glutamicum using porin. Applied Microbiology and Biotechnology. 2009;84(4):733–739. doi: 10.1007/s00253-009-2021-9. [DOI] [PubMed] [Google Scholar]


