Abstract
Background:
Several recent studies demonstrated that microRNAs are stably detectable in plasma/serum. We tested whether miR-18a, which is located in the miR-17-92 cluster and reported to be highly expressed in tissues of oesophageal squamous cell carcinoma (ESCC), served as a plasma biomarker in patients with ESCC.
Methods:
This study was divided into three steps: (1) confirmation of higher miR-18a levels in primary ESCC tissues and cell lines than normal ESCC tissues and a human fibroblast cell line. (2) Evaluation of the plasma miR-18a assay using quantitative RT–PCR by comparing results from 106 consecutive patients with ESCC and 54 healthy volunteers. (3) Evaluation of the assay for monitoring tumour dynamics in patients with ESCC.
Results:
(1) Expression of miR-18a was significantly higher in ESCC tissues (P=0.0020) and ESCC cell lines (P=0.0121) than normal tissues and fibroblasts. (2) Plasma concentrations of miR-18a were significantly higher in ESCC patients than healthy volunteers (P<0.0001; ESCC patients vs healthy volunteers (mean±s.d.): 11.77±13.45 vs 0.73±0.54 amol μl−1). The value of the area under the receiver-operating characteristic (ROC) curve (AUC) was 0.9449. Furthermore, the ROC curves to detect early ESCC such as pTis-1 and pStage0-I showed AUCs of 0.9479 and 0.9642, respectively. (3) Plasma levels of miR-18a were significantly lower in postoperative samples than preoperative samples (P=0.0076).
Conclusion:
Plasma miR-18a may be a very useful biomarker for cancer detection and the monitoring of tumour dynamics in patients with ESCC.
Keywords: esophageal cancer, plasma, microRNA, biomarker
Oesophageal squamous cell carcinoma (ESCC) is the eighth most common cancer in the world and is the sixth leading cause of cancer-related death (Enzinger and Mayer, 2003). In Asian countries, ESCC accounts for ∼90% of oesophageal carcinomas (Hiyama et al, 2007) and remains one of the most aggressive carcinomas in the gastrointestinal tract. Recent improvements in surgical techniques and perioperative management have reduced operation-related deaths during perioperative periods; however, ESCC still has extremely poor prognosis. Therefore, primary tumours must be detected at an early stage, and recurrent disease must be diagnosed when it is still minimal or clinically occult, in order to improve the prognosis of patients with ESCC.
To identify biological factors involved in the malignant behaviour of ESCC, several recent studies have elucidated that certain molecules, such as p53, cyclin D1, and FAS, have important roles in tumorigenesis and the development of ESCC (Hollestein et al, 1990; Adélaide et al, 1995; Gratas et al, 1998). In clinical settings, however, few molecules have been assayed as therapeutic and/or prognostic biomarkers. Conventional serum tumour markers, such as carcinoembryonic antigen (CEA) and squamous cell carcinoma antigen (SCC), have been used as convenient diagnostic assays (Kosugi et al, 2004; Mroczko et al, 2008) for early detection and monitoring the tumour dynamics of ESCC. These serum tumour markers, however, lack sufficient sensitivity and specificity to facilitate early detection of cancer. Therefore, the significance of detecting novel biomarkers using a less invasive diagnostic assay for ESCC should be emphasised.
In recent years, several studies have shown that microRNAs (miRNAs), which are involved in carcinogenesis and development of various cancers (He et al, 2005; Lu et al, 2005; Calin and Croce, 2006; He et al, 2007), are detectable in plasma/serum (Calin and Croce, 2006; Chen et al, 2008; Filipowics et al, 2008; Ichikawa et al, 2012). Tumour-derived miRNAs are resistant to endogenous ribonuclease activity because these may bind to proteins, such as the Argonaute 2 protein and high-density lipoprotein (Arroyo et al, 2011; Vickers et al, 2011), or are packaged by secretory particles including apoptotic bodies and exosomes in plasma/serum (Hasselmann et al, 2001; Mitchell et al, 2008; Cocucci et al, 2009; Kosaka et al, 2010). Therefore, miRNAs can be present in a remarkably stable form (Mitchell et al, 2008; Hongyan and Guo-Chang, 2011) and the expression level of serum miRNAs is reproducible and consistent among individuals (Chen et al, 2008; Mitchell et al, 2008). Furthermore, secretory vesicles including miRNAs can function as intercellular transmitters (Valadi et al, 2007; Skog et al, 2008; Rechavi et al, 2009). These findings should open up a new and interesting field in the screening and monitoring of ESCC patients. However, to date, there have been few reports on the role of circulating miRNAs in plasma/serum of patients with ESCC (Zhang et al, 2010; Komatsu et al, 2011).
In this study, we tested miR-18a, which was located in the miR-17-92 cluster and was previously reported to be highly expressed in the tissues of SCC (Chang et al, 2008; Avissar et al, 2009; Gao et al, 2011; Liu et al, 2011). This cluster has been reported to have a potential oncogenic function in various tumours. Therefore, we hypothesised that miR-18a could be detected in plasma and may be a useful plasma biomarker for patients with ESCC. Consequently, we demonstrated that miR-18a was very useful for measuring plasma miRNA in ESCC among previous reported plasma/serum miRNAs. Our results provide evidence that plasma levels of miR-18 contribute to cancer detection and can be used to monitor ESCC patients to a clinically satisfactory degree of sensitivity and specificity.
Patients and Methods
Patients and samples
This study was approved by the Institutional Review Board of Kyoto Prefectural University of Medicine and each subject provided signed informed consent. Between November 2008 and June 2011, 106 consecutive preoperative plasma samples were collected from consecutive patients with ESCC, who underwent radical oesophagectomy (R0 or R1) at Kyoto Prefectural University of Medicine. Patient characteristics with respect to age, sex, venous invasion, lymphatic invasion, T-stage, N-stage, stage of disease, and resection status are described in Table 1. Preoperative plasma samples were collected from 106 patients with ESCC. All patients were pathologically diagnosed as having ESCC using surgical specimens and biopsies. Ten ESCC specimens were collected from patients undergoing oesophagectomy, and eleven normal oesophageal tissue specimens of the abdominal oesophagus were collected from patients undergoing total gastrectomy for gastric cancer because the non-cancerous oesophageal tissues of ESCC patients may exhibit dysplasia or potentially be cancerous tissue. As a control, plasma was collected from 54 healthy volunteers. Volunteers were medical personnel and patients with benign disease such as cholelithiasis. They underwent medical examinations and did not have any oesophageal disease or other cancerous disease. The stage of tumours was assessed according to the Union of International Control of Cancer (UICC) classification (Sobin et al, 2009).
Table1. ESCC patients characteristics and plasma miR-18a concentrations.
| Variable | Patient (n=106) | Plasma miR-18a mean concentration (amol μl−1)a | P-valueb |
|---|---|---|---|
|
Age (years) | |||
| <65 | 50 (47%) | 13.4 | 0.3308 |
| 65≦ |
56 (53%) |
10.3 |
|
|
Sex | |||
| Male | 87 (82%) | 12.7 | 0.1399 |
| Female |
19 (18%) |
7.4 |
|
|
Venous invasionc | |||
| v0 | 67 (63%) | 12.6 | 0.2993 |
| v1 | 22 (21%) | 7.3 | |
| v2–3 |
17 (16%) |
14.5 |
|
|
Lymphatic invasionc | |||
| ly0 | 61 (58%) | 14.4 | 0.1014 |
| ly1 | 30 (28%) | 7.2 | |
| ly2–3 |
15 (14%) |
10.4 |
|
|
T-stagec | |||
| Tis | 5 (5%) | 26.7 | 0.1560 |
| T1 | 37 (35%) | 15.0 | |
| T2 | 13 (12%) | 10.0 | |
| T3 | 42 (40%) | 8.9 | |
| T4 |
9 (8%) |
6.3 |
|
|
N-stagec | |||
| N0 | 43 (41%) | 12.2 | 0.3147 |
| N1 | 33 (31%) | 14.0 | |
| N2 | 19 (18%) | 10.9 | |
| N3 |
11 (10%) |
4.9 |
|
|
Stagec | |||
| 0 | 2 (2%) | 32.3 | 0.2793 |
| I | 28 (26%) | 12.1 | |
| II | 28 (26%) | 14.6 | |
| III | 37 (35%) | 8.3 | |
| IV |
11 (11%) |
11.5 |
|
|
Resection status | |||
| R0 | 77 (73%) | 12.4 | 0.5301 |
| R1 | 29 (27%) | 10.1 | |
Abbreviations: ESCC=oesophageal squamous cell carcinoma; ROC=receiver-operating characteristic.
P-values were considered significant at 0.05.
Cutoff value of plasma miR-18a concentrations in healthy volunteers (ROC curve with Younden Index): 1.99 amol μl−1.
The Mann–Whitney U-test and Kruskal–Wallis H-test were performed to compare plasma miRNA concentrations.
TNM classification.
Peripheral blood (7 ml) was obtained from each patient at the same time before the initial skin incision for oesophagectomy, and healthy volunteer controls. Immediately after collection, blood samples were subjected to isolation of cell-free nucleic acids using a 3-spin protocol (1500 r.p.m. for 30 min, 3000 r.p.m. for 5 min, 4500 r.p.m. for 5 min) to prevent contamination by cellular nucleic acids. Plasma samples were stored at −80 °C until further processing. Resected specimens were fixed in formalin and embedded in paraffin for pathological diagnosis. Macroscopic and microscopic classification of tumours were based on the UICC/TNM staging system (Sobin et al, 2009).
RNA extraction
Total RNA was extracted from 400 μl of plasma using a mirVana PARIS Kit (Ambion, Austin, TX, USA), and finally eluted into 100 μl of preheated (95 °C) Elution Solution according to the manufacturer's protocol. Using formalin-fixed paraffin-embedded tissues, total RNA was extracted from four slices of 15 μm thick (total 60 μm in thickness) using a Recover All Total Nucleic Acid Isolation Kit (Ambion), and then eluted into 60 μl of Elution Solution according to the manufacturer's protocol.
Protocol for the detection of microRNA
The amounts of miRNAs in plasma were quantified by qRT-PCR using human TaqMan MicroRNA Assay Kits (Applied Biosystems, Foster City, CA, USA). The reverse transcription reaction was carried out with a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) in 15 μl containing 5 μl of RNA extract, 0.15 μl of 100 mℳ dNTPs, 1 μl of MultiScribe Reverse Transcriptase (50 U μl−1), 1.5 μl of 10 × Reverse Transcription Buffer, 0.19 μl of RNase inhibitor (20 U μl−1), 1 μl of gene-specific primer (has-miR-18a; Assay ID: 002422, RNU6B; Assay ID: 001093), and 4.16 μl of Nuclease-free water. For the synthesis of cDNA, reaction mixtures were incubated at 16 °C for 30 min, at 42 °C for 30 min, and at 85 °C for 5 min and then held at 4 °C. Next, 1.33 μl of cDNA solution was amplified using 10 μl of TaqMan 2 × Universal PCR Master Mix with no AmpErase UNG (Applied Biosystems), 1 μl of gene-specific primer/probe, and 7.67 μl of nuclease-free water in a final volume of 20 μl. Quantitative PCR was run on a 7300 Real-time PCR system (Applied Biosystems) and reaction mixtures were incubated at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. The cycle threshold (Ct) values were calculated with SDS 1.4 software (Livak and Schmittgen, 2001).
The amounts of miRNAs in plasma were calculated on a standard curve constructed with the use of synthetic miRNAs, the mirVana miRNA Reference Panel (Ambion). Standard reference miRNAs were amplified for each reaction. However, the expression of miRNAs from tissue samples was normalised using the 2-ΔΔCT method relative to U6 small nuclear RNA (RNU6B). ΔCt was calculated by subtracting Ct values of RNU6B from Ct values of the miRNAs of interest. ΔΔCt was then calculated by subtracting ΔCt of normal ESCC tissue from ΔCt of ESCC tissues. The change in gene expression was calculated with the equation 2-ΔΔCt (Akobeng, 2007; Sobin).
ESCC cell lines and culture
ESCC cell lines, such as TE2, TE9, TE13, and KYSE70 and fibroblast cell line WI-38, were purchased from RIKEN Cell Bank (Tsukuba, Japan). WI-38 cell was cultured in Dulbecco's minimum essential medium: F12 medium and the others in Roswell Park Memorial Institute (RPMI)-1640 medium (Sigma, St Louis, MO, USA). All media were purchased from Sigma and supplemented with 100 ml l−1 FBS (Trace Scientific, Melbourne, Australia). All cell lines were cultured in 50 ml l−1 carbon dioxide at 37 °C in a humidified chamber.
Statistical analysis
The Mann–Whitney test was used to compare differences in plasma miRNA concentrations and miRNA ratios between the cancer group and healthy group, and the Wilcoxon test was used to compare the paired plasma samples before and 1 month after oesophagectomy. A P-value of 0.05 was considered significant. Receiver-operating characteristic (ROC) curves and area under the curve (AUC) were used to assess the feasibility of using plasma miRNA concentrations as a diagnostic tool for detecting ESCC. The Younden Index was used to determine the cutoff value for plasma miRNA concentrations (Akobeng, 2007).
Results
Study design to develop a novel biomarker of plasma miRNA
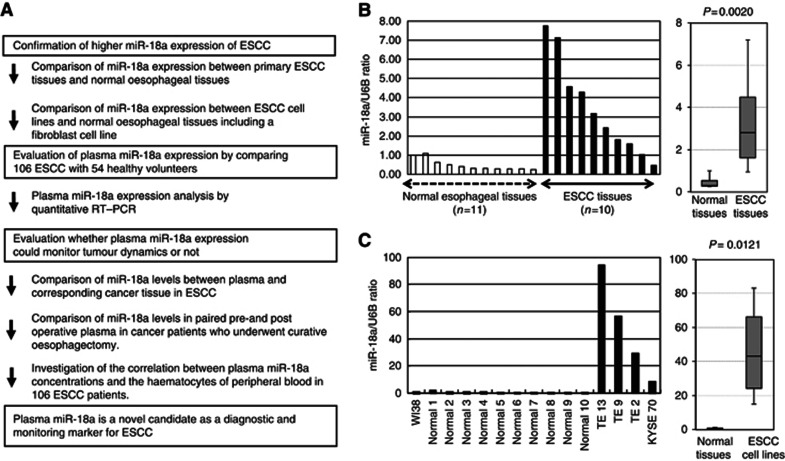
This study was divided into three parts: (1) Confirmation of higher miR-18a levels in primary ESCC tissue and cell lines than normal ESCC tissues and human fibroblasts. (2) Evaluation of the miR-18a assay using quantitative RT–PCR by comparing results from 106 consecutive patients with ESCC and 54 healthy volunteers. (3) Evaluation for monitoring tumour dynamics by the plasma miR-18a assay in patients with ESCC (Figure 1A).
Figure 1.
(A) Study design to develop a novel biomarker of plasma microRNA. (B) The expression of miR-18a in ESCC tissues. Differential expressions of miR-18a in ESCC tissues were compared with those of normal tissues by a waterfall plot (A). Expression levels of miR-18a were significantly higher in ESCC tissues than normal oesophageal tissues (P=0.0020). The upper and lower limits of the boxes and lines inside the boxes indicate the 75th and 25th percentiles and the median, respectively. Upper and lower horizontal bars denote the 90th and 10th percentiles. (C) The expression of miR-18a in ESCC cell lines. Differential expressions of miR-18a in ESCC cell lines were compared with those of a fibroblast cell line and normal tissues. In ESCC cell lines, expression levels of miR-18a were significantly higher than in both a fibroblast cell line and normal oesophageal tissues (P=0.0121). *Normal means normal tissues including the normal fibroblast cell line, such as WI-38.
MiR-18a expression in primary ESCC tissues and ESCC cell lines
To confirm whether miR-18a is highly expressed in primary ESCC, expression of miR-18a in 10 ESCC tissues and 11 normal oesophageal tissues was determined by quantitative RT–PCR. Also, human ESCC cell lines TE-2, TE-9, TE-13, and KYSE70 and a human fibroblast cell line WI-38 were evaluated by quantitative RT–PCR. Results are shown in Figures 1B and C after normalisation to control U6 expression. Levels of miR-18a were significantly higher in ESCC tissues than normal ESCC tissues (P=0.0020; Figure 1B). Also, in ESCC cell lines, levels of miR-18a were significantly higher than in the fibroblast cell line and normal oesophageal tissues (P=0.0121; Figure 1C). Consequently, expression of miR-18a was significantly increased in most ESCC tissues and cell lines, but not in normal tissues and fibroblasts.
Evaluation of plasma miR-18a expression using quantitative RT–PCR in ESCC patients
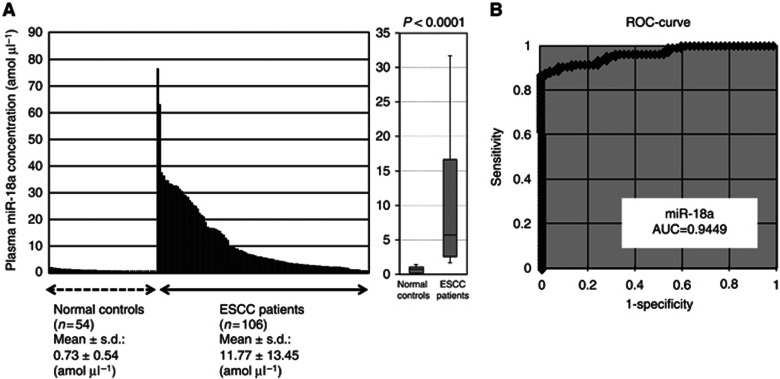
Next, we hypothesised that higher miR-18a expression in primary tumours would influence plasma levels of miR-18a in ESCC patients. To evaluate the appropriateness of this assay, we first conducted amplification by RT–PCR of a 10-fold serial dilution of the mirVana miRNA Reference Panel. The linearity of quantitative RT–PCR was confirmed from concentrations of 1 fmol to 0.0001 fmol of each synthetic miRNA, such as miR-18a (R2=0.9919) between the logarithm of the amount of input miRNA and Ct values (Supplementary Figure 1S). Although some investigators have determined quantities of plasma miRNAs by comparing internal control miRNAs (Ng et al, 2009), it remains controversial as to which miRNAs are suitable as internal controls for plasma assays. Therefore, we confirmed a linear correlation between the logarithm of the amount of input synthetic miRNA and the cycle threshold value on real-time PCR, as well as the feasibility of extracting total RNA and amplifying specific miRNA in plasma samples. Based on these findings, we utilised the absolute concentration for measuring plasma miRNA in this study (Supplementary Figure 1S).
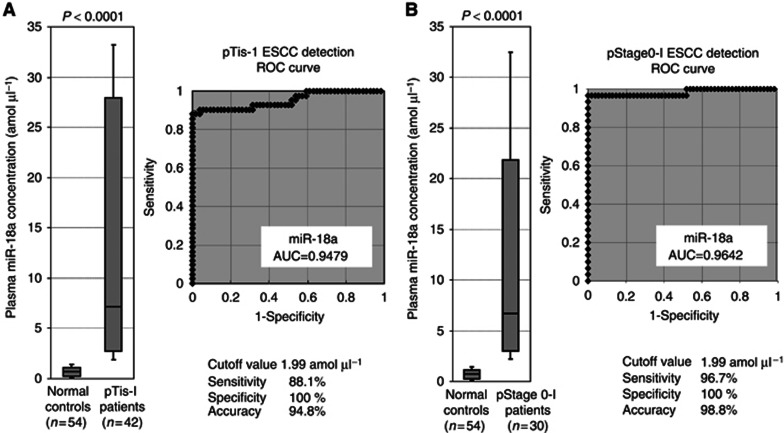
Using this assay, circulating miRNA such as miR-18a was detectable in all samples from 106 consecutive ESCC patients and 54 healthy volunteers. The differential expression of plasma miR-18a in ESCC patients was compared with that in normal healthy volunteers by a waterfall plot (Figure 2A). Concentrations of miR-18a were significantly higher in plasma from ESCC patients than in healthy volunteers (P<0.0001; ESCC patients vs healthy volunteers (mean±s.d.): 11.77±13.45 vs 0.73±0.54 amol μl−1; Figure 2A). Representation of the data using an ROC plot showed strong separation between the two groups, with an AUC of 0.9449 (Figure 2B). To detect cutoff points that could discriminate cancer patients from healthy volunteers, we utilised ROC curves with the Younden index (Akobeng, 2007). An optimal cutoff point was indicated at 1.99 amol μl−1 with a sensitivity of 86.8% and a specificity of 100%. Furthermore, plasma miR-18a concentrations in patients with early ESCC such as pathological (p) Tis-1* or pStage0-I** were significantly higher than those of healthy volunteers (P<0.0001*, P<0.0001** pTis-1* vs pStage0-I** vs healthy volunteers (mean): 16.36 vs 13.46 vs 0.73 amol μl−1; Figures 3A and B). The values of AUC used to detect early ESCC such as pTis-1 and pStage0-I were 0.9479 and 0.9642, respectively. The diagnostic sensitivity, specificity, and accuracy of early ESCC at the cutoff value of 1.99 amol μl−1 using ROC curves were 88.1%, 100%, and 94.8% in pT0-1 or 96.7%, 100%, and 98.8% in pStage0-I, respectively (Figures 3A and B). Indeed, the sensitivity of plasma miR-18a for ESCC detection was much higher than that of conventional tumour markers including SCC, CEA, and SYFRA at our institute (pT0-1: SCC, CEA, and SYFRA: 25.0%, 21.1%, and 12.5% pStage0-I: SCC, CEA, and SYFRA: 24.1%, 25.0%, and 3.4% details not shown). Our results provided evidence that plasma levels of miR-18a can be used to distinguish early ESCC patients from healthy volunteers to a clinically satisfactory degree of sensitivity and specificity.
Figure 2.
(A) Plasma miR-18a concentrations in 106 consecutive ESCC patients and 54 healthy volunteers. Using a real-time RT–PCR assay, circulating miRNA, such as miR-18a, was detectable in all samples from 106 consecutive ESCC patients and 54 healthy volunteers. Differential expressions of plasma miR-18a with ESCC patients were compared with those of normal healthy volunteers by a waterfall plot. Concentrations of miR-18a were significantly higher in plasma from ESCC patients than those in healthy volunteers (P<0.0001). The upper and lower limits of the boxes and lines inside the boxes indicate the 75th and 25th percentiles and the median, respectively. Upper and lower horizontal bars denote the 90th and 10th percentiles. (B) Receiver-operating characteristic (ROC) curve analysis in the miR-18a assay for detecting ESCC patients. Receiver-operating characteristic analysis showed the greatest AUC of 0.9449 for miR-18a.
Figure 3.
(A) Plasma miR-18a concentrations in 42 pathological Tis-T1 ESCC patients and 54 healthy volunteers. Concentrations of miR-18a were significantly higher in plasma from pathological Tis-T1 ESCC patients than in healthy volunteers (P<0.0001). The value of AUC to detect pTis-1 ESCC was 0.9479. (B) Plasma miR-18a concentrations in 30 pathological Stage0-1 ESCC patients and 54 healthy volunteers. Concentrations of miR-18a were significantly higher in plasma from pathological Stage0-I ESCC patients than in healthy volunteers (P<0.0001). The value of AUC to detect pTis-1 ESCC was 0.9642.
Also, we examined the association of plasma miR-18a concentrations with clinicopathological factors in 106 consecutive ESCC patients. Table 1 shows expression levels of miR-18a in plasma. However, we cannot present any significant correlations between the miR-18a level and clinical factors.
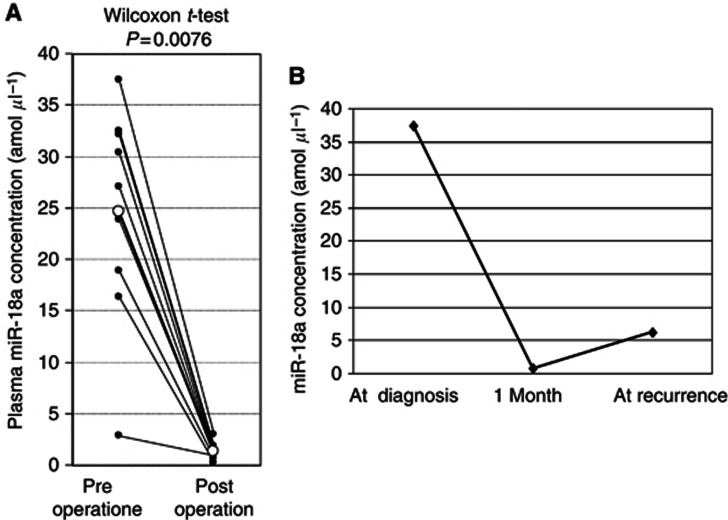
Evaluation of the use of this assay for monitoring tumour dynamics in patients with ESCC
We divided ESCC patients into two groups with high or low plasma miR-18a concentrations. We utilised a cutoff value of 1.99 amol μl−1, which is the plasma miR-18a concentration detected by ROC curves using the Younden index to distinguish ESCC patients from healthy volunteers. To clarify the correlation of miR-18a expression between plasma and ESCC tissue, we selected 13 patients with high plasma miR-18a concentrations and compared miR-18a expression between plasma and ESCC tissues in each patient. As a result, in all high plasma miR-18a concentration patients analysed, miR-18a showed higher expression in primary ESCC tissues (100%) than the cutoff value of 1.66 for the tissue miR-18a/U6B ratio based on the mean value+2 s.d. in normal oesophageal tissues (Supplementary Table 1). Moreover, the concentration of miR-18a was analysed in paired pre- and postoperative plasma samples from ESCC patients who underwent curative oesophagectomy, and was found to be significantly reduced in postoperative samples (P=0.0076; Figure 4A). These findings indicated that the level of miR-18a in plasma might reflect the expression in the tumour. In one patient, re-elevation of plasma miR-18a concentration was found at haematogenous recurrence after surgery (Figure 4B). Solitary liver metastasis, whose tumour size was 5 cm diameter, was detected at 6 months after curative oesophagectomy. These results clearly showed that detection of miR-18a in plasma should provide a new complementary tumour marker for ESCC.
Figure 4.
(A) Comparison of plasma miR-18a concentrations between pre- and postoperative samples from ESCC patients. Concentrations of plasma miR-18a were significantly lower in postoperative samples compared than those in preoperative samples (P=0.0076). (B) In one patient, re-elevation of plasma miR-18a concentrations was found at recurrence after surgery.
Correlation between plasma miR-18a concentrations and the haematocytes of peripheral blood
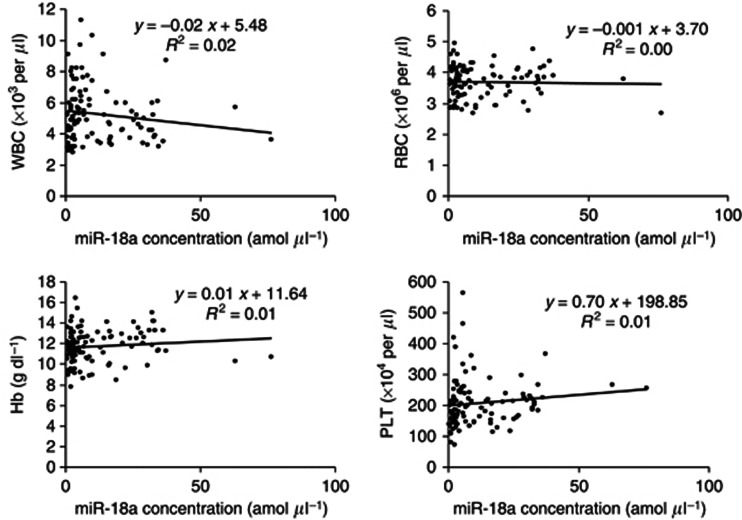
A recent report demonstrated that some circulating miRNAs may be derived from peripheral blood cells (Pritchard et al, 2011). Therefore, we determined the correlation between plasma miR-18a concentrations and the haematocytes of peripheral blood in 106 ESCC patients. The results showed that there was no significant correlation between plasma miR-18a concentrations and any type of haematocyte in peripheral blood (Figure 5). Therefore, these findings imply that plasma miR-18a may not be derived from any type of haematocyte.
Figure 5.
Correlation between plasma miR-18a concentrations and the haematocytes of peripheral blood in ESCC patients. There was no significant correlation between plasma miR-18a concentrations and any type of peripheral haematocyte and plasma.
Discussion
During the last decade, non-coding RNAs, so-called miRNAs, have been demonstrated to regulate gene expression by targeting mRNAs for translational repression or cleavage. MicroRNAs have recently been proven to contribute to carcinogenesis and the progression of various cancer tumours and may provide new therapeutic strategies such as biomarkers and therapeutic targets for cancers (He et al, 2005; Lu et al, 2005; Calin and Croce, 2006; He et al, 2007). Furthermore, miRNAs were demonstrated to be present in a remarkably stable form in plasma/serum and the expression level of serum/plasma miRNAs is reproducible and consistent among individuals (Chen et al, 2008; Mitchell et al, 2008). In fact, accumulating reports have suggested that the presence of circulating miRNAs and their potential use as novel biomarkers for cancers, such as prostate cancer (Mitchell et al, 2008), leukemia (Lawrie et al, 2008), oral cancer (Wong et al, 2008), pancreatic cancer (Wong et al, 2008; Morimura et al, 2011), colorectal cancer (Wang et al, 2009), lung cancer (Hu et al, 2010), breast cancer (Heneghan et al, 2010), and gastric cancer (Tsujiura et al, 2010: Konishi et al, 2012).
Concerning ESCC, there have been only two reports on the role of circulating miRNAs in plasma/serum patients with ESCC (Zhang et al, 2010: Komatsu et al, 2011). Zhang investigated serum miRNA profiles in patients with ESCC using Solexa sequencing. Among 25 selected candidate miRNAs analysed, they identified seven serum miRNAs (miR-10a, 22, 100, 148b, 223, 133a, and 127-3p) as ESCC biomarkers. The area under the ROC curve for selected miRNAs ranged from 0.817 to 0.949. The AUCs using ROC curves for selected miRNAs were higher than for conventional serum tumour markers. Also, we analysed several candidate miRNAs in plasma of patients with ESCC, whose expression analyses are frequently reported in SCC tissues (Chang et al, 2008; Avissar et al, 2009). As a result, miR-21 and miR-375 were stably detectable in plasma and reflected tumour dynamics as a tumour marker for ESCC (Komatsu et al, 2011). Moreover, concentrations of plasma miR-21 were significantly lower in postoperative samples than those in preoperative samples, and re-elevation of plasma miR-21 concentrations were found at recurrence after surgery despite the lack of an increase in conventional serum tumour markers. These findings could open up a new and promising field in the screening and monitoring of ESCC. However, these reports did not always warrant that these miRNAs were only candidate biomarkers for ESCC. Therefore, more sensitive and promising candidate miRNAs should be detected in clinical settings.
In this study, we tested miR-18a, which was located in the miR-17-92 cluster and was previously reported to be highly expressed in the tissues of SCC (Chang et al, 2008; Avissar et al, 2009; Gao et al, 2011; Liu et al, 2011), as a new candidate miRNA in the plasma of patients with ESCC. miR-18a was expressed as part of the miR-17-92 cluster, which consists of seven miRNAs: miR-17-3p, miR-17-5p, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a. This cluster has been reported to have a potential oncogenic function in various tumours, with elevations in this cluster being caused by genome amplification or transcriptional activation by MYC (Woods et al, 2007). miR-18a has been shown to be significantly upregulated in gastric cancer (Yao et al, 2009), diffuse large B-cell lymphoma (Alencar et al, 2011), urothelial carcinomas of the bladder (Ayala de la Pena et al, 2011), nasopharyngeal carcinoma (Li et al, 2011), hepatocellular carcinoma, pancreatic carcinoma, and colorectal carcinomas (Motoyama et al, 2009). In colorectal cancer tissues, patients with overexpressed miR-18a had a poorer clinical prognosis. Also, miR-18a was confirmed to directly target ER-α and showed higher levels of expression in ER-α-negative clinical tumours (Leivonen et al, 2009). Interestingly, miR-18a had a proliferative effect on hepatocellular carcinoma cells, but an inhibitory effect on breast cancer cells (Leivonen et al, 2009).
Previously, we reported that the presence of markedly elevated miR-18a levels in plasma could provide new complementary tumour markers for pancreatic cancer (Morimura et al, 2011). Namely, plasma concentrations of miR-18a were significantly higher in pancreatic cancer patients than in controls. The value of the area under the ROC curve (AUC) was 0.9369. Therefore, we hypothesised that miR-18a may also be a useful biomarker in the plasma of patients with ESCC. As a result, we confirmed that expression of miR-18a was significantly higher in ESCC tissues and ESCC cell lines than normal tissues and fibroblasts. Plasma concentrations of miR-18a were significantly higher in ESCC patients than controls. The AUC was 0.9449, which was markedly higher than other miRNAs in plasma of cancer patients (Tsujiura et al, 2010; Komatsu et al, 2011; Morimura et al, 2011; Konishi et al, 2012). Furthermore, the ROC curve used to detect early ESCC such as pTis-1 or pStage0-I showed an AUC of 0.9479 or 0.9642, respectively, suggesting that plasma miR-18a concentrations may be a useful biomarker for the detection of ESCC.
Next, we investigated whether plasma miR-18a concentrations could reflect tumour dynamics in ESCC by three different analyses. One is the comparison between expression of miR-18a in plasma and primary tumour tissue, which demonstrated that plasma and primary ESCC tissue samples showed that high levels of plasma miR-18a represented higher expressions in primary ESCC tissues than in normal mucosa in all patients analysed (100%) (Supplementary Table 1). Some patients, however, showed a different pattern of miRNA levels, low plasma miR-18a with a high expression in ESCC tissue (data not shown). The reasons for these discrepancies in some patients remain to be identified; however, one possible explanation for this finding is the heterogeneity of primary tumours. Second analysis involves comparison of plasma miR-18a concentrations in paired plasma obtained before and after surgery. As a result, concentrations of miR-18a were significantly reduced postoperatively in patients with high preoperative plasma miR-18a (Figure 4A). These findings were similar to those in patients with ESCC, gastric, and pancreatic cancer in our previous report (Tsujiura et al, 2010; Komatsu et al, 2011; Morimura et al, 2011; Konishi et al, 2012). Third analysis was to determine that re-elevation of plasma miR-18a concentrations were found at recurrence after surgery even in one ESCC patient as well as our previous reports (Komatsu et al, 2011; Morimura et al, 2011) (Figure 4B). These findings clearly demonstrated that plasma concentrations of miR-18a reflect tumour dynamics and are available as a new plasma biomarker for monitoring tumour status such as residue and recurrence of ESCC.
Recently, Pritchard CC, Tewari M, and his colleagues reported the caution in a cancer biomarker study of circulating miRNAs because circulating miRNAs may be derived from peripheral blood cells (Pritchard et al, 2011). Therefore, we evaluated the correlation between plasma miR-18a concentrations and haematocytes of peripheral blood in 106 consecutive ESCC patients. As a result, there was no significant association between plasma miR-18a concentrations and any types of peripheral haematocyte (Figure 5). This result indicated that miR-18a expression in plasma may reflect tumour dynamics in ESCC patients. However, as secretion, kinetics, and metabolism in plasma miRNAs have not been clearly elucidated, this issue is currently under evaluation.
Taken together, we clearly demonstrated that plasma levels of miR-18a may potentially be useful for cancer screening and monitoring tumour dynamics in ESCC patients postoperatively. Nevertheless, many issues must be addressed before these findings can be translated into a clinically useful, non-invasive screening strategy for ESCC patients. We will prospectively confirm the usefulness of plasma miR-18a in a large number of patients and report on these results in the near future. Furthermore, if possible, more sensitive biomarkers in plasma miRNAs should be detected for translation into the clinical setting of ESCC. These issues are also currently under evaluation in an array-based study using large candidate numbers of miRNA and will be reported in the near future.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Adélaide J, Monges G, Dérdérian C, Seitz JF, Birnbaum D. Oesophageal cancer and amplification of the human cyclin D gene CCND1/PRAD1. Br J Cancer. 1995;71:64–68. doi: 10.1038/bjc.1995.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akobeng AK. Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Paediatr. 2007;96:644–647. doi: 10.1111/j.1651-2227.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- Alencar AJ, Malumbres R, Kozloski GA, Advani R, Talreja N, Chinichian S, Briones J, Natkunam Y, Sehn LH, Gascoyne RD, Tibshirani R, Lossos IS. MicroRNAs are independent predictors of outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Clin Cancer Res. 2011;17:4125–4135. doi: 10.1158/1078-0432.CCR-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute 2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avissar M, Christensen BC, Kelsey KT, Marsit CJl. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala de la Pena F, Kanasaki K, Kanasaki M, Tangirala N, Maeda G, Kalluri R. Loss of p53 and acquisition of angiogenic microRNA profile are insufficient to facilitate progression of bladder urothelial carcinoma in situ to invasive carcinoma. J Biol Chem. 2011;286:20778–20787. doi: 10.1074/jbc.M110.198069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Chang SS, Jiang WW, Smith I, Poeta LM, Begum S, Glazer C, Shan S, Westra W, Sidransky D, Califano JA. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;12:2791–2797. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Zen K, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight. Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gao W, Shen H, Liu L, Xu J, Xu J, Shu Y. MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol. 2011;4:557–566. doi: 10.1007/s00432-010-0918-4. [DOI] [PubMed] [Google Scholar]
- Gratas C, Tohma Y, Barnas C, Taniere P, Hainaut P, Ohgaki H. Up-regulation of Fas (APO-1/CD95) ligand and down-regulation of Fas expression in human esophageal cancer. Cancer Res. 1998;58:2057–2062. [PubMed] [Google Scholar]
- Hasselmann DO, Rappl G, Tilgen W, Reinhold U. Extracellular tyrosinase mRNA within apoptotic bodies is protected from degradation in human serum. Clin Chem. 2001;47:1488–1489. [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and esophageal cancer risk. Int J Cancer. 2007;121:1643–1658. doi: 10.1002/ijc.23044. [DOI] [PubMed] [Google Scholar]
- Hollstein MC, Metcalf RA, Welsh JA, Montesano R, Harris CC. Frequent mutation of the p53 gene in human esophageal cancer. Proc Natl Acad Sci USA. 1990;87:9958–9961. doi: 10.1073/pnas.87.24.9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongyan Zhu, Guo-Chang Fan. Extracellular/circulating microRNAs and their potential role in cardiovascular disease. Am J Cardiovasc Dis. 2011;2:138–149. [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- Ichikawa D, Komatsu S, Konishi H, Otsuji E. Circulating MicroRNA in Digestive Tract Cancers. Gastroenterology. 2012;142:1074–1078. doi: 10.1053/j.gastro.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105:104–111. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi H, Ichikawa D, Komatsu S, Shiozaki A, Tsujiura M, Takeshita H, Morimura R, Nagata H, Arita T, Kawaguchi T, Hirashima S, Fujiwara H, Okamoto K, Otsuji E. Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br J Cancer. 2012;4:740–747. doi: 10.1038/bjc.2011.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17442. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Nishimaki T, Kanda T, Nakagawa S, Ohashi M, Hatakeyama K. Clinical significance of serum carcinoembryonic antigen, carbohydrate antigen 19-9, and squamous cell carcinoma antigen levels in esophageal cancer patients. World J Surg. 2004;28:680–685. doi: 10.1007/s00268-004-6865-y. [DOI] [PubMed] [Google Scholar]
- Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- Leivonen SK, Makela R, Ostling P, Kohonen P, Haapa-Paananen S, Kleivi K, Enerly E, Aakula A, Hellstrom K, Sahlberg N, Kristensen VN, Borresen-Dale AL, Saviranta P, Perala M, Kallioniemi O. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene. 2009;28:3926–3936. doi: 10.1038/onc.2009.241. [DOI] [PubMed] [Google Scholar]
- Li T, Chen JX, Fu XP, Yang S, Zhang Z, Chen KhH, Li Y. microRNA expression profiling of nasopharyngeal carcinoma. Oncol Rep. 2011;25:1353–1363. doi: 10.3892/or.2011.1204. [DOI] [PubMed] [Google Scholar]
- Liu M, Wang Z, Yang S, Zhang W, He S, Hu C, Zhu H, Quan L, Bai J, Xu N. TNF-α is a novel target of miR-19a. Int J Oncol. 2011;4:1013–1022. doi: 10.3892/ijo.2011.924. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–440. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura R, Komatsu S, Ichikawa D, Takeshita H, Tujiura M, Nagata H, Konishi H, Shiozaki A, Ikoma H, Okamoto K, Ochiai T, Taniguchi H, Otsuji E. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 2011;11:1733–1740. doi: 10.1038/bjc.2011.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, Sugihara K, Mori M. Over- and under-expressed microRNAs in human colorectal cancer. Int J Oncol. 2009;34:1069–1075. doi: 10.3892/ijo_00000233. [DOI] [PubMed] [Google Scholar]
- Mroczko B, Kozłowski M, Groblewska M, Łukaszewicz M, Nikliński J, Jelski W, Laudański J, Chyczewski L, Szmitkowski M. The diagnostic value of the measurement of matrix metalloproteinase 9 (MMP-9), squamous cell cancer antigen (SCC) and carcinoembryonic antigen (CEA) in the sera of esophageal cancer patients. Clin Chim Acta. 2008;389:61–66. doi: 10.1016/j.cca.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res. 2011;5:492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O, Erlich Y, Amram H, Flomenblit L, Karginov FV, Goldstein I, Hannon GJ, Kloog Y. Genes Dev. 2009. pp. 1971–1979. [DOI] [PMC free article] [PubMed]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr., Carter BS, Krichevsky AM, Breakefield XO. Nat Cell Biol. 2008. pp. 1470–1476. [DOI] [PMC free article] [PubMed]
- Sobin LH, Gospodarowicz MK, Wittekind C.2009TNM classification of Malignant Tumors7th ednWiley-liss: New York [Google Scholar]
- Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–1179. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Nat Cell Biol. 2007. pp. 654–659. [DOI] [PubMed]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila Pa) 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microrna of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L, Zhang WG, Nan KJ, Song TS, Huang C. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2:963–970. doi: 10.3892/mmr_00000199. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, Dai J, Hu Z, Zhou X, Chen L, Zhang Y, Li Y, Qiu H, Xing J, Liang Z, Ren B, Yang C, Zen K, Zhang CY. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–1879. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.