Abstract
Background:
MicroRNAs (miRNAs) are involved in essential biological activities, and have been reported to exhibit differential expression profiles in various cancers. Our previous study demonstrated that intercellular adhesion molecule-2 (ICAM2) inhibition induces radiosensitisation in oral squamous cell carcinoma (OSCC) cells. Thus, we hypothesised that certain miRNAs play crucial roles in radioresistance in OSCC by regulating ICAM2 expression.
Methods:
Because predicted target gene analyses revealed that microRNA-125b (miR-125b) potentially regulates ICAM2 mRNA expression, we examined the association between miR-125b and radioresistance. The expression of miR-125b was investigated by real-time quantitative reverse transcriptase–PCR. For a functional analysis, miR-125b was transfected to OSCC-derived cells.
Results:
A downregulated expression of miR-125b was found in OSCC-derived cell lines and OSCC samples. The miR-125b-transfected cells showed a decreased proliferation rate, enhanced radiosensitivity to X-ray irradiation and diminished ICAM2 mRNA expression. Moreover, miR-125b expression correlated with OSCC tumour staging and survival.
Conclusion:
These findings suggested that the downregulated miR-125b expression was associated with proliferation and radioresistance mechanisms, probably through ICAM2 signalling. Thus, controlling the expression or activity of miR-125b might contribute to suppressing proliferation and overcoming radioresistance in OSCC.
Keywords: oral squamous cell carcinoma (OSCC), microRNA, miR-125b, intercellular adhesion molecule 2 (ICAM2), radioresistance
MicroRNAs (miRNAs) are involved in essential biological activities (Harfe, 2005; Carleton et al, 2007). Recent studies have revealed differential expression of miRNAs in various cancers, including oral cancer, suggesting that certain miRNAs play roles in tumourigenesis (Garzon et al, 2009; Wu et al, 2011). In addition, previous studies have revealed that some miRNAs are closely associated with clinical outcomes (Schetter et al, 2008). Moreover, miRNA expression may predict the efficacy of therapies, including radiotherapy (Niemoeller et al, 2011). In fact, miRNAs have been shown to modulate the radiosensitivity of primary human dermal microvascular endothelial cells (Wagner-Ecker et al, 2010), lung cancer cells (Weidhaas et al, 2007) and breast cancer cells (Kato et al, 2009). However, irradiation-induced alterations in miRNA profiles have not been analysed in oral squamous cell carcinoma (OSCC). Thus, this study aimed to identify miRNAs associated with radioresistance in OSCC and reveal their biological function and clinical significance.
Materials and Methods
Analysis of predicted target genes
To identify crucial miRNAs for intercellular adhesion molecule-2 (ICAM2) expression, we examined miRNAs that had binding sites like ICAM2 and had been reported to be related to human cancer, especially proliferation, radioresistance and chemoresistance, with publicly available algorithms (TargetScan (http://www.targetscan.org/), EMBL-EBI (http://www.ebi.ac.uk/) or microRNA.org (http://www.microrna.org/microrna/)).
Cell lines and tissue specimens
The OSCC-derived cell lines used were HSC-2, HSC-3, HSC-4, SCC4, HO-1-N-1 and Ca9-22 (Human Science Research Resources Bank, Osaka, Japan). Five independent human normal oral keratinocyte (HNOK) cell lines were cultured and maintained in defined keratinocyte-serum-free medium (Life Technologies Corporation, Grand Island, NY, USA) (Shiiba et al, 2010). Tissue samples from 50 unrelated Japanese patients with primary OSCC who were treated at Chiba University Hospital (Supplementary Table 1) were collected after obtaining informed consents under a protocol that was approved by the Chiba University institutional review board.
RNA extraction and reverse transcription
Total RNA was extracted with TRIzol Reagent (Life Technologies Corporation) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesised from total RNA using Ready-To-Go You-Prime First-Strand Beads (GE Healthcare, Little Chalfont, UK), an oligo(dT) primer (Sigma-Aldrich Co. LLC, St Louis, MO, USA) for ICAM2 and the NCode VILO miRNA cDNA Synthesis Kit (Life Technologies Corporation) for miRNA, according to the manufacturers' protocols.
Real-time quantitative reverse transcriptase–PCR (qRT–PCR) analysis
Real-time qRT–PCR was performed to validate miRNA and mRNA expression with the NCode EXPRESS SYBR GreenER microRNA qRT–PCR Kit (Life Technologies Corporation) for miRNA and a LightCycler FastStart DNA Master SYBR Green 1 Kit (Roche Diagnostics GmbH, Mannheim, Germany) for mRNA. Normalised expression was calculated with 18S ribosomal RNA for miRNA and glyceraldehyde-3-phosphate dehydrogenase for mRNA. Transcript amount was estimated from standard curves and normalised. The nucleotide sequences of the specific primers for miRNAs and ICAM2 are shown in Supplementary Table 2.
Transfection with miR-125b
The HSC-2 and HSC-3 cells (2 × 105) were transfected with mirVana miRNA mimic (microRNA-125b) and mirVana miRNA Mimic Negative Control #1 (Life Technologies Corporation) at a final concentration of 50 nℳ. The cells were transfected with miRNA duplexes with Lipofectamine 2000 reagent (Life Technologies Corporation) by following the manufacturer's protocol. The pmax-green fluorescent protein (GFP) plasmid (Lonza Group Ltd, Basel, Switzerland) was co-transfected simultaneously, and the transfection efficiency was determined by analysing GFP-expressing cells with flow cytometry.
Construction of reporter plasmids and luciferase reporter assays
The full-length 3′-untranslated region (UTR) of ICAM2 containing putative miR-125b-binding sites was subcloned into a pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega Corporation, Madison, WI, USA) located 3′ to the firefly luciferase translational stop codon. A mutant 3′-UTR of ICAM2 with a mutated sequence (5′-…AACTCAGTGTGACTCATCGATCTTGAGGTCC…-3′) in the complementary site for the miR-125b seed region was generated with fusion PCR (mutated sequence is expressed as italic and underlined). For the luciferase reporter assay, HSC-2 cells (5 × 104) were placed in 24-well plates and co-transfected with 0.2 μg of pmirGLO-3′-UTR-WT (ICAM2-WT) or pmirGLO-3′-UTR-MUT (ICAM2-Mut) and miR-125b or negative control with Lipofectamine 2000 (Life Technologies Corporation). Assays were performed 24 h after transfection with the Dual-Luciferase Reporter Assay System (Promega Corporation). Transfections were done three times in independent experiments.
Cell proliferation assay
Transfectant and parental cells were seeded in 12-well plates (1 × 104 viable cells per well). Cells were harvested 1, 2, 3 and 4 days later, and the cells were trypsinised and counted with a haemocytometer in triplicate.
X-ray irradiation and clonogenic survival assay
Transfectant and parental cells were irradiated with four single-radiation doses (2, 4, 6 and 8 Gy) with X-ray-irradiation equipment (MBR-1520R-3; Hitachi, Tokyo, Japan) at a source-to-target distance of 55 cm when the cells reached 70–80% confluence (Higo et al, 2005; Ishigami et al, 2008). The colonies were stained with crystal violet (Sigma-Aldrich Co. LLC), and colonies of ⩾50 cells were counted (Ishigami et al, 2008). Each experiment was repeated at least three times.
Statistical analysis
Clinicopathological profiles were assessed by χ2 tests and Fisher's exact tests. Overall survival rate was investigated by Kaplan–Meier analyses, and the difference was analysed with log-rank tests. Student's t-tests were used for other analyses. All tests were two sided. P<0.05 was considered significant.
Results
Analysis of the predicted target gene
We initially performed predicted target gene analyses of ICAM2 mRNA with publicly available algorithms to find miRNA that potentially regulated ICAM2 expression. The databases showed that miR-125b had complementary sequences for ICAM2 mRNA, suggesting that miR-125b could directly regulate ICAM2 activity through imperfect base pairing with the ICAM2 mRNA 3′-UTR (Supplementary Figure 1A–C). Altered miR-125b expression has been reported in various human cancers, and miR-125b-induced cell cycle arrest has been reported for several cancers (Guan et al, 2011; Huang et al, 2011; Zhang et al, 2011). We hypothesised that miR-125b plays a role in OSCC by regulating ICAM2 expression. Thus, miR-125b was analysed further.
The miR-125b levels in OSCC-derived cell lines and OSCC samples
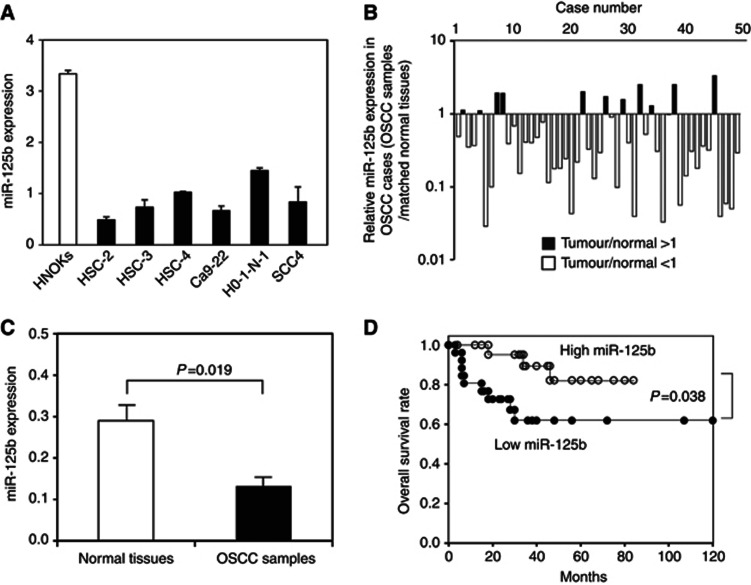
The expression of miR-125b was less than twice that of HNOK cells in all cell lines (Figure 1A). Lower expression levels were observed in 39 of 50 (78%) OSCC samples compared with matched normal tissues (Figure 1B). The median miR-125b expression level was 0.280 and 0.132 in normal tissues and OSCC samples, respectively (Figure 1C), which was significant (P=0.019; Student's t-test).
Figure 1.
Expression profiles of miR-125b in oral squamous cell carcinoma (OSCC)-derived cell lines and OSCC samples. The quantification of miR-125b expression levels in OSCC-derived cell lines (A) and OSCC samples (B) was performed by real-time qRT–PCR analyses. The data are expressed as means±s.d. Significantly lower miR-125b expression levels were detected in primary OSCCs compared with matched normal tissues (P=0.019, Student's t-test) (C). The data are expressed as means±s.d. The overall survival rate was investigated by Kaplan–Meier analysis (D). Lower miR-125b expression levels were significantly associated with poor outcome in patients with OSCC. The log-rank test revealed significant P-values (P=0.038).
The relationship between clinical factors and miR-125b expression was evaluated (Table 1). The expression of miR-125b was significantly lower in advanced stages (III+IV) than in early stages (I+II) (P=0.037).
Table 1. Correlation between miR-125b expression and clinicopathological profiles.
| |
|
MicroRNA-125b expression |
|
|
|---|---|---|---|---|
| Clinicopathological profiles | n | Low (<median) | High (>median) | P-value |
|
Tumour size (T) | ||||
| T1 | 5 | 1 (20%) | 4 (80%) | P=0.086a |
| T2 | 14 | 5 (36%) | 9 (64%) | |
| T3 | 10 | 8 (80%) | 2 (20%) | |
| T4 |
21 |
11 (53%) |
10 (47%) |
|
| T1+T2 | 19 | 6 (32%) | 13 (68%) | P=0.079b |
| T3+T4 |
31 |
19 (61%) |
12(39%) |
|
|
Regional lymph node metastasis (N) | ||||
| Negative | 33 | 16 (48%) | 17 (52%) | P=0.765a |
| Positive |
17 |
9 (53%) |
8 (47%) |
|
|
Stage classification | ||||
| I | 5 | 1 (20%) | 4 (80%) | P=0.100a |
| II | 6 | 1 (17%) | 5 (83%) | |
| III | 7 | 5 (71%) | 2 (29%) | |
| IV |
32 |
18 (56%) |
14 (44%) |
|
| I+II | 11 | 2 (18%) | 9 (82%) | P=0.037b |
| III+V |
39 |
23 (59%) |
16 (41%) |
|
|
Differentiation | ||||
| Well | 28 | 13 (46%) | 15 (54%) | P=0.776b |
| Moderate+poor | 22 | 12 (55%) | 10 (45%) | |
The χ2 test.
Fisher's exact test.
Prognostic significance of miR-125b expression in OSCC
Tumours with significant miR-125b downregulation (median <0.34) were considered low miR-125b (n=25). Survival curves for miR-125b expression are shown in Figure 1D. Kaplan–Meier analyses indicated that miR-125b expression was significantly associated with overall survival (P=0.038; Figure 1D), suggesting that decreased miR-125b expression in patients with OSCCs was significantly associated with poor prognoses.
Effects of miR-125b overexpression on cellular proliferation and radiosensitivity
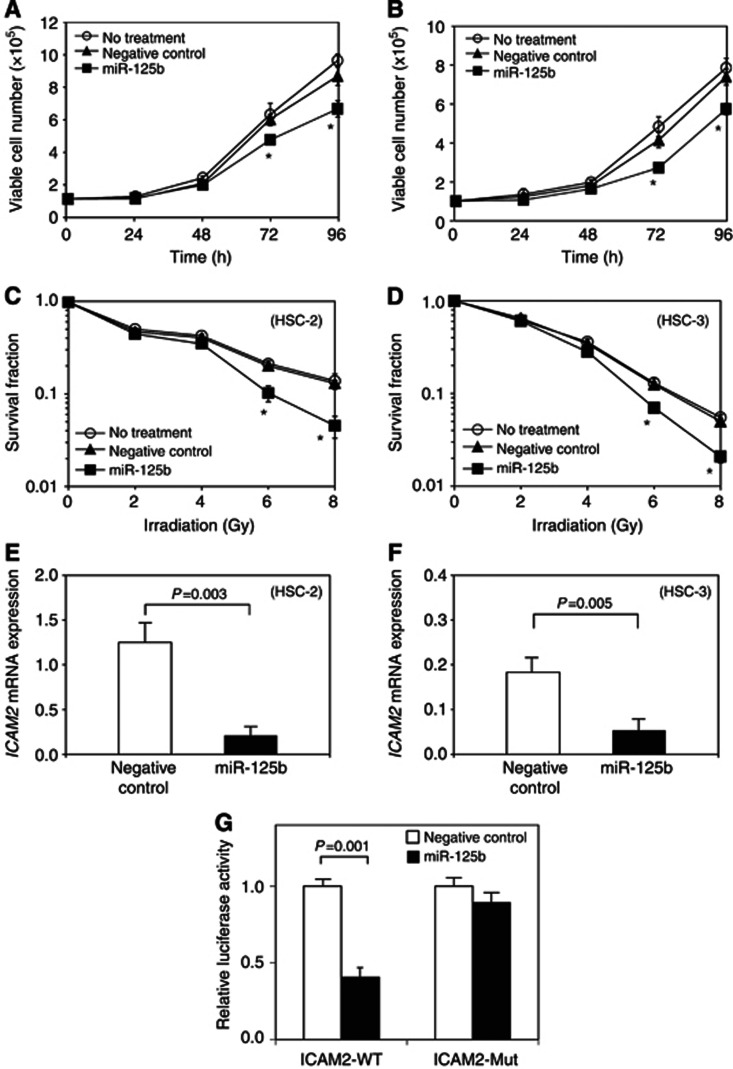
After validating miR-125b overexpression (Supplementary Figure 2A and B) with appropriate transfection efficiency, miR-125b-transfected cells were subjected to functional assays. The proliferation rate was significantly lower in miR-125b-transfected HSC-2 and HSC-3 cells after 72 and 96 h compared with negative controls (Figures 2A and B). In HSC-2 and HSC-3 cells, the survival rates were significantly decreased in 6-Gy-irradiated and 8-Gy-irradiated miR-125b transfectants compared with negative controls (Figures 2C and D).
Figure 2.
Effect of miR-125b overexpression on OSCC-derived cell lines. A cellular proliferation assay was performed to investigate the effects of the overexpression of miR-125b on the OSCC-derived cell lines HSC-2 (A) and HSC-3 (B). *Significant differences between transfectants and negative controls (NCs; P<0.05, Student's t-test). The data are expressed as means±s.d. Cell survival after irradiation was examined with a clonogenic survival assay (C and D). After exposure to 2, 4, 6 and 8 Gy of X-ray irradiation, cells were seeded and cultured. The significance of the differences between transfectant and NC cells is indicated with asterisks (*) (P<0.05, Student's t-test). The data are expressed as means±s.d. The ICAM2 mRNA expression in miR-125b-transfected cells and NC cells in HSC-2 (E) and HSC-3 (F) is shown. The expression levels of ICAM2 mRNA were decreased after the introduction of miR-125b to both cell lines (P<0.01, Student's t-test). The data are expressed as means±s.d. The luciferase activity of the reporter gene containing the wild-type 3′-UTR of the ICAM2 gene (ICAM2-WT) or the mutated 3′-UTR of the ICAM2 gene (ICAM2-Mut) was determined (G). The data are expressed as means±s.d. Relative luciferase activity was significantly suppressed in the cells with ICAM2-WT when miR-125b was co-transfected (P=0.001, Student's t-test).
The miR-125b overexpression suppressed ICAM2 expression in OSCC-derived cell lines
We examined whether altered radiosensitivity was associated with the ICAM2-related signalling pathway. The expression of ICAM2 was downregulated in miR-125b-transfected OSCC cells (Figure 2E and F).
MiR-125b targets ICAM2
The relative luciferase activity of the reporter gene containing wild-type ICAM2 3′-UTR was significantly suppressed when miR-125b was co-transfected. However, decreased luciferase activity was not observed in cells with the reporter gene containing mutated ICAM2 3′-UTR (Figure 2G). This suggested that the ICAM2 3′-UTR was a miR-125b target and that miR-125b suppressed ICAM2 expression through binding.
Discussion
We have previously identified genes, including ICAM2, that play important roles in OSCC radioresistance (HSC-2 and HSC-3) (Ishigami et al, 2007, 2008). The ICAM2 has been suggested to facilitate an apoptotic-blocking survival signal by activating the PI3K/AKT pathway (Perez et al, 2002). Another study has reported that ICAM2 expression deficiencies result in impaired angiogenesis in vitro and in vivo, defective in vitro migration and increased apoptosis (Huang et al, 2005). The downregulation of ICAM2 siRNA enhanced OSCC radiosensitivity and increased apoptosis through AKT phosphorylation and caspase-3 activation (Ishigami et al, 2008). Moreover, ICAM2 overexpression induced greater OSCC resistance to X-ray irradiation (Ishigami et al, 2007).
In this study, decreased ICAM2 expression was observed in both miR-125b-transfected HSC-2 and miR-125b-transfected HSC-3 cells; moreover, radiosensitivity towards X-ray irradiation was enhanced in miR-125b-transfected HSC-2 and HSC-3 cells. Luciferase reporter assays showed that the ICAM2 3′-UTR was a miR-125b target. Therefore, the data strongly suggest that miR-125b is associated with radiation response through regulating ICAM2 expression in OSCC.
The importance of miR-125b as an anticancer agent was demonstrated by the impact of its expression on cell proliferation. We showed that OSSC-derived cells proliferated less rapidly when miR-125b expression was increased by transfection, suggesting that miR-125b could regulate cell proliferation. This finding may be associated with cell cycle regulation as miR-125b-induced cell cycle arrest has been reported for several cancers (Guan et al, 2011; Huang et al, 2011; Zhang et al, 2011).
In conclusion, controlling miR-125b expression or activity could contribute to suppressing cell proliferation and overcoming radioresistance in OSCC, which could be useful in developing a cure for OSCC. Moreover, miR-125b was significantly correlated with survival and radiotherapy response in OSCC and could be used as a prognostic marker.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6:2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Guan Y, Yao H, Zheng Z, Qiu G, Sun K. MiR-125b targets BCL3 and suppresses ovarian cancer proliferation. Int J Cancer. 2011;128:2274–2283. doi: 10.1002/ijc.25575. [DOI] [PubMed] [Google Scholar]
- Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev. 2005;15:410–415. doi: 10.1016/j.gde.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Higo M, Uzawa K, Kouzu Y, Bukawa H, Nimura Y, Seki N, Tanzawa H. Identification of candidate radioresistant genes in human squamous cell carcinoma cells through gene expression analysis using DNA microarrays. Oncol Rep. 2005;14:1293–1298. [PubMed] [Google Scholar]
- Huang L, Luo J, Cai Q, Pan Q, Zeng H, Guo Z, Dong W, Huang J, Lin T. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int J Cancer. 2011;128:1758–1769. doi: 10.1002/ijc.25509. [DOI] [PubMed] [Google Scholar]
- Huang MT, Mason JC, Birdsey GM, Amsellem V, Gerwin N, Haskard DO, Ridley AJ, Randi AM. Endothelial intercellular adhesion molecule (ICAM)-2 regulates angiogenesis. Blood. 2005;106:1636–1643. doi: 10.1182/blood-2004-12-4716. [DOI] [PubMed] [Google Scholar]
- Ishigami T, Uzawa K, Fushimi K, Saito K, Kato Y, Nakashima D, Higo M, Kouzu Y, Bukawa H, Kawata T, Ito H, Tanzawa H. Inhibition of ICAM2 induces radiosensitization in oral squamous cell carcinoma cells. Br J Cancer. 2008;98:1357–1365. doi: 10.1038/sj.bjc.6604290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigami T, Uzawa K, Higo M, Nomura H, Saito K, Kato Y, Nakashima D, Shiiba M, Bukawa H, Yokoe H, Kawata T, Ito H, Tanzawa H. Genes and molecular pathways related to radioresistance of oral squamous cell carcinoma cells. Int J Cancer. 2007;120:2262–2270. doi: 10.1002/ijc.22561. [DOI] [PubMed] [Google Scholar]
- Kato M, Paranjape T, Muller RU, Nallur S, Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB, Slack FJ. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemoeller OM, Niyazi M, Corradini S, Zehentmayr F, Li M, Lauber K, Belka C. MicroRNA expression profiles in human cancer cells after ionizing radiation. Radiat Oncol. 2011;6:29. doi: 10.1186/1748-717X-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez OD, Kinoshita S, Hitoshi Y, Payan DG, Kitamura T, Nolan GP, Lorens JB. Activation of the PKB/AKT pathway by ICAM-2. Immunity. 2002;16:51–65. doi: 10.1016/s1074-7613(02)00266-2. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiiba M, Nomura H, Shinozuka K, Saito K, Kouzu Y, Kasamatsu A, Sakamoto Y, Murano A, Ono K, Ogawara K, Uzawa K, Tanzawa H. Down-regulated expression of SERPIN genes located on chromosome 18q21 in oral squamous cell carcinomas. Oncol Rep. 2010;24:241–249. doi: 10.3892/or_00000852. [DOI] [PubMed] [Google Scholar]
- Wagner-Ecker M, Schwager C, Wirkner U, Abdollahi A, Huber PE. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat Oncol. 2010;5:25. doi: 10.1186/1748-717X-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, Gillespie E, Slack FJ. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BH, Xiong XP, Jia J, Zhang WF. MicroRNAs: new actors in the oral cancer scene. Oral Oncol. 2011;47:314–319. doi: 10.1016/j.oraloncology.2011.03.019. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ, Huang MY, Hou JH, Wu QL, Zeng MS, Huang WL, Zeng YX, Shao JY. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011;71:3552–3562. doi: 10.1158/0008-5472.CAN-10-2435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.