Abstract
In this study, we reported thirty-nine endophytic fungi identified as Colletotrichum spp. associated with Brazilian pepper tree or aroeira (Schinus terebinthifolius Raddi. Anacardiaceae) in Paraná state, Brazil. These endophytes were identified by morphological and molecular methods, using PCR taxon-specific with CaInt/ITS4, CgInt/ITS4, and Col1/ITS4 primers, which amplify specific bands in C. acutatum, C. gloeosporioides lato sensu, and Colletotrichum boninensis, respectively, and by DNA sequence analysis of the nrDNA internal transcribed spacer region (ITS1, 5.8S, ITS2). We also assayed the presence of dsRNA particles in Colletotrichum spp. isolates. Combining both morphological characters and molecular data, we identified the species C. gloeosporioides, C. boninense, and C. simmondsii. However, we found a high genetic variability intraspecific in C. gloeosporioides which suggests the existence of several other species. Bands of double-stranded RNA (dsRNA) were detected in three of thirty-nine isolates. Identity of these bands was confirmed by RNAse, DNAse, and S1 nuclease treatments for the isolates LGMF633, LGMF726, and LGMF729. This is the first study reporting these particles of dsRNA in C. gloeosporioides.
1. Introduction
Potential sources of new natural products have been explored in medicine, agriculture, and industry. Endophytic fungi have been recognized as useful sources of bioactive secondary metabolites [1], especially those isolated from medicinal plants [2, 3]. Various important characteristics are currently attributed, such as the increase in resistance to stressful conditions; alteration in physiological properties; production of phytohormones, toxins, medicinal substances, immunosuppressants, antitumor agents, and compounds of biotechnological interest such as enzymes [1, 4–13].
Brazilian pepper tree also called aroeira (Schinus terebinthifolius Raddi-Anacardiaceae) is native to Argentina, Brazil, and Paraguay [14]. In Brazil, the bark leaves and fruits have been used in popular medicine due to their medicinal properties [15–18]. Actions anti-inflammatory and antiseptic for treatment of wounds, urinary, and respiratory infections are listed as medicinal properties popularly known [19]. Studies showed antimicrobial activity [15, 18, 20–24], antifungal activity [25, 26], as antioxidant [15, 27], and antitumor [18, 28]. Despite its importance, there are a few records about the endophytic community in this plant.
Colletotrichum has been isolated from numerous plant species especially as symptomatic pathogens but can be found as asymptomatic endophytes. The genus has wide geographic distribution, being more important in the tropics. Studies involving the complex C. gloeosporioides and C. boninense revealed high genetic variability and molecular diversity [21, 29–31]. There is significant interest in developing a fast, simple, and efficient method to identify species of Colletotrichum. Several authors have described new species and morphological characteristics associated to species of Colletotrichum [32–36]. Afanador-Kafuri et al. [32] developed specific primers to C. boninense [34]. The species C. acutatum was also organized and divided into three species, C. acutatum, C. fioriniae comb. et stat. nov., and C. simmondsii sp. nov. [36].
Several authors have investigated the influence of viral particles on fungi [37]. Fungal virus genomes are commonly composed of dsRNA that can modulate plant-fungal symbioses [38]. The associations between fungal viruses and their hosts are similar to those involved in plant-endophyte interactions [37]. Changes in morphological characteristics and increased production of conidia have been reported as associated with the presence of dsRNA in Beauveria bassiana [39], Metarhizium anisopliae [40, 41], and Nectria radicicola [34].
In the present study, we isolated endophytic fungi from leaves of medicinal tree called aroeira (Schinus terebinthifolius Raddi). These endophytes were identified by morphological and molecular methods. We also assayed the presence of dsRNA particles in Colletotrichum spp. isolates.
2. Material and Methods
2.1. Fungal Isolates
Isolates were obtained from leaves of plants of Brazilian pepper tree (S. terebinthifolius Raddi), located in the campus of the University of Paraná, Paraná, Brazil. The isolates were obtained as described by Petrini [42] and identified by macroscopic and microscopic reproductive structures after growth on PDA medium. The cultures are permanently stored in the fungal collection of the Laboratory of Microorganisms (LabGeM-UFPR), Paraná, Brazil.
3. Molecular Characterization
3.1. DNA Extraction
Colletotrichum isolates were grown on PDA medium for 3 days at 28°C. The mycelium was harvested, lyophilized for 24 h, and ground with a mortar and pestle under liquid nitrogen. Genomic DNA was obtained according to methods described by Raeder and Broda [43], modified by Glienke-Blanco et al. [44].
3.2. Species-Specific PCR
Species-specific amplifications were performed using the primer ITS4 [45] with specific primers for C. gloeosporioides complex (CgInt: 5′-GGCCTCCCGCCTCCGGGCGG-3′) [46], C. boninense (Col1: 5′-GCCGTCCCCTGAAAAG) [47], or C. acutatum complex (CaInt2: 5′-GGGGAAGCCTCTCGCGG-3′) [32].
According to the method described by Pileggi et al. [47], PCR reactions were performed in a total volume of 25 μL, containing 1X buffer solution, 1.5 mM MgCl2, 0.2 mM of each dNTP (Invitrogen, CA, USA), 0.5 μM primer, 1.5 Unit of Taq DNA polymerase (Invitrogen, CA, USA), and 20 ng of genomic DNA. Amplifications were carried using the following conditions: an initial denaturation at 95°C for 5 min, followed by 40 cycles of 30 s at 95°C, 30 s at 65°C, and 1.5 min at 72°C, and a final extension at 72°C for 3 min.
To identify C. acutatum and C. gloeosporioides complex, the PCR reactions were performed as previously described. Amplifications were carried out in a gradient thermocycler with an initial denaturation period of 5 min at 95°C, followed by one cycle of 30 sec at 94°C, 45 seconds at 62°C, 90 seconds at 72°C, one cycle of 30 seconds at 94°C, 45 seconds at 60°C, 90 seconds at 72°C, followed by 33 cycles of 30 seconds at 94°C, 45 seconds at 58°C e 90 seconds at 72°C, and a final extension period of 3 minutes at 72°C.
Genomic DNA of positive control of C. gloeosporioides (Col11), C. boninense (Col7), is obtained from Pileggi et al. [47]. Genomic DNAs of C. acutatum strains FDC89A08 and FDC31A08 were obtained from Fundecitrus, SP.
3.3. DNA Analysis and Sequencing
The primers V9G [48] and ITS4 [45] were used to amplify the internal transcribed spacer region (ITS) of the nuclear ribosomal RNA operon, including the 3′ end of the 18S rRNA, the first internal transcribed spacer region, the 5.8S rRNA gene; the second internal transcribed spacer region and the 5′ end of the 28S rRNA gene. PCR was performed in total reaction volume of 50 μL, which was composed of 1 × PCR Buffer (Applied Biosystems, Foster City, USA), 2 mM MgCl2, 40 μM dNTPs, 0.08 μM of each forward and reverse primer, 0.5 U of Taq DNA polymerase (Roche Diagnostics, Indianapolis, USA), and 1–10 ng of genomic DNA. Thirty cycles were performed: 94°C for 30 s, 56°C for 1 min, 72°C for 1 min, and 2 min initial and terminal delay. The second condition had a total reaction volume of 12.5 μL, which was composed of 1 × PCR Buffer (Bioline GmbH, Luckenwalde, Germany), 5.6% DMSO (v/v), 2 mM MgCl2, 20 μM dNTPs, 0.2 μM of each forward and reverse primer, 0.25 U of BioTaq Taq DNA polymerase (Bioline GmbH, Luckenwalde, Germany), and 1–10 ng of genomic DNA. The PCR cycle conditions were 5 min of 94°C, followed by 40 cycles of 94°C for 30 s, 52°C for 30 s, 72°C for 30 s, and a final elongation step at 72°C for 7 min.
Amplified rDNA fragments were cleaned with 50 μL 20% PEG and resuspended in 15 μL of ultrapure water. To confirm the presence of DNA in the sample, 1 μL was applied on a 1.4% agarose gel. rDNA Internal Transcribed Spacer (ITS) was sequenced with primers ITS4 and ITS1 [45]. PCR was performed in 10 μL volumes of a reaction mixture containing sterile distilled water, 0.5 μL PCR buffer (10x, Applied Biosystems), 0.5 μL of primer (50 pmol), 0.5 μL of Big Dye (Applied Biosystems), and 1 μL PCR products. Thirty five cycles were performed: 96°C for 10 s (denaturation), 50°C for 5 s (annealing), 60°C for 4 min (extension), and 60 s initial and terminal delay. Sequencing was performed on an ABI 3130 automatic sequencer (Perkin-Elmer, Massachusetts, USA).
3.4. Sequence Assembly and Alignment
Sequences were edited using BioEdit 7.0 [40]. ITS sequences were aligned on the basis of similarity by means of the sequence editor CLUSTAL-W 1.7 [49]. Sequence analysis was performed using the sequence alignment software BLASTn run against the NCBI database (National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/)).
Maximum likelihood tree search was done with GARLI version 2.0 [50]. The algorithm settings was the default, with 1.000 bootstraps (50 runs of 20 repetitions). Bayesian tree search was done with parallel MrBayes version 3.1.2 [51, 52], using 20.000.000 generations and 4 independents runs. The Model, SYM+I, was selected with jMODELTEST v0.1.1 [53] using Bayesian Information Criterion (BIC). The traces were analysed with Tracer v1.5 [54] and AWTY [55] to evaluate the stationary phase, setting the burn-in to 2.000.000 generations. To merge bootstrap replicates and posterior probabilities and summarize the consensus tree, SumTrees [36] from DendroPy package version 3.7.0 [23] was used.
3.5. dsRNA Analysis
After genomic DNA extraction was performed electrophoresis on 0.7% agarose gel to observe the occurrence of bands of dsRNA. For confirmation, the total DNA of the isolates was submitted separately to the treatment of enzymatic digestion with RNAse, DNAse, and S1 Nuclease. Three digestions were performed as described by Azevedo et al. [1].
4. Results
4.1. Species-Specific PCR
The isolates were investigated in PCR with specific primers for C. acutatum, C. gloeosporioides, and C. boninense (Table 1). Isolates LGMF625, 666, 667, and 738 did not amplify the expected band with CgInt/ITS4 primers. LGMF666 showed amplification using primer Col1/ITS4, that amplifies a specific band for C. boninense and LGMF738 amplifies a specific band with CaInt/ITS4 primer, and was identified as C. acutatum complex (Table 1).
Table 1.
Identification of Colletotrichum using primers taxon-specific CaInt-ITS4, CgInt-ITS4 and Col-ITS4.
| Species | Strain number | PCR identification | ITS | ||
|---|---|---|---|---|---|
| CaInt-ITS4 | CgInt-ITS4 | Col-ITS4 | |||
| C. boninense | LGMF625 | − | − | − | C. boninense |
| LGMF666 | − | − | + | na | |
| C. acutatum | LGMF738 | + | − | − | C. acutatum |
| C. gloeosporioides | LGMF621 | − | + | − | na |
| LGMF622 | − | + | − | C. gloeosporioides | |
| LGMF623 | − | + | − | C. gloeosporioides | |
| LGMF626 | − | + | − | C. gloeosporioides | |
| LGMF627 | − | + | − | C. gloeosporioides | |
| LGMF633* | − | + | − | na | |
| LGMF638 | − | + | − | na | |
| LGMF667 | − | - | − | C. gloeosporioides | |
| LGMF669 | − | + | − | na | |
| LGMF672 | − | + | − | C. gloeosporioides | |
| LGMF678 | − | + | − | na | |
| LGMF680 | − | + | − | C. gloeosporioides | |
| LGMF682 | − | + | − | na | |
| LGMF686 | − | + | − | na | |
| LGMF687 | − | + | − | C. gloeosporioides | |
| LGMF688 | − | + | − | C. gloeosporioides | |
| LGMF689 | − | + | − | C. gloeosporioides | |
| LGMF721 | − | + | − | C. gloeosporioides | |
| LGMF722 | − | + | − | C. gloeosporioides | |
| LGMF723 | − | + | − | C. gloeosporioides | |
| LGMF726* | − | + | − | na | |
| LGMF727 | − | + | − | C. gloeosporioides | |
| LGMF728 | − | + | − | C. gloeosporioides | |
| LGMF729* | − | + | − | na | |
| LGMF730 | − | + | − | na | |
| LGMF731 | − | + | − | C. gloeosporioides | |
| LGMF732 | − | + | − | C. gloeosporioides | |
| LGMF733 | − | + | − | na | |
| LGMF734 | − | + | − | C. gloeosporioides | |
| LGMF735 | − | + | − | C. gloeosporioides | |
| LGMF736 | − | + | − | C. gloeosporioides | |
| LGMF740 | − | + | − | na | |
| LGMF741 | − | + | − | C. gloeosporioides | |
| LGMF742 | − | + | − | C. gloeosporioides | |
| LGMF743 | − | + | − | na | |
| LGMF744 | − | + | − | C. gloeosporioides | |
(*) dsRNA confirmed after analysis digestion with RNAse, DNAse, and S1 Nuclease. na: not available.
4.2. DNA Analysis and Sequencing
Twenty five isolates were sequenced for the ITS1-5.8S-ITS2 of rDNA, generating fragments between 600 and 700 bp. Phylogenetic analysis grouped the Colletotrichum isolates into three clades (Figure 1). The first clade included the C. acutatum complex species, with 100% bootstrap support. The isolate LGMF738 clustered with C. simmondsii (GU183331) holotype strain. All Colletotrichum isolates that clustered in clade II included isolates of the C. gloeosporioides from GenBank (AB439815; AB273196; EU734587; EU552111; EU687190) with 84% bootstrap support. The third clade clustered the isolate LGMF625 with C. boninense species (EU822802 and AB051401).
Figure 1.
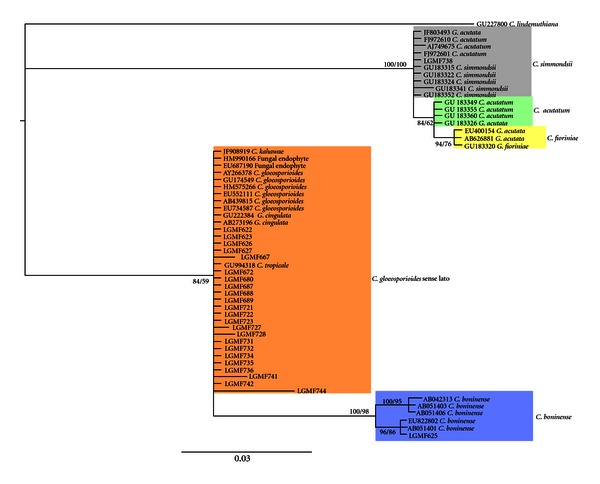
Phylogenetic relationships among the studied strains based on maximum likelihood and Bayesian inference. Values below branches represent either bootstrap support values (maximum likelihood tree) or posterior probabilities (Bayesian inference).
4.3. dsRNA Analyses
After electrophoresis of genomic DNA was observed bands of approximately 3000 bp indicating the presence of dsRNA in six isolates of C. gloeosporioides complex LGMF633, 638, 689, 726, 729, and LGMF736 (Table 1).
5. Discussion
The taxonomy of Colletotrichum is confused, both for the anamorphic species and its teleomorph Glomerella. The combined use of molecular diagnostic tools along with traditional morphological techniques is at present an appropriate approach for studying Colletotrichum species complexes [6, 47].
Afanador-Kafuri et al. [32] proposed the use of two pairs of primers for the identification of Colletotrichum species, one for C. gloeosporioides (CgInt) and another for Colletotrichum sp (Col1). Moriwaki et al. [30] proposed the classification of isolates originally identified as C. gloeosporioides as belonging to a new species, they called C. boninense. Pileggi et al. [47] suggested that the primers pair developed by Afanador-Kafuri et al. [32] for Colletotrichum sp amplify isolates of the new species C. boninense proposed by Moriwaki et al. [30]. Moriwaki et al. [30] showed that the ITS1 region of C. boninense was 190 bp, whereas, for C. gloeosporioides, this region was 171 bp. Consequently, the difference between C. boninense and C. gloeosporioides should reflect interspecific relationships and should be further investigated.
In this paper, we proposed PCR identification species-specific anticipated results, enabling identification of approximately 70% of isolates without the need for extensive morphological analysis. ITS sequence analysis confirmed species-specific results and resolved the identification of C. boninense (LGMF625) and C. gloeosporioides complex. However, the PCR species-specific mistakenly identified the isolate LGMF738 as C. acutatum when in fact it belongs to the new species C. simmondsii (Table 1).
The analyzed Brazilian pepper trees were colonized by three different species of Colletotrichum and showed high genetic diversity; including the species C. gloeosporioides sense lato, C. boninense, and C. simmondsii. The ecological roles of endophytes are diverse and varied. Colletotrichum gloeosporioides complex is a worldwide plant pathogen that infects many plant species. These isolates will need more examination to ensure the correct identification. Zou et al. [27] reported one endophytic isolate of C. gloeosporioides from stem of Artemisia mongolica that produced the colletotric acid, with antimicrobial activity against Bacillus subtilis, Staphylococcus aureus, Sarcina lutea, and Helminthosporium sativum [27].
Many studies discriminate Colletotrichum complex using ribosomal ITS sequence data; however, due to the limited number of informative sites been identified, other regions of the Genome, such as the β-tubulin gene, have been identified suitable for the phylogenetic reconstruction [56]. Hyde et al. [57] suggest epitypification and use of multilocus phylogeny to delimit species and gain a better understanding of the genus. Our data corroborated the existence of more than one species in the C. gloeosporioides complex. Also, our data corroborated the reassessment of Colletotrichum acutatum complex and the new species C. simmondsii introduced by Shivas and Tan [58]. It was the first report of C. simmondsii as an endophyte from Schinus terebinthifolius. The host range and host specificity of C. simmondsii are not clear [36].
This paper is the first study describing the existence of dsRNA particles in C. gloeosporioides isolates. The presence of dsRNA in entomopathogenic fungi is described in a long time [41, 59–63]. Dalzoto et al. [39] described the horizontal transfer and hypovirulence associated with dsRNA in the fungus B. bassiana. The authors suggest the increased production of conidia in strains without dsRNA when compared with the strains positive to dsRNA particles.
Morphological changes of colonies associated with the presence of the dsRNA particles were described in Chalara elegans [64], Metarhizium anisopliae [40, 41], Diaporthe ambigua [62], and Nectria radicicola [34].
Double-stranded RNA viruses have been described for a long time in a wide variety of filamentous fungi and yeasts [9, 37, 65–67]. Marquez et al. [38] suggested the associations between fungal viruses and their hosts are similar to plant-endophyte associations. In this study, Marquez et al. [38] found no differences in colony morphology among isolates with dsRNA and those free dsRNA. Also, the authors did not find any association between the presences of dsRNA and genetically different groups. In the Colletotrichum genus, it is not yet known the influence that these particles can have on fungi morphology or physiology. So, we suggest the investigation by scanning electron microscopy and also by the study of these strains after cure (elimination) of dsRNA.
Acknowledgments
The authors thank the Brazilian agency CNPq Brazil for financial support. They are grateful to High Performance Computing Center and Professor Sergio Scheer/PRPPG/UFPR/Brazil.
References
- 1.Azevedo JL, Maccheroni W, Pereira JO, de Araújo WL. Endophytic microorganisms: a review on insect control and recent advances on tropical plants. Electronic Journal of Biotechnology. 2000;3(1):40–65. [Google Scholar]
- 2.Huang Z, Cai X, Shao C, et al. Chemistry and weak antimicrobial activities of phomopsins produced by mangrove endophytic fungus Phomopsis sp. ZSU-H76. Phytochemistry. 2008;69(7):1604–1608. doi: 10.1016/j.phytochem.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Zhang HW, Song YC, Tan RX. Biology and chemistry of endophytes. Natural Product Reports. 2006;23(5):753–771. doi: 10.1039/b609472b. [DOI] [PubMed] [Google Scholar]
- 4.Backman PA, Sikora RA. Endophytes: an emerging tool for biological control. Biological Control. 2008;46(1):1–3. [Google Scholar]
- 5.Bills G, Dombrowsky A, Pelaez F, Polishook J. Tropical Mycology: Micromycetes. Vol. 2. New York, NY, USA: CABI; 2002. Recent and future discoveries of pharmacologically active metabolites from tropical fungi; pp. 165–194. [Google Scholar]
- 6.Cai L, Wu CD. Compounds from Syzygium aromaticum possessing growth inhibitory activity against oral pathogens. Journal of Natural Products. 1996;59(10):987–990. doi: 10.1021/np960451q. [DOI] [PubMed] [Google Scholar]
- 7.Esposito E, Azevedo JL. Fungos: Uma Introdução à Biologia, Bioquímica e Biotecnologia. Caxias do Sul, Brazil: EDUSC; 2004. [Google Scholar]
- 8.Rodrigues KF, Hesse M, Werner C. Antimicrobial activities of secondary metabolites produced by endophytic fungi from Spondias mombin . Journal of Basic Microbiology. 2000;40:261–267. doi: 10.1002/1521-4028(200008)40:4<261::AID-JOBM261>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 9.Soares DGS, de Oliveira CB, Leal C, et al. Susceptibilidade in vitro de bactérias bucais a tinturas de fitoterápicos. Revista Odonto Ciência. 2006;21(53):232–238. [Google Scholar]
- 10.Strobel G, Daisy B, Castillo U, Harper J. Natural products from endophytic microorganisms. Journal of Natural Products. 2004;67(2):257–268. doi: 10.1021/np030397v. [DOI] [PubMed] [Google Scholar]
- 11.Tejesvi MV, Kini KR, Prakash HS, Subbiah V, Shetty HS. Genetic diversity and antifungal activity of species of Pestalotiopsis isolated as endophytes from medicinal plants. Fungal Diversity. 2007;24:37–54. [Google Scholar]
- 12.Velázquez E, Tournier HA, Mordujovich de Buschiazzo P, Saavedra G, Schinella GR. Antioxidant activity of Paraguayan plant extracts. Fitoterapia. 2003;74(1-2):91–97. doi: 10.1016/s0367-326x(02)00293-9. [DOI] [PubMed] [Google Scholar]
- 13.Yu H, Zhang L, Li L, et al. Recent developments and future prospects of antimicrobial metabolites produced by endophytes. Microbiological Research. 2010;165(6):437–449. doi: 10.1016/j.micres.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Mytinger L, Williamson GB. The invasion of Schinus into saline communities of Everglades National Park. Florida Scientist. 1987;50:7–12. [Google Scholar]
- 15.Degáspari CH, Waszczynskyj N, Pardo MRM. Atividade antimicrobiana de Schinus terebentifolius Raddi. Ciência e Agrotecnologia. 2005;29(3):617–622. [Google Scholar]
- 16.Guerra MJM, Barreiro ML, Rodriguez ZM, Rubalcaba Y. Actividad antimicrobiana de um extracto fluido al 80% de Schinus terebenthifolius Raddi (copal) Revista Cubana de Plantas Medicinales. 2000;5(1):23–25. [Google Scholar]
- 17.Lorenzi H. Árvores Brasileiras—Manual de Identificação e Cultivo de Plantas Arbóreas Nativas do Brasil. 4th edition. Vol. 1. Instituto Plantarum; 2002. [Google Scholar]
- 18.Ribas MO, Sousa MH, Sartoretto J, et al. Efeito da Schinus terebenthifolius Raddi sobre o processo de reparo tecidual das lesões ulceradas induzidas na mucosa bucal do rato. Revista Odonto Ciência/PUCRS. 2006;21(53):245–252. [Google Scholar]
- 19.de Lima MRF, de Souza Luna J, Dos Santos AF, et al. Anti-bacterial activity of some Brazilian medicinal plants. Journal of Ethnopharmacology. 2006;105(1-2):137–147. doi: 10.1016/j.jep.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman I, Coutinho LS, Torres OJM, et al. Efeito do extrato hidroalcoólico da Aroeira (Schinus terebentifolius Raddi) na cicatrização de anastomoses colônicas. Estudo experimental em ratos. Acta Cirurgica Brasileira. 2006;21(3):49–54. doi: 10.1590/s0102-86502006000900008. [DOI] [PubMed] [Google Scholar]
- 21.Johann S, Pizzolatti MG, Donnici CL, Resende MA. Atividade antifúngica de plantas utilizadas na medicina tradicional brasileira contra fungos de relevância clínica. Brazilian Journal of Microbiology. 2007;38(4):632–637. [Google Scholar]
- 22.Nunes JAT, Jr., Ribas-Filho JM, Malafaia O, et al. Evaluation of the hydro-alcoholic Schinus terebinthifolius Raddi (Aroeira) extract in the healing process of the alba linea in rats. Acta Cirurgica Brasileira. 2006;21(3):8–15. doi: 10.1590/s0102-86502006000900003. [DOI] [PubMed] [Google Scholar]
- 23.Sukumaran J, Holder MT. DendroPy: a Python library for phylogenetic computing. Bioinformatics. 2010;26(12):1569–1571. doi: 10.1093/bioinformatics/btq228. Article ID btq228. [DOI] [PubMed] [Google Scholar]
- 24.Talhinhas P, Sreenivasaprasad S, Neves-Martins J, Oliveira H. Genetic and morphological characterization of Colletotrichum acutatum causin anthracnose of luppins. Phythopathology. 2002;92:986–996. doi: 10.1094/PHYTO.2002.92.9.986. [DOI] [PubMed] [Google Scholar]
- 25.Fenner R, Betti AH, Mentz LA, et al. Plantas utilizadas na medicina popular brasileira com potencial atividade antifúngica. Revista Brasileira de Ciências Farmacêuticas. 2006;42(3):369–394. [Google Scholar]
- 26.Soares DGS, Oliveira CB, Leal C, et al. Atividade Antibacteriana in vitro da Tintura de Aroeira (Schinus terebinthifolius) na Descontaminação de Escovas Dentais Contaminadas pelo S. mutans . Pesquisa Brasileira em Odontopediatria e Clínica Integrada. 2007;7(3):253–257. [Google Scholar]
- 27.Zou WX, Meng JC, Lu H, et al. Metabolites of Colletotrichum gloeosporioides, an endophytic fungus in Artemisia mongolica . Journal of Natural Products. 2000;63(11):1529–1530. doi: 10.1021/np000204t. [DOI] [PubMed] [Google Scholar]
- 28.Queires LCS, Fauvel-Lafève F, Terry S, et al. Polyphenols purified from the Brazilian aroeira plant (Schinus terebinthifolius, Raddi) induce apoptotic and autophagic cell death of DU145 cells. Anticancer Research. 2006;26(1 A):379–387. [PubMed] [Google Scholar]
- 29.Lu G, Cannon PF, Reid A, Simmons CM. Diversity and molecular relationships of endophytic Colletotrichum isolates from the Iwokrama Forest Reserve, Guyana. Mycological Research. 2004;108(1):53–63. doi: 10.1017/s0953756203008906. [DOI] [PubMed] [Google Scholar]
- 30.Moriwaki J, Sato T, Tsukiboshi T. Morphological and molecular characterization of Colletotrichum boninense sp . nov. from Japan. Mycoscience. 2003;44(1):47–53. [Google Scholar]
- 31.Rojas EI, Rehner SA, Samuels GJ, et al. Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panamá: multilocus phylogenies distinguish host-associated pathogens from asymptomatic endophytes. Mycologia. 2010;102(6):1318–1338. doi: 10.3852/09-244. [DOI] [PubMed] [Google Scholar]
- 32.Afanador-Kafuri L, Minz D, Maymon M, Freeman S. Characterization of Colletotrichum isolates from tamarillo, passiflora, and mango in Colombia and identification of a unique species from the genus. Phytopathology. 2003;93(5):579–587. doi: 10.1094/PHYTO.2003.93.5.579. [DOI] [PubMed] [Google Scholar]
- 33.Freeman S, Katan T, Shabi E. Characterization of Colletotrichum species responsible for anthracnose diseases of various fruits. Plant Disease. 1998;82(6):596–605. doi: 10.1094/PDIS.1998.82.6.596. [DOI] [PubMed] [Google Scholar]
- 34.Ahn IP, Lee YH. A viral double-stranded RNA up regulates the fungal virulence of Nectria radicicola . Molecular Plant-Microbe Interactions. 2001;14(4):496–507. doi: 10.1094/MPMI.2001.14.4.496. [DOI] [PubMed] [Google Scholar]
- 35.Schulz BJE. Mutualistic interactions with fungal root endophytes. In: Schulz BJE, Boyle CJC, Sieber TN, editors. Microbial Root Endophytes. Berlin, Germany: Springer; 2006. pp. 261–280. [Google Scholar]
- 36.Sukumaran J, Holder MT. SumTrees: summarization of split support on phylogenetic trees. Version 1.0.2. Part of the DendroPy Phylogenetic Computation Library Version 3.7.0, 2010, http://packages.python.org/DendroPy/
- 37.Herrero N, Sánchez Márquez S, Zabalgogeazcoa I. Mycoviruses are common among different species of endophytic fungi of grasses. Archives of Virology. 2009;154(2):327–330. doi: 10.1007/s00705-008-0293-5. [DOI] [PubMed] [Google Scholar]
- 38.Marquez LM, Redman RS, Rodriguez RJ, Roossinck MJ. A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science. 2007;315(5811):513–515. doi: 10.1126/science.1136237. [DOI] [PubMed] [Google Scholar]
- 39.Dalzoto PR, Glienke-Blanco C, Kava-Cordeiro V, Ribeiro JZ, Kitajima EW, Azevedo JL. Horizontal transfer and hypovirulence associated with double-stranded RNA in Beauveria bassiana . Mycological Research. 2006;110(12):1475–1481. doi: 10.1016/j.mycres.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 41.Melzer MJ, Bidochka MJ. Diversity of double-stranded RNA viruses within populations of entomopathogenic fungi and potential implications for fungal growth and virulence. Mycologia. 1998;90(4):586–594. [Google Scholar]
- 42.Petrini O. Taxonomy of endophytic fungi of aerial plant tissues. In: Fokkema NJ, van den Heuvel J, editors. Microbiology of the Phyllosphere. Cambridge, UK: Cambridge University Press; 1986. pp. 175–187. [Google Scholar]
- 43.Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Letters in Applied Microbiology. 1985;1(1):17–20. [Google Scholar]
- 44.Glienke-Blanco C, Aguilar-Vildoso CI, Carneiro Vieira ML, Barroso PAV, Azevedo JL. Genetic variability in the endophytic fungus Guignardia citricarpa isolated from citrus plants. Genetics and Molecular Biology. 2002;25(2):251–255. [Google Scholar]
- 45.White JF, Jr., Morrow AC. Endophyte-host associations in forage grasses. XII. A fungal endophyte of Trichachne insularis belonging to Psedocercosporella . Mycologia. 1990;82(2):218–226. [Google Scholar]
- 46.Mills PR, Sreenivasaprasad S, Brown AE. Detection and differentiation of Colletotrichum gloeosporioides isolates using PCR. FEMS Microbiology Letters. 1992;98(1–3):137–143. doi: 10.1016/0378-1097(92)90145-e. [DOI] [PubMed] [Google Scholar]
- 47.Pileggi SAV, Oliveira SFV, Waculicz-Andrade CE, et al. Molecular and Morphological Identification of Colletotrichum gloeosporioides and Colletotrichum boninense isolated from Maytenus ilicifolia . Canadian Journal of Microbiology. 2009;55:1076–1088. doi: 10.1139/w09-059. [DOI] [PubMed] [Google Scholar]
- 48.de Hoog GS, Gerrits van den Ende AHG. Molecular diagnostics of clinical strains of filamentous Basidiomycetes. Mycoses. 1998;41(5-6):183–189. doi: 10.1111/j.1439-0507.1998.tb00321.x. [DOI] [PubMed] [Google Scholar]
- 49.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zwickl D. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin, Tex, USA: University of Texas at Austin; 2006. Ph.D. thesis. [Google Scholar]
- 51.Huelsenbeck JP, Ronquist F. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 52.Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20(3):407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- 53.Posada D. jModeltest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 54.Rambaut A, Drummond AJ. Tracer v1.5. 2009, http://beast.bio.ed.ac.uk/Tracer.
- 55.Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. Awty (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24(4):581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- 56.Crous PW, Kang JC, Schoch CL, Mchau GRA. Phylogenetic relationships of Cylindrocladium pseudogracile and Cylindrocladium rumohrae with morphologically similar taxa, based on morphology and DNA sequences of internal transcribed spacers and β-tubulin. Canadian Journal of Botany. 1999;77(12):1813–1820. [Google Scholar]
- 57.Hyde KD, CAI L, Cannon PF, et al. Colletotrichum—names in current use. Fungal Diversity. 2009;39:147–182. [Google Scholar]
- 58.Shivas RG, Tan YP. A taxonomic reassessment of Colletotrichum acutatum, introducing C. fioriniae comb. et stat. nov. and C. simmondsii sp. nov . Fungal Diversity. 2009;39:111–122. [Google Scholar]
- 59.Castro M, Kramer K, Valdivia L, Ortiz S, Benavente J, Castillo A. A new double-stranded RNA mycovirus from Botrytis cinerea . FEMS Microbiology Letters. 1999;175(1):95–99. doi: 10.1111/j.1574-6968.1999.tb13606.x. [DOI] [PubMed] [Google Scholar]
- 60.Howitt RLJ, Beever RE, Pearson MN, Forster RLS. Presence of double-stranded RNA and virus-like particles in Botrytis cinerea . Mycological Research. 1995;99(12):1472–1478. [Google Scholar]
- 61.Papp T, Nyilasi I, Fekete C, Ferenczy L, Vagvolgyi C. Presence of double-stranded RNA and virus-like particles in Rhizopus isolates. Canadian Journal of Microbiology. 2001;47(5):443–447. doi: 10.1139/w01-020. [DOI] [PubMed] [Google Scholar]
- 62.Preisig O, Moleleki N, Smit WA, Wingfield BD, Wingfield MJ. A noval RNA mycovirus in a hypovirulent isolate of the plant pathogen Diaporthe ambigua . Journal of General Virology. 2000;81(12):3107–3114. doi: 10.1099/0022-1317-81-12-3107. [DOI] [PubMed] [Google Scholar]
- 63.Robinson HL, Deacon JW. Double-stranded RNA elements in Rhizoctonia solani AG3. Mycological Research. 2002;106(1):12–22. [Google Scholar]
- 64.Punja ZK. Influence of double-stranded RNAs on growth, sporulation, pathogenicity, and survival of Chalara elegans . Canadian Journal of Botany. 1995;73(7):1001–1009. [Google Scholar]
- 65.Dawe AL, Nuss DL. Hypoviruses and chestnut blight: exploiting viruses to understand and modulate fungal pathogenesis. Annual Review of Genetics. 2001;35:1–29. doi: 10.1146/annurev.genet.35.102401.085929. [DOI] [PubMed] [Google Scholar]
- 66.Hollings M. Mycoviruses and plant pathology. Plant Disease. 1982;66:1106–1112. [Google Scholar]
- 67.Pearson MN, Beever RE, Boine B, Arthur K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Molecular Plant Pathology. 2009;10(1):115–128. doi: 10.1111/j.1364-3703.2008.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


