Abstract
The primary underlying cause of cervical cancer is infection with one or more high-risk (HR) types of the human papilloma virus (HPV). Detection and typing of HPV have been commonly carried out by PCR-based assays, where HPV detection and typing are two separate procedures. Here, we present a multiplex PCR-based HPV typing assay that detects 20 HPV types (15 HR, 3 probably HR and 2 low risk) using type-specific primers and agarose gel electrophoresis. 46 cervical, urethral, and biopsy samples were analyzed by both Multiplex PCR and PGMY09/11 consensus PCR, and results were compared. 611 samples were further analyzed by Multiplex PCR, 282 were positive for HR HPV, and 101 showed multiple HR HPV infections. The relatively ease and economic accessibility of the method and its improved ability to detect high-risk HPV types in multiple HPV-infected samples make it an attractive option for HPV testing.
1. Introduction
Cervical cancer is the second most common cancer in women worldwide [1] and is the most common cancer in women from low-income countries, where an estimated 80% of cases occur [2]. 16,000 cases of cervical cancer are newly detected every year in Mexico, resulting in a high incidence rate (50 cases per 100,000 women) [3, 4]. The primary underlying cause of cervical cancer is infection with one or more high-risk (HR) types of the human papilloma virus (HPV) [5–10]. 15 HR types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82) have been proposed, including 3 types (26, 53, and 66) that should be considered probably carcinogenic [11, 12].
Detection and typing of HPV have been commonly carried out by PCR-based assays, where HPV DNA is amplified by consensus primers and then typed by restriction enzyme analysis (RFLP), hybridization with type-specific probes, or direct sequencing of the amplicons, among the most common methods [13]. Recently, methods that use multiplex PCR amplification with type-specific primers have been reported, where detection and typing are deducted from the amplification pattern of capillary electrophoresis [14].
Here, we present a multiplex PCR-based HPV typing assay that detect 20 HPV types (15 HR), 3 probably HR and 2 low risk (LR) using type-specific primers and agarose gel electrophoresis.
2. Materials and Methods
2.1. Sample Preparation
611 samples of cervical (232) and urethral (164) scrapes and paraffin-embedded tissue biopsies (215) submitted for HPV assessment were collected for Multiplex PCR HPV analysis. A subset of 46 cervical, 16 urethral, and 21 tissue biopsies were randomly selected for additional analysis with PGMY09/11 consensus primer PCR. DNA extraction of samples was performed using the Dneasy Tissue Kit (Qiagen, Germany), following manufacturer's instructions.
2.2. Primers Design
DNA sequence files for HPV types 6, 11, 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, and 82 were obtained from Genbank (http://www.ncbi.nih.gov/genbank/). Primers were designed for each HPV type, and unique specificity was confirmed by BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/). Primer selection for each reaction tube mix was carried out in silico [15] and experimentally to ensure primer compatibility, and a primer pair specific for β-globin was included as positive control [16]. Sequences of primers included in each reaction mix, with predicted amplification product size and digestion product sizes with indicated restriction enzymes are shown in Table 1.
Table 1.
Multiplex PCR primer list.
| Name | Forward primer sequence | Reverse primer sequence | lane | size bp | R.E. | Digested fragment sizes |
|---|---|---|---|---|---|---|
| 6-1 | acgtggccttgtgcggtacagtc | agagacgagtcaggcaatgc | 4 | 757 | HaeIII | 259, 250, 101, 72, 68, 7 |
| 6-2 | tgtcccatctgcgcaccgaagac | cgtacactgtttgtgggcgcttc | 4 | 592 | AluI | 366, 129, 97 |
| 11-1 | agttccgtagatgccaagggca | tgcctcaggtgaggcccaatgc | 3 | 528 | RsaI | 194, 172, 94, 68 |
| 11-2 | tggtaccccctacacagggtgg | acagaatgttggacagggtcagg | 3 | 730 | HaeIII | 433, 160, 137 |
| 16-1 | ttaggcagcacttggccaacca | taatccgtcctttgtgtgagct | 3 | 207 | MspI | 110, 97 |
| 16-2 | actgcaatgtttcaggacccac | cgaagcgtagagtcacacttgc | 2 | 661 | MspI | 405, 199, 57 |
| 18-1 | tcgcgtcctttatcacagggcga | tgcccaggtacaggagactgtg | 1 | 536 | AluI | 235, 200, 101 |
| 18-2 | tccgtggtgtgcatcccagcag | cacttgtgcatcattgtggacc | 2 | 274 | RsaI | 185, 48, 41 |
| 26-1 | tggtatacaacgagtgtcagctcc | ggggcaatgatggccatgtcg | 7 | 635 | MspI | 425, 210 |
| 31-1 | aggcacggttggtgaatcggtc | tagatgctgagggtgcactacg | 1 | 683 | HaeIII | 481, 202 |
| 31-2 | catgaactaagctcggcattgg | tccaacatgctatgcaacgtcc | 2 | 385 | RsaI | 233, 152 |
| 33-1 | agcttagaggtgtggctttgtg | tgcagttagttgcagtacgtgc | 2 | 493 | RsaI | 211, 145, 122, 15 |
| 33-2 | tgacccacctacagctgcaatc | gggtgtgtacattatccacatcg | 2 | 899 | RsaI | 615, 277, 7 |
| 35-1 | ccaccaagtggttccaacgcag | tgtaggcgtgtagctgtgtagc | 3 | 488 | RsaI | 216, 193, 79 |
| 35-2 | gtcctgttggaaaccaacacgt | acacacagacgtagtgtcgcct | 3 | 251 | AluI | 135, 81, 35 |
| 39-1 | acacaaacggtgtattccgtgcca | tgtgcagttggagatttgggatcc | 5 | 200 | RsaI | 129, 71 |
| 39-2 | tgtgcagtaccagtgacggatcg | atttttggcgttgtgactctgtg | 6 | 438 | HaeIII | 221, 217 |
| 45-1 | ggacatcacacctaccgtggac | ctgtgaggtggacacacggacc | 4 | 298 | RsaI | 126, 69, 59, 26, 18 |
| 45-2 | acctgcacaattgcaacctggt | caactgccaggggtttcacgca | 4 | 345 | RsaI | 179, 105, 61 |
| 51-1 | aattgctggcaacgtacacgac | acacttgaacacctgcaacacg | 4 | 255 | RsaI | 190, 51, 14 |
| 51-2 | cctactccaggggttagtcgca | taaggagggcaactgcctagac | 4 | 504 | HaeIII | 270, 234 |
| 52-1 | cccaagtgtaacgtcatgcgtg | agggttgtttatagccgtgcac | 3 | 323 | AluI | 215, 108 |
| 52-2 | acctccgcagtgtccgtgggtg | aagagcggcctaagcactgcac | 1 | 601 | RsaI | 143, 126, 96, 95, 66, 41, 34 |
| 53-1 | ttgttcagtgtacggggctagc | gtgacgccattgcagttatcgcct | 6 | 549 | MspI | 389, 160 |
| 53-2 | ttctgcagtaagctatgagggcat | aaccactgtcgatttcggtgtt | 7 | 449 | MspI | 270, 179 |
| 56-1 | ctgggcactaggtcaaagcctgct | caaccacgcgtaaaagcactcat | 6 | 307 | AluI | 278, 29 |
| 58-1 | ggtagtaccccaccgtctgagg | agacgtgacattgccactgtca | 1 | 414 | MspI | 289, 125 |
| 58-2 | accagactccagagacaacacc | tcacctttgtcatcactggtcc | 1 | 264 | RsaI | 165, 62, 25, 12 |
| 59-1 | agacaccgttacatgagctgct | tcattctcggagtcggagtcag | 1 | 320 | AluI | 213, 91, 16 |
| 59-2 | tctaacgccatctgcagcaagg | acagtagtccactgacacgctg | 4 | 438 | HaeIII | 339, 99 |
| 66-1 | tgcggtagtatccttgggcagtg | tacaataagggctacacgccaa | 5 | 388 | RsaI | 135, 131, 122 |
| 68-1 | ggtactgcttggaacacgcctg | ggcccccagacatagggacctt | 6 | 368 | RsaI | 307, 57, 4 |
| 68-2 | gtcaaaaagacgcccctgcaccta | cacaccttagggtagggctacaa | 5 | 490 | HaeIII | 329, 161 |
| 73-1 | acaggctattagttgccaacgtc | ttcttaggtgtggcacttgtg | 6 | 222 | AluI | 115, 62, 45 |
| 73-2 | ggggtgggcaaaggtaggtagc | acaatccaggggcctctggtccga | 7 | 322 | RsaI | 189, 108, 25 |
| 82-1 | tgtccgtggacacctgcgacca | gtagttaaaggtgatgtggcaacc | 7 | 546 | RsaI | 275, 215, 56 |
| 82-2 | cccaaaaccaatacacgtgctgaa | aacatcctgttggtcgttgcca | 5 | 270 | HaeIII | 189, 81 |
| β-globin | gaagagccaaggacaggtac | caacttcatccacgttcacc | 7 | 268 |
2.3. Multiplex PCR
The QIAGEN Multiplex PCR kit (Qiagen, Germany) was used in all Multiplex PCR reactions, following manufacturer instructions. Each PCR was carried out in a DNA thermal cycler (MaxyGene Gradient Thermal Cycler, Axygen Scientific, USA) with the following conditions: initial denaturing step at 95°C for 15 min, 10 cycles of 30 s at 94°C, 90 s at 65°C, and 90 s at 72°C, followed by 30 cycles of 30 s at 94°C, 90 s at 63°C, and 90 s at 72°C, with a final extension at 72°C for 10 min. PCR products were analyzed by electrophoresis on a 2% agarose gel stained with ethidium bromide, band sizes were estimated by comparison with a 100 bp molecular weight marker (GeneRuler 100 bp DNA Ladder, Fermentas International, Canada), and gels were photographed in a UV transilluminator (UVP, USA) with a Canon PowerShot A60 digital camera (Canon, USA). HPV type was assigned based on the amplification pattern. In cases where band interpretation was not clear, an additional PCR amplification with specific primers was performed to confirm. Selected PCR amplified fragments were cloned into pGem-T vector (Promega, USA), each cloned product was sequenced with universal forward and reverse primers to confirm fragment identity. Additionally, selected amplified fragments were digested with restriction enzymes AluI, HaeIII, RsaI, or MspI (New England Biolabs, USA), and digestion patterns were observed in a 2% agarose gel to also confirm fragment identity.
2.4. PGMY09/11 Consensus PCR
HPV consensus PCR was performed using primers PGMY09/PGMY11 designed to amplify a fragment of the HPV L1 gene of approximately 450 bp as previously described [17]. HPV genotype was assigned by sequencing of amplified fragments using primers PGMY11.
3. Results and Discussion
Polymerase Chain reaction (PCR) with consensus primers can potentially detect most mucosal HPV types [18]. There are several consensus primers described, GP5/6 and their improved GP5+/6+ [19, 20], SPF [21], My09/11 and their improved PGMY09/11 [17, 18, 22], L1C1 with L1C2 and L1C2M [23], pU-1M/pU-2R and their enhanced pU-1M-L and pU-2R-N [24]. Typing of amplified fragments is usually performed by different techniques such as, hybridization to specific probes [25], restriction fragment length polymorphisms [26, 27], or direct DNA sequencing [28–30].
In this report, we present an assay based in HPV DNA amplification with type-specific primers in a Multiplex PCR format to detect and type single or multiple HR HPV infections in samples of different sources. Primers specific to each of 15 high-risk, 3 probably high-risk, and 2 low-risk HPV types were included in seven independent Multiplex PCR reactions. Typing was assigned based on the amplification pattern. As a result of having specific primers, stringent PCR conditions can be set to increase the clarity of results by reducing the presence of amplification artifacts, and all HPV types in a sample are amplified by their specific primer pair. Also, detection and HPV typing are accomplished at the same time, without the need of an additional protocol for typing after HPV detection (see examples of HPV detection in Figure 1).
Figure 1.
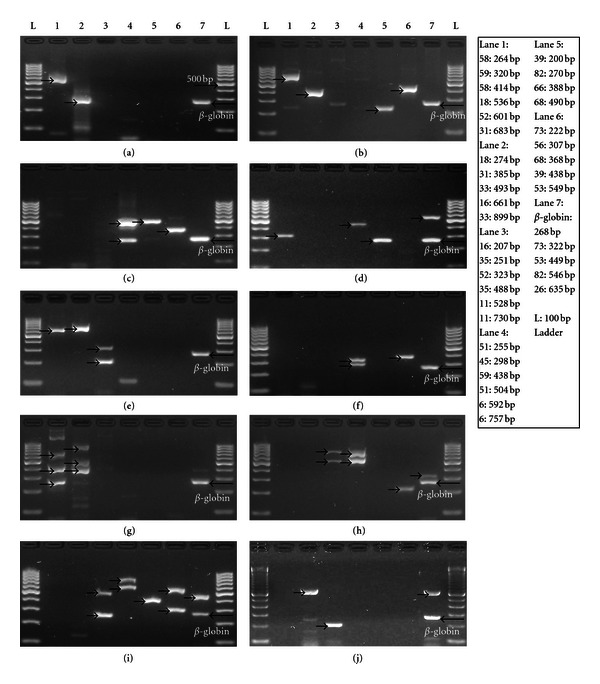
HPV detection and typing by Multiplex PCR. (a) Amplification pattern shows two bands in lanes 1 and 2 (arrows), consistent with the expected amplification pattern of HPV 18. (b) Four bands are observed in lanes 1, 2, 5, and 6 (arrows), consistent with HPV 31 and 39. (c) Bands in lanes 4, 5, and 6 (arrows), consistent with HPV 51 and 68. (d) Detection of HPV 59 and 82, (e) HPV 16 and 52, and (f) HPV 45 and 56. (g) Detection of HPV 31, 33, and 58, and (h) HPV 6, 11, and 73. (i) Five HR HPV types are observed: HPV 6, 35, 53, 56, and 66. (j) Detection of HPV 16 and 26, only one case of HPV 26 was detected in 611 samples.
In order to evaluate the newly developed HPV detection assay, 83 samples (46 cervical and 16 urethral scrapes and 21 tissue biopsies) were analyzed with Multiplex PCR and PGMY09/11 consensus PCR. Positive high risk is determined when a HR or probable HR HPV type is detected, and negative high risk is determined when a LR HPV type or no HPV infection is detected (Table 2). 47 samples reported negative results by both Multiplex PCR (Multiplex) and by PCR with PGMY09/11 primers (PGMY), while no samples were reported positive by PGMY and negative by Multiplex. 19 samples were reported positive by both Multiplex and PGMY, but 17 samples were reported positive by Multiplex and negative by PGMY. Of those, 12 samples did not produce any amplified fragment (no detection) and 5 samples reported LR HPV types (failed to detect the HR HPV type also present in the sample). These results suggest that Multiplex PCR can detect HR HPV as well as PGMY PCR, and that Multiplex PCR can potentially detect HR HPV infections not reported by PGMY PCR, due to the presence of LR types that are preferentially amplified over HR types in multiple infections, as observed in 5 samples.
Table 2.
83 samples analyzed by Multiplex PCR and PGMY consensus PCR.
| Detection comparison of HR HPV | Multiplex PCR | |
|---|---|---|
| PGMY PCR | Positive | Negative |
| Positive | 13 cervical | 0 cervical |
| 4 urethral | 0 urethral | |
| 2 biopsies | 0 biopsies | |
|
| ||
| Negative | 10 cervical | 23 cervical |
| 2 urethral | 10 urethral | |
| 5 biopsies | 14 biopsies | |
Particularly important is the capacity of detection of HPV multiple infections. A total of 611 samples (232 cervical and 164 urethral scrapes and 215 tissue biopsies) were analyzed by Multiplex PCR, including 83 samples mentioned above. 324 (53.03%) samples were negative for HR HPV, and 282 (46.15%) samples were positive for HR HPV. Only 5 (0.82%) samples (1 cervical and 4 paraffin-embedded tissue biopsies) failed to amplify the β-globin control gene and were reported as being not informative. 35% (101) of HR HPV positive samples had infections with two or more HR HPV types, representing 16% of the total number of samples analyzed. Detecting all HR HPV present in a sample is important in patient treatment to asses prevalence of infection and response to treatment.
Multiplex PCR HPV detection and typing are simple and potentially affordable. After DNA extraction and Multiplex PCR amplification, detection and typing of HPV are deduced from the amplification pattern observed in an agarose gel electrophoresis. This is particularly important in low-income countries. According to a study in Peru [31], simple, effective, and cost-efficient HPV testing is the best option for primary cervical screening. The entire cost in Mexico of the equipment and reagents for DNA extraction, amplification, agarose gel electrophoresis, and documentation is approximately $22,000 USD, and many research laboratories in Mexico already have all the necessary equipments.
4. Conclusions
Multiplex PCR HPV can detect single or multiple HR HPV infections in cervical and urethral scrapes and paraffin-embedded tissue biopsies. The relatively ease and economic accessibility of the method can potentially have an impact in HPV screening in low-income countries like Mexico, and its improved ability to detect high-risk HPV types in multiple HPV-infected samples makes it an attractive option for HPV testing.
Conflict of Interests
Authors declare no conflict of interests.
References
- 1.Parkin DM, Bray F. Chapter 2: the burden of HPV-related cancers. Vaccine. 2006;24(3):S11–S25. doi: 10.1016/j.vaccine.2006.05.111. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. International Agency for Research on Cancer. GLOBOCAN database 2002. CANCERMondial, 2002, http://www-dep.iarc.fr/
- 3.Lazcano-Ponce EC, Najera P, Alonso de Ruiz P, Buiatti E, Hernandez-Avila M. Programa de detección oportuna del cáncer cervical en México. I. Diagnóstico situacional. Revista del Instituto Nacional de Cancerología. 1996;42(3):123–140. [Google Scholar]
- 4.Secretaría de Salud (Méx) Registro Histopatológico de Neoplasias en México. México D. F., Mexico: Secretaría de Salud; 1999. [Google Scholar]
- 5.Bosch FX, Lorincz A, Muñoz N, Meijer CJLM, Shah KV. The causal relation between human papillomavirus and cervical cancer. Journal of Clinical Pathology. 2002;55(4):244–265. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chichareon S, Herrero R, Muñoz N, et al. Risk factors for cervical cancer in Thailand: a case-control study. Journal of the National Cancer Institute. 1998;90(1):50–57. doi: 10.1093/jnci/90.1.50. [DOI] [PubMed] [Google Scholar]
- 7.Koutsky LA, Holmes KK, Critchlow CW, et al. A cohort study of the risk of cervical intraepithelial neoplasia grade 2 or 3 in relation to papillomavirus infection. The New England Journal of Medicine. 1992;327(18):1272–1278. doi: 10.1056/NEJM199210293271804. [DOI] [PubMed] [Google Scholar]
- 8.Lehtinen M, Luukkaala T, Wallin KL, et al. Human papillomavirus infection, risk for subsequent development of cervical neoplasia and associated population attributable fraction. Journal of Clinical Virology. 2001;22(1):117–124. doi: 10.1016/s1386-6532(01)00172-x. [DOI] [PubMed] [Google Scholar]
- 9.Schiffman MH, Bauer HM, Hoover RN, et al. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. Journal of the National Cancer Institute. 1993;85(12):958–964. doi: 10.1093/jnci/85.12.958. [DOI] [PubMed] [Google Scholar]
- 10.Wright TC, Thomas Cox J, Stewart Massad L, Twiggs LB, Wilkinson EJ. 2001 consensus guidelines for the management of women with cervical cytological abnormalities. Journal of the American Medical Association. 2002;287(16):2120–2129. doi: 10.1001/jama.287.16.2120. [DOI] [PubMed] [Google Scholar]
- 11.Muñoz N, Bosch FX, De Sanjosé S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. The New England Journal of Medicine. 2003;348(6):518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 12.De Villiers EM, Fauquet C, Broker TR, Bernard HU, Zur Hausen H. Classification of papillomaviruses. Virology. 2004;324(1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 13.Molijn A, Kleter B, Quint W, Van Doorn LJ. Molecular diagnosis of human papillomavirus (HPV) infections. Journal of Clinical Virology. 2005;32:S43–S51. doi: 10.1016/j.jcv.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Nishiwaki M, Yamamoto T, Tone S, et al. Genotyping of human papillomaviruses by a novel one-step typing method with multiplex PCR and clinical applications. Journal of Clinical Microbiology. 2008;46(4):1161–1168. doi: 10.1128/JCM.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalendar R, Lee D, Schulman AH. FastPCR software for PCR primer and probe design and repeat search. Genes, Genomes and Genomics. 2009;3(1):1–14. [Google Scholar]
- 16.Bell DA, Taylor JA, Paulson DF, Robertson CN, Mohler JL, Lucier GW. Genetic risk and carcinogen exposure: a common inherited defect of the carcinogen-metabolism gene glutathione S-transferase M1 (GSTM1) that increases susceptibility to bladder cancer. Journal of the National Cancer Institute. 1993;85(14):1159–1164. doi: 10.1093/jnci/85.14.1159. [DOI] [PubMed] [Google Scholar]
- 17.Gravitt PE, Peyton CL, Alessi TQ, et al. Improved amplification of genital human papillomaviruses. Journal of Clinical Microbiology. 2000;38(1):357–361. doi: 10.1128/jcm.38.1.357-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gravitt PE, Jamshidi R. Diagnosis and management of oncogenic cervical human papillomavirus infection. Infectious Disease Clinics of North America. 2005;19(2):439–458. doi: 10.1016/j.idc.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 19.De Roda Husman AM, Walboomers JMM, Van den Brule AJC, Meijer CJLM, Snijders PJF. The use of general primers GP5 and GP6 elongated at their 3’ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. Journal of General Virology. 1995;76(4):1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs MV, Snijders PJF, Voorhorst FJ, et al. Reliable high risk HPV DNA testing by polymerase chain reaction: an intermethod and intramethod comparison. Journal of Clinical Pathology. 1999;52(7):498–503. doi: 10.1136/jcp.52.7.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleter B, Van Doorn LJ, Schrauwen L, et al. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. Journal of Clinical Microbiology. 1999;37(8):2508–2517. doi: 10.1128/jcm.37.8.2508-2517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manos MM, Ting Y, Wright DK, Lewis AJ, Broker TR, Wolinski SM. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells. 1989;7:209–214. [Google Scholar]
- 23.Yoshikawa H, Kawana T, Kitagawa K, Mizuno M, Yoshikura H, Iwamoto A. Detection and typing of multiple general human papillomaviruses by DNA amplification with consensus primers. Japanese Journal of Cancer Research. 1991;82(5):524–531. doi: 10.1111/j.1349-7006.1991.tb01882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Astori G, Arzese A, Pipan C, De Villiers EM, Botta GA. Characterization of a putative new HPV genomic sequence from a cervical lesion using L1 consensus primers and restriction fragment length polymorphism. Virus Research. 1997;50(1):57–63. doi: 10.1016/s0168-1702(97)00054-3. [DOI] [PubMed] [Google Scholar]
- 25.Van den Brule AJC, Pol R, Fransen-Daalmeijer N, Schouls LM, Meijer CJLM, Snijders PJF. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. Journal of Clinical Microbiology. 2002;40(3):779–787. doi: 10.1128/JCM.40.3.779-787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lungu O, Wright TC, Silverstein S. Typing of human papillomaviruses by polymerase chain reaction amplification with L1 consensus primers and RFLP analysis. Molecular and Cellular Probes. 1992;6(2):145–152. doi: 10.1016/0890-8508(92)90059-7. [DOI] [PubMed] [Google Scholar]
- 27.Kay P, Meehan K, Williamson AL. The use of nested polymerase chain reaction and restriction fragment length polymorphism for the detection and typing of mucosal human papillomaviruses in samples containing low copy numbers of viral DNA. Journal of Virological Methods. 2002;105(1):159–170. doi: 10.1016/s0166-0934(02)00100-3. [DOI] [PubMed] [Google Scholar]
- 28.Iftner T, Villa LL. Chapter 12: human papillomavirus technologies. Journal of the National Cancer Institute. Monographs. 2003;(31):80–88. doi: 10.1093/oxfordjournals.jncimonographs.a003487. [DOI] [PubMed] [Google Scholar]
- 29.Feoli-Fonseca JC, Oligny LL, Brochu P, Simard P, Falconi S, Yotov WV. Human papillomavirus (HPV) study of 691 pathological specimens from Quebec by PCR-direct sequencing approach. Journal of Medical Virology. 2001;63(4):284–292. doi: 10.1002/1096-9071(200104)63:4<284::aid-jmv1003>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 30.Speich N, Schmitt C, Bollmann R, Bollmann M. Human papillomavirus (HPV) study of 2916 cytological samples by PCR and DNA sequencing: genotype spectrum of patients from the west German area. Journal of Medical Microbiology. 2004;53(2):125–128. doi: 10.1099/jmm.0.05447-0. [DOI] [PubMed] [Google Scholar]
- 31.Almonte M, Ferreccio C, Winkler JL, et al. Cervical screening by visual inspection, HPV testing, liquid-based and conventional cytology in Amazonian Peru. International Journal of Cancer. 2007;121(4):796–802. doi: 10.1002/ijc.22757. [DOI] [PubMed] [Google Scholar]


