Abstract
Poly(ADP-ribosyl)ation is a modification of nuclear proteins that regulates DNA replication, repair and transcription. In order to investigate the biological effects of degradation of poly(ADP-ribose), knockdown of the poly(ADP-ribose) glycohydrolase (PARG) gene was performed by introducing a short interfering RNA (siRNA)-pool into HeLa S3 cells. Notably, poly(ADP-ribosyl)ated proteins did not accumulate in the cells. Western blotting, quantitative RT-PCR analysis and a transient transfection assay revealed that poly(ADP-ribose) polymerase 1 (PARP1) gene/protein expression and its promoter activity were reduced in the PARG knockdown cells. These results suggest that the amount of poly(ADP-ribose) in a cell is regulated under the control of PARP1/PARG gene expression balance. Furthermore, in this study, we showed that PARG-siRNA enhanced cell death induced by staurosporine (STS). Thus, we propose a PARG-siRNA utilizing gene-therapy for cancer treatment.
Keywords: poly(ADP-ribose), poly(ADP-ribose) glycohydrolase, poly(ADP-ribose) polymerase, short interfering RNA, Sp1
Introduction
Poly(ADP-ribosyl)ation is a NAD+-dependent post-transcriptional modification of chromosomal proteins mediated by poly(ADP-ribose) polymerases (PARPs) and poly(ADP-ribose) glycohydrolase (PARG) (1). Reversible poly(ADP-ribosyl)ation of chromosomal proteins has been suggested to play important roles in various biological processes, including DNA replication (2,3), repair (4–6), spindle assembly (7), transcription (8), telomere and chromosomal maintenance (9) and epigenetic gene regulation (10). Previous studies have suggested that poly(ADP-ribose) metabolism is associated with differentiation and proliferation (11,12), cell death (13) and apoptosis (14,15). Thus, poly(ADP-ribosyl)ation is significantly involved in various biological activities, suggesting that it requires precise controlling systems to adjust the amount and length of poly(ADP-ribose) in eukaryotic cells.
Previous studies indicated that several transcription factors, including Sp1 (16), YY1 (17), and NF-κB (18), are poly(ADP-ribosyl)ated and reduce the transcription activity (16). If there are binding sites for these transcription factors in the promoter region, transcription might be affected by the poly(ADP-ribosyl)ation. To date, the promoter regions of the human PARP1(19) and PARG(20,21) genes have been isolated and characterized. In this study, we investigated the effect of short interfering RNAs (siRNAs) for the human PARG cDNA on poly(ADP-ribosyl)ation and PARP1 gene expression in HeLa S3 cells. The results showed that expression and promoter activity of the PARP1 gene were reduced by the knockdown of the PARG gene and that the amounts of poly(ADP-ribose) in the cells did not increase compared to the control cells. Moreover, PARG knockdown cells showed stronger cell death sensitivity to staurosporine (STS) than the control cells, suggesting that retarded turnover of poly(ADP-ribose)-NAD+ metabolism might induce intracellular apoptosis signals.
It is well known that PARP1 activity is downregulated by its augmented auto-poly(ADP-ribosyl)ation (22,23), and artificially accumulated poly(ADP-ribose) induces apoptosis (13). Collectively, our results indicate that reduced poly(ADP-ribose) degradation subsequently suppresses transcription of the PARP1 gene to escape excessive poly(ADP-ribose) accumulation, thereby achieving a balance in poly(ADP-ribose) levels for cell survival. Therefore, poly(ADP-ribose) may act as a dual regulator for PARP1 activity not only at the post-translational level but also at the transcriptional level. Hence, we propose a molecular mechanism that prevents cells from accumulating excess amounts of poly(ADP-ribose) by regulating transcription of the PARP1 gene.
Materials and methods
Cell culture
Human cervical carcinoma (HeLa S3) cells (24) were grown in Dulbecco’s modified Eagle’s medium (DMEM; Nacarai, Tokyo, Japan), supplemented with 10% fetal bovine serum (FBS) (Sanko Pure Chemicals, Tokyo, Japan) and penicillin-streptomycin at 37°C in a humidified atmosphere with 5% CO2.
Transfection of siRNA
The ON-TARGETplus SMARTpool siRNAs used for knockdown of the human PARG gene were purchased from Thermo Fisher Scientific Inc. (Lafayette, CO, USA). They were introduced into HeLa S3 cells with DharmaFECT Transfection reagent following the manufacturer’s protocol (Thermo Fisher Scientific). In brief, 2 μM siRNA (50 μl) were added to serum-free DMEM (50 μl) in one tube, and DharmaFECT1 (1.5 μl) was added to 98.5 μl of serum-free medium in the other tube. They were gently mixed and incubated for 5 min at room temperature, and were then combined, mixed and further incubated for 20 min at room temperature. Subsequently, complete medium (800 μl) was added and cells were cultivated with the medium in a 35-mm culture dish.
Cell viability MTS assay
An MTS assay was performed according to the manufacturer’s instructions. In brief, mock- or siRNA-transfected cells were cultured in microtiter plate wells. MTS solution (20 μl) (Promega, Madison, WI, USA) was added to each well (containing 100 μl of cell culture) and incubated for 3 h in a 37°C, 5% CO2-humidified incubator. Then, the absorbance at 492 nm was measured by a microtiter plate reader (Thermo Electron Corp., Vantaa, Finland) and normalized by the absorbance at 630 nm.
Reverse transcriptase and quantitative real-time polymerase chain reaction (RT-qPCR)
RT-qPCR was carried out as previously described (24). First-strand cDNAs were synthesized with ReverTra Ace (Toyobo Corp., Tokyo, Japan), random primers (Takara, Kyoto, Japan) and total RNAs were extracted from HeLa S3 cells. A primer pair to amplify the human GAPDH cDNAs was previously reported (24), and those for amplifying the PARP1 and PARG cDNAs were: hPARP1S514, 5′-GCAGAGTATGCCAAGTCCAACAG-3′ and hPARP1AS813, 5′-ATCCACCTCATCGCCTTTTC-3′; and hPARG-S, 5′-ATGTGTAAGTGGCAAAATGAAGGG-3′ and hPARG-A952, 5′-CTTCTCTGGCCTGTTCATCTTC-3′, respectively. Real-time PCR analysis was carried out using the Mx3000P Real-Time QPCR system (Stratagene, La Jolla, CA, USA) as previously described (24). For PCR amplification, cDNAs were amplified using SYBR-Green real-time PCR Master Mix (Toyobo) and 0.3 μM of each primer pair. Amplification of the PARP1 cDNA was carried out, starting with an initial step for 1 min at 94°C, followed by 42 cycles (94°C 30 sec, 55°C 30 sec and 72°C 1 min). The conditions for amplification of the PARG and GAPDH cDNAs were 1 min at 94°C, followed by 42 cycles (94°C 15 sec, 55°C 10 sec, and 72°C 15 sec).
Western blot analysis
Western blotting was carried out as previously described (24,25) with antibodies against PARP1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and PAR (Calbiochem, Darmstadt, Germany) followed by the addition of horseradish peroxidase (HRP)-conjugated secondary antibody (Calbiochem). Signal intensities were quantified with a LAS4000 system and Multi Gauge Software (Fuji Film, Tokyo, Japan).
Construction of luciferase (Luc) reporter plasmids
Luc reporter plasmids carrying 75-bp of the human PARG promoter regions were designated pKBST-Δ6 (21). The 5′-flanking regions of the human PARP1 gene were obtained by PCR with PrimeStar Taq polymerase (Takara) and the template genomic DNA from HeLa S3 cells as previously described (26). The sense and antisense primers used for PCR were: hPARP1-2660, 5′-TCGGTACCGGGTCCTCCAAAGAGCTAC-3′; and AhPARP1-2895, 5′-ATCTCGAGCCGCCACCGAACACGC CGC-3′, respectively. The amplified DNA fragments were digested with KpnI and XhoI and ligated into the MCS of the pGL4-basic (pGL4.10[luc2]), vector (Promega) to make pGL4-PARP1. Deletion derivatives, pGL4-PARP1Δ1 and pGL4-PARP1Δ2, were generated by PCR with pGL4-PARP1 as the template and primer sets: hPARP1-2851, 5′-TCGGTA CCGCCAGGCATCAGCAATCTA-3′ and AhPARP1-2895; and hPARP1-2660 and AhPARP1-2851, 5′-ATCTCGAGTA GATTGCTGATGCCTGGC-3′, respectively. The nucleotide sequences of the PCR products were determined by a DNA Sequencing system (Applied Biosystems, Foster City, CA, USA) with Rv (5′-TAGCAAAATAGGCTGTCCCC-3′) and GL (5′-CTTTATGTTTTTGGCGTCTTCC-3′) primers.
Transient transfection and Luc assays
Plasmid DNAs were transfected into HeLa S3 cells by the DEAE-dextran method (24–26). After a further 24 h of incubation, cells were collected and lysed with 100 μl of 1X cell culture lysis reagent, mixed and centrifuged at 12,000 × g for 5 sec. The supernatant was stored at −80°C. The Dual Luciferase assay was performed with the Dual Luciferase assay system (Promega), as previously described (21).
Results
Decrease in the amounts of PARG gene transcripts after introducing its siRNAs
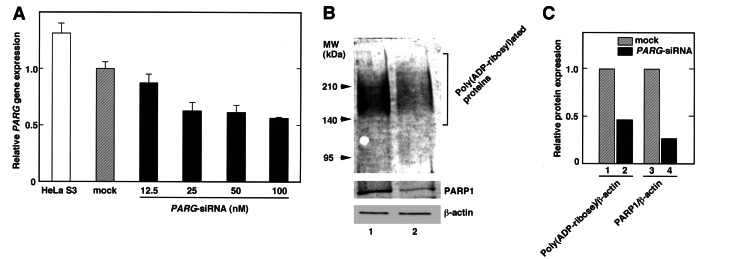
In order to suppress PARG gene expression, ON-TARGETplus SMARTpool siRNAs (PARG-siRNA) were transfected into HeLa S3 cells with DharmaFECT 1 reagent. The relative amounts of PARG transcripts were reduced by PARG-siRNA treatment (Fig. 1A). In 100 nM of PARG-siRNA-treated cells, the PARG gene expression level decreased to approximately half of that in the mock-transfected cells. Since this treatment did not affect viability of cells compared with the mock-transfected cells (data not shown), further experiments were performed with 100 nM of PARG-siRNA.
Figure 1.
The effect of knockdown of the PARG gene by introducing siRNA into HeLa S3 cells. (A) Quantitative real-time PCR analysis was performed with total RNAs isolated from HeLa S3 cells after 48 h of transfection with 0 (mock), 12.5, 25, 50 or 100 nM of PARG-siRNA. Results show the relative expression of the PARG gene compared to the GAPDH gene. The data are shown as the means ± SEM of three independent experiments. (B) HeLa S3 cells were transfected with 0 or 100 nM of PARG-siRNA (lanes 1 and 2, respectively). After 48 h of incubation, proteins were extracted from cells and separated by SDS-PAGE, then western blotting was performed with antibodies against poly(ADP-ribose), PARP1 and β-actin (upper, middle and lower panels, respectively). (C) Each band was quantified and results show relative PAR/β-actin and PARP1/β-actin expression ratios compared with those of the mock-transfected cells.
Treatment of PARG-siRNA reduces the amounts of poly(ADP-ribose) and PARP1
As the PARG gene encodes the main poly(ADP-ribose) degrading enzyme, the level of poly(ADP-ribosyl)ated proteins was assumed to increase following the introduction of PARG-siRNA into HeLa S3 cells. Western blotting revealed that the amounts of poly(ADP-ribose) decreased in the PARG-siRNA-transfected HeLa S3 cells (Fig. 1B). In addition, the decrease of poly(ADP-ribose) in the whole-cell extracts was accompanied by a decrease in PARP1 protein levels (Fig. 1C), suggesting that the PARG-siRNA diminished poly(ADP-ribose) and the PARP1 protein.
PARP1 gene expression and its promoter activity are downregulated by transfection with PARG-siRNA
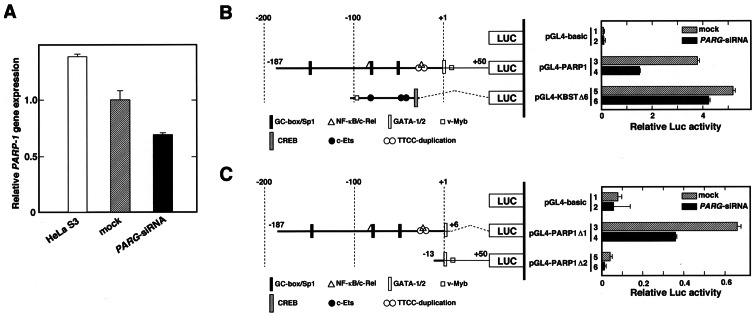
To examine whether the decrease in PARP1 protein level is caused by changes in gene expression and promoter activity, RT-qPCR analyses and transient transfection experiments were performed. As shown in Fig. 2A, the relative PARP1 gene expression of PARG-siRNA-transfected cells was 60% that of the control (mock) cells. In addition, the transient transfection and Luc reporter assay indicated that the PARP1 promoter activity decreased in the PARG-siRNA-transfected cells compared to the control cells (Fig. 2B). We also tested whether the 75-bp core promoter of the PARG gene (21) responds to PARG-siRNA treatment (Fig. 2B, bars 5 and 6), but only a 10% decrease was observed. To limit the PARG-siRNA responsive element(s), we constructed two deletion plasmids that contained −187 to +6 and −13 to +50 bp of the human PARP1 promoter (Fig. 2C). The results showed that the 193-bp fragment, which contains the GC-box/Sp1, NF-κB/c-Rel, GATA-1 and GATA-2 binding sequences, responded to the PARG-siRNA. The promoter activity obtained from pGL4-PARP1Δ2-transfected cells was lower than that of the pGL4-basic vector-transfected cells, indicating that the 63-bp sequence from −13 to +50 is not essential for transcription of the PARP1 gene. These results suggest that the transfection of PARG-siRNA leads to suppression of PARP1 gene expression through the 193-bp sequence of the PARP1 promoter region.
Figure 2.
PARG knockdown leads to downregulation of PARP1 gene expression and its promoter activity. (A) HeLa S3 cells were transfected with 100 nM of the PARG-siRNA. After 48 h of incubation, total RNAs were isolated and subjected to real-time qPCR analysis with PARP1- or GAPDH-specific primer pairs. Histograms show relative expression of the PARP1 gene compared to the GAPDH gene. The data are shown as the means ± SEM of three independent experiments. (B) HeLa S3 cells were transfected with 100 nM of the PARG-siRNA. After 48 h of incubation, cells were transfected with Luc-reporter plasmids, pGL4-basic (bars 1 and 2), pGL4-PARP1 (bars 3 and 4), and pGL4-KBSTΔ6 (bars 5 and 6). After a further 24 h of incubation, cells were harvested and a dual Luc assay was carried out. (C) Similar experiments were carried out by transfecting pGL4-basic (bars 1 and 2), pGL4-PARP1Δ1 (bars 3 and 4), and pGL4-PARP1Δ2 (bars 5 and 6). (B and C) Histograms show the relative Luc activities compared to cells transfected with the pGL3-promoter vector. The data are shown as the means + SEM of three independent experiments.
Viability of PARG siRNA-introduced HeLa S3 cells following N-methyl-N′ nitro-N-nitrosoguanidine (MNNG) and STS treatments
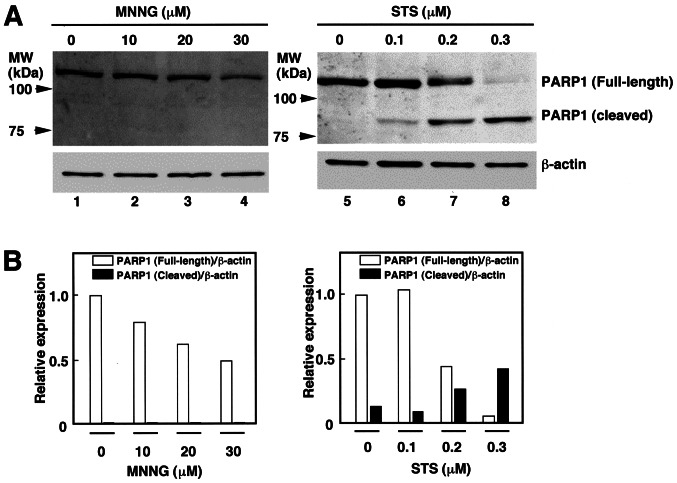
It has been demonstrated that poly(ADP-ribose) metabolism is associated with cell death or apoptosis (13–15). To examine the effects of MNNG and STS on PARP1 in HeLa S3 cells, western blot analysis was performed (Fig. 3). The treatment with STS (0.1–0.3 μM) induced cleavage of PARP1, suggesting that caspases were activated and cells underwent apoptosis. Although the relative amount of PARP1 was diminished to 50%, cleaved forms of PARP1 were not detected by the treatment with 30 μM of MNNG (Fig. 3).
Figure 3.
Effect of N-methyl-N′ nitro-N-nitrosoguanidine (MNNG) and staurosporine (STS) on PARP1 cleavage in HeLa S3 cells. (A) HeLa S3 cells (1×106) were treated with 10, 20 and 30 μM of MNNG (lanes 2, 3 and 4, respectively), or 0.1, 0.2 and 0.3 μM of STS (lanes 6, 7 and 8). After 24 h of incubation, cells were harvested, and whole protein extract was prepared with RIPA buffer, which was then subjected to SDS-PAGE. Western blotting was performed with antibodies against PARP1 (upper panels) and β-actin (lower panels). Lanes 1 and 5 represent untreated control cells. (B) Signal intensities of full-length (open columns) and cleaved-form (closed columns) PARP1 were quantified and normalized to β-actin. Histograms show the relative protein level compared to that of the full-length PARP1/β-actin ratio estimated from control cells.
Subsequently, the effect of MNNG and STS on cell viability was analyzed following the transfection of PARG-siRNA. Although 40 μM of MNNG treatment caused severe damage to the cells, transfection of PARG-siRNA did not further increase cell death (Fig. 4A). On the other hand, PARG-siRNA strongly impaired cell viability when cells were treated with 0.3 and 0.4 μM of STS (Fig. 4B). These results suggest that PARG-siRNA treatment enhances cell death in conditions that cause PARP1 cleavage.
Figure 4.
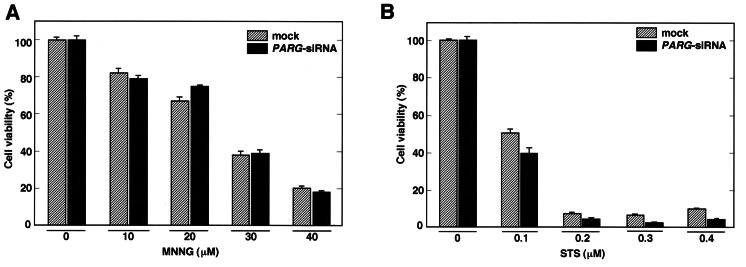
Effect of PARG knockdown on the sensitivity to cell death-inducing reagents. After 48 h of transfection with PARG-siRNA (100 nM), (A) MNNG or (B) STS was added to the culture medium. After a further 24 h of incubation, an MTS assay was carried out. The data are shown as the means ± SEM of three independent experiments. MNNG, N-methyl-N’ nitro-N-nitrosoguanidine; STS, staurosporine.
Discussion
Several experiments concerning expression of the PARG gene that have utilized PARG-siRNA or PARG-shRNA overexpressing systems have been reported (27). Constitutive suppression of PARG gene expression by introducing shRNA into HeLa S3 cells caused accumulation of poly(ADP-ribose) and led to enhancement of mitotic catastrophe by X-ray irradiation (28). It was reported that poly(ADP-ribose) accumulation occurred after a PARG shRNA expression vector was stably introduced into A549 cells, a lung adenocarcinoma cell line (29). Of note, these PARG-suppressed A549 cells exhibited reduced PARP activity. The decrease in poly(ADP-ribose) was reported in mouse 3T3 embryonic fibroblasts, PARG-Δ2,3 cells, which express PARG60 but not full-length PARG110 (30). In the present study, a PARG-siRNA pool that contains sequences located upstream of exon 5 was transfected into HeLa S3 cells. Therefore, PARG-siRNA used here may selectively reduce full-length PARG expression but may not affect PARG60. In addition, the suppressing effect by the introduction of 25–100 nM of the PARG-siRNA was approximately 50% of the control (Fig. 1A). This low efficiency might be caused by the reduced poly(ADP-ribose) level in the cells (Fig. 1B).
It has been suggested that poly(ADP-ribosyl)ation of Sp1 impairs activation of the PARP1 promoter (16). It has also been reported that PARP1 binds to its own promoter to suppress transcription (31). Since PARP1 physically interacts with Sp1 (16), both may cooperatively regulate transcription of the PARP1 gene. In our study, the introduction of PARG-siRNA into HeLa S3 cells caused suppression of the PARP1 promoter activity and led to subsequent reduction of its gene/protein expression. These data suggest a possible mechanism that prevents cells from accumulating excess poly(ADP-ribose) by suppressing PARP1 promoter activity. It is noteworthy that inverted repeats or putative cruciform-like structures are found both in the PARP1(31) and PARG(21) promoters. Since PARP1 has been suggested to recognize and bind to the cruciform-like structures of DNA (31), the PARG promoter is possibly one of the targets for PARP1. Moreover, the PARP1 promoter harbors overlapped TTCC motifs as 5′-GCGGGTT CCGTGGGCGTTCCCGCGG-3′ (Fig. 2B and C). The duplicated GGAA (TTCC) motifs, which are also found in the human PARG promoter region (21), have been suggested to be one of the determinants of TSS in TATA-less promoters with responses to various stimuli, including cytokine and differentiation inducing signals (32–34). In addition, previous reports suggested that PARP1 and PARG interact with each other (14) and coordinately regulate global patterns of gene expression, and that the binding of PARP1 to the promoters in the genome is dependent on the presence of PARG (27). Collectively, these observations indicate that the location of PARP1 with PARG on these promoters may play an important role in the regulation of poly(ADP-ribose) metabolism at the transcriptional level. This might be relevant to the cooperative function of PARP1 and PARG to repair single-strand break of DNA (35).
In this study, we showed that the introduction of PARG-siRNA enhances sensitivity to STS but not to MNNG (Fig. 4). Since PARP1 cleavage was detected in STS-treated cells (Fig. 3), the introduction of PARG-siRNA may cause elevation of cell damage by reducing PARP1 expression and lead to complete loss of native PARP1, and induce cell death by apoptosis. On the other hand, cell death induced by MNNG was not further affected by the PARG-siRNA, suggesting that the signals induced by the reduction of PARP1 have been adequately saturated by the MNNG treatment. However, this possibility remains to be elucidated in future analyses.
In conclusion, the data presented in this study provide a new theory that PARP1 gene expression is regulated by the amount of PARG transcripts, suggesting a mechanism to avoid accumulation of excess amounts of poly(ADP-ribose) in a cell by regulating their transcription. Since poly(ADP-ribose) metabolism also controls NAD+ levels, local changes in the concentrations of NAD+ might affect transcription of the PARP1 and PARG genes. Markedly, in accordance with the development of PARG inhibitors (36), the use of the PARG-siRNA may contribute to a treatment of cancer with a reduced dose of anti-cancer drugs or ionizing radiation.
Acknowledgements
The authors thank T. Sato and A. Asanuma for their technical assistance. This study was supported in part by a Research Fellowship from the Research Center for RNA Science, RIST, Tokyo University of Science.
References
- 1.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 2.Tanuma S, Kanai Y. Poly(ADP-ribosyl)ation of chromosomal proteins in the HeLa S3 cell cycle. J Biol Chem. 1982;257:6565–6570. [PubMed] [Google Scholar]
- 3.Lönn U, Lönn S. Accumulation of 10-kilobase DNA replication intermediates in cells treated with 3-aminobenzamide. Proc Natl Acad Sci USA. 1985;82:104–108. doi: 10.1073/pnas.82.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283:593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- 5.Kreimeyer A, Wielckens K, Adamietz P, Hilz H. DNA repair-associated ADP-ribosylation in vivo. Modification of histone H1 differs from that of the principal acceptor proteins. J Biol Chem. 1984;259:890–896. [PubMed] [Google Scholar]
- 6.Tanuma S, Yagi T, Johnson GS. Endogenous ADP ribosylation of high mobility group proteins 1 and 2 and histone H1 following DNA damage in intact cells. Arch Biochem Biophys. 1985;237:38–42. doi: 10.1016/0003-9861(85)90251-6. [DOI] [PubMed] [Google Scholar]
- 7.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 8.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 9.d’Adda di Fagagna F, Hande MP, Tong WM, Lansdorp PM, Wang ZQ, Jackson SP. Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat Genet. 1999;23:76–80. doi: 10.1038/12680. [DOI] [PubMed] [Google Scholar]
- 10.Zampieri M, Passananti C, Calabrese R, Perilli M, Corbi N, De Cave F, Guastafierro T, Bacalini MG, Reale A, Amicosante G, Calabrese L, Zlatanova J, Caiafa P. Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PloS One. 2009;4:e4717. doi: 10.1371/journal.pone.0004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simbulan-Rosenthal CM, Rosenthal DS, Hilz H, Hickey R, Malkas L, Applegren N, Wu Y, Bers G, Smulson ME. The expression of poly(ADP-ribose) polymerase during differentiation-linked DNA replication reveals that it is a component of the multiprotein DNA replication complex. Biochemistry. 1996;35:11622–11633. doi: 10.1021/bi953010z. [DOI] [PubMed] [Google Scholar]
- 12.Uchiumi F, Ikeda D, Tanuma S. Changes in the activities and gene expressions of poly(ADP-ribose) glycohydrolases during the differentiation of human promyelocytic leukemia cell line HL-60. Biochim Biophys Acta. 2004;1676:1–11. doi: 10.1016/j.bbaexp.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keil C, Gröbe T, Oei SL. MNNG-induced cell death is controlled by interactions between PARP-1, poly(ADP-ribose) glycohydrolase, and XRCC1. J Biol Chem. 2006;281:34394–34405. doi: 10.1074/jbc.M606470200. [DOI] [PubMed] [Google Scholar]
- 15.García S, Gómez-Reino JJ, Conde C. Poly(ADP-ribose) polymerase suppression protects rheumatoid synovial fibroblast from Fas-induced apoptosis. Rheumatology (Oxford) 2009;48:483–489. doi: 10.1093/rheumatology/ken502. [DOI] [PubMed] [Google Scholar]
- 16.Zaniolo K, Desnoyers S, Leclerc S, Guérin SL. Regulation of poly(ADP-ribose) polymerase-1 (PARP-1) gene expression through the post-translational modification of Sp1: a nuclear target protein of PARP-1. BMC Mol Biol. 2007;8:96. doi: 10.1186/1471-2199-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oei SL, Griesenbeck J, Schweiger M, Babich V, Kropotov A, Tomilin N. Interaction of the transcription factor YY1 with human poly(ADP-ribosyl) transferase. Biophem Biophys Res Commun. 1997;240:109–111. doi: 10.1006/bbrc.1997.7621. [DOI] [PubMed] [Google Scholar]
- 18.Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-kappaB transcriptional activation. Biol Chem. 1999;380:953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama Y, Kawamoto T, Mitsuuchi Y, Kurosaki T, Toda K, Ushiro H, Terashima M, Sumimoto H, Kuribayashi I, Yamamoto Y, Maeda T, Ikeda H, Sagara Y, Shizuta Y. Human poly(ADP-ribose) polymerase gene. Cloning of the promoter region. Eur J Biochem. 1990;194:521–526. doi: 10.1111/j.1432-1033.1990.tb15647.x. [DOI] [PubMed] [Google Scholar]
- 20.Meyer RG, Meyer-Ficca ML, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase (PARG) gene and the common promoter sequence it shares with inner mitochondrial membrane translocase 23 (TIM23) Gene. 2003;314:181–190. doi: 10.1016/s0378-1119(03)00738-8. [DOI] [PubMed] [Google Scholar]
- 21.Uchiumi F, Sakakibara G, Sato J, Tanuma S. Characterization of the promoter region of the human PARGgene and its response to PU.1 during differentiation of HL-60 cells. Genes Cells. 2008;13:1229–1248. doi: 10.1111/j.1365-2443.2008.01240.x. [DOI] [PubMed] [Google Scholar]
- 22.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 23.Benjamin RC, Gill DM. ADP-ribosylation in mammalian cell ghosts. Dependence of poly(ADP-ribose) synthesis on strand breakage in DNA. J Biol Chem. 1980;255:10493–10501. [PubMed] [Google Scholar]
- 24.Zhou B, Ikejima T, Watanabe T, Iwakoshi K, Idei Y, Tanuma S, Uchiumi F. The effect of 2-deoxy-D-glucose on Werner syndrome RecQ helicase gene. FEBS Lett. 2009;583:1331–1336. doi: 10.1016/j.febslet.2009.03.033. [DOI] [PubMed] [Google Scholar]
- 25.Uchiumi F, Enokida K, Shiraishi T, Masumi A, Tanuma S. Characterization of the promoter region of the human IGHMBP2(Sμbp-2) gene and its response to TPA in HL-60 cells. Gene. 2010;463:8–17. doi: 10.1016/j.gene.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 26.Uchiumi F, Watanabe T, Tanuma S. Characterization of various promoter regions of the human DNA helicase-encoding genes and identification of duplicated ets(GGAA) motifs as an essential transcription regulatory element. Exp Cell Res. 2010;316:1523–1534. doi: 10.1016/j.yexcr.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, Sauve AA, Kraus WL. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009;284:33926–33938. doi: 10.1074/jbc.M109.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amé JC, Fouquerel E, Gauthier LR, Biard D, Boussin FD, Dantzer F, de Murcia G, Schreiber V. Radiation-induced mitotic catastrophe in PARG-deficient cells. J Cell Sci. 2009;122:1990–2002. doi: 10.1242/jcs.039115. [DOI] [PubMed] [Google Scholar]
- 29.Erdélyi K, Bai P, Kovács I, Szabó E, Mocsár G, Kakuk A, Szabó C, Gergely P, Virág L. Dual role of poly(ADP-ribose) glycohydrolase in the regulation of cell death in oxidatively stressed A549 cells. FASEB J. 2009;23:3553–3563. doi: 10.1096/fj.09-133264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao H, Coyle DL, Meyer-Ficca M, Meyer RG, Jacobson EL, Wang ZQ, Jacobson MK. Altered poly(ADP-ribose) metabolism impairs cellular responses to genotoxic stress in a hypomorphic mutant poly(ADP-ribose) glycohydrolase. Exp Cell Res. 2007;313:984–996. doi: 10.1016/j.yexcr.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 31.Soldatenkov VA, Chasovskikh S, Potaman VN, Trofimova I, Smulson ME, Dritschilo A. Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J Biol Chem. 2002;277:665–670. doi: 10.1074/jbc.M108551200. [DOI] [PubMed] [Google Scholar]
- 32.Uchiumi F, Miyazaki S, Tanuma S. The possible functions of duplicated ets(GGAA) motifs located near transcription start sites of various human genes. Cell Mol Life Sci. 2011;68:2039–2051. doi: 10.1007/s00018-011-0674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchiumi F, Larsen S, Tanuma S. Biological systems that control transcription of DNA repair-and telomere maintenance-associated genes. In: Chen C, editor. DNA Repair-New Research Directions. InTech-Open Access Publisher, Inc; Rijeka, Croatia: (In press) [Google Scholar]
- 34.Uchiumi F, Larsen S, Masumi A, Tanuma S. Introduction to Sequence and Genome Analysis. iConcept Press Ltd; Hong Kong: The implications of duplicated GGAA-motifs located in the human interferon regulated genes (ISGs) (iConcept ed) (In press) [Google Scholar]
- 35.Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okita N, Ashizawa D, Ohta R, Abe H, Tanuma S. Discovery of novel poly(ADP-ribose) glycohydrolase inhibitors by a quantitative assay system using dot-blot with anti-poly(ADP-ribose) Biochem Biophys Res Commun. 2010;392:485–489. doi: 10.1016/j.bbrc.2010.01.044. [DOI] [PubMed] [Google Scholar]