Abstract
Background
Infections caused by methicillin-resistant Staphylococcus aureus (MRSA) in Thailand occur most frequently in healthcare facilities. However, reports of community-associated MRSA are limited.
Methods
We characterized 14 MRSA isolates from outpatients (O-1 to O-14) by phenotypic and genotypic methods and compared them with 5 isolates from inpatients (I-1 to I-5). Thai MRSA isolates from a healthcare worker (N-1) and a pig (P-1) were also included as ST9 MRSA strains from other sources.
Results
All MRSA isolates from the outpatients and inpatients were multidrug-resistant (resistant to ≥3 classes of antimicrobials). All of them except strains O-2 and I-3 carried type III SCCmec and belonged to agrI, coagulase IV, spa type t037 or t233, which related to ST239. The strain O-2 (JCSC6690) carried type IX SCCmec and belonged to agrII, coagulaseXIc, spa type t337 and ST9, whereas the strain I-3 carried a type III SCCmec and belonged to ST1429. Nucleotide sequence determination revealed that the type IX SCCmec element in strain O-2 was distinct from that in a Thai ST398 strain (JCSC6943) previously identified in 2011; nucleotide identities of ccrA and ccrB were 93 and 91%, respectively and several open reading frames (ORFs) at the joining regions were different. PCR experiments suggested that strain O-2 and N-1 carried similar SCCmec element, whereas that of strain P-1 was different, suggesting that distinct ST9-MRSA–IX clones might be spreading in this province.
Conclusions
The SCCmecIX-ST9 MRSA clones of distinct SCCmec subtypes might have emerged in the Thai community and might also have disseminated into the hospital.
Keywords: S. aureus, CA-MRSA, SCCmec, ST9, Livestock
Background
Staphylococcus aureus is an important human pathogen that causes a broad spectrum of infections, from mild to life-threatening. The organism becomes methicillin-resistant Staphylococcus aureus (MRSA) by acquiring the mecA gene. Expression of the mecA results in production of a special penicillin-binding protein, PBP2a, which has reduced affinity to β-lactam antibiotics [1] and confers resistance to all practically used β-lactam antimicrobials. The mecA gene is usually carried on a mobile genetic element called the staphylococcal cassette chromosome mec (SCCmec), which contains two main parts: the ccr gene complex (ccr) and the mec gene complex (mec). The ccr gene complex comprises ccr gene (s), ccrA and ccrB, or ccrC, which encode site-specific recombinases responsible for the mobility of SCCmec, and surrounding open reading frames (ORFs). The mec gene complex comprises the mecA gene, its regulatory genes, and insertion sequences upstream or downstream of the mecA gene [1]. SCCmec elements identified in S. aureus have been classified according to the combination of ccr allotypes with the mec gene complex: 11 types (I-XI) have been reported to date [2,3].
MRSA strains were regarded to be pathogens that cause hospital-acquired (HA) infections; however, MRSA infections in the community have been increasingly reported [4,5]. MRSA strains that colonized or caused infections in persons who meet the following criteria are considered to be CA-MRSA: healthy persons; person with no prior history of a healthcare-associated risk factors such as a recently (within the past year) hospitalized or had a medical procedure (such as dialysis, surgery, catheters) [5,6]. Recently, MRSA strains from livestock (e.g., pigs) or their products have emerged throughout Europe, America and Asia. Most of livestock-associated MRSA (LA-MRSA) strains belonged to clonal complex (CC) 398 as defined by multilocus sequence typing [7], although the other clones, e.g., CC5, CC9, CC30, and CC97, have been occurred [8].
Most of MRSA infection in Thailand was hospital-associated whereas CA-MRSA infection was very rare [9]. The hospital-associated MRSA strains usually carried types III or II SCCmec elements [10,11], while characteristics of CA-MRSA strains were not fully investigated. We aimed to characterize CA-MRSA strains disseminating in the Thai community compared with HA-MRSA strains from a teaching hospital in Thailand. Thai MRSA strains from other sources such as hospital staff, pig were also included and compared to a previously identified type IX SCCmec MRSA strain from a Thai veterinarian [12].
Methods
Bacterial strains
MRSA isolates (O-1 to O-14) from 14 outpatients at Srinagarind Hospital, Khon Kaen University, Thailand, were collected between September 2005 and March 2006: 11 isolates from skin or tissue infection samples and 3 from respiratory tracts. These strains met the category of CA-MRSA since they were isolated from samples collected from the outpatient, who did not admit to hospitals within a year. Five MRSA strains (I-1 to I-5) isolated from inpatients >72 hours after admission were also used as HA-MRSA control strains. In addition, 2 MRSA strains with type IX SCCmec (N-1 and P-1) were examined to investigate distribution of MRSA clone in the Thai community: the N-1 strain (isolated from a nasal swab of a nurse) was chosen from 19 S. aureus strains isolated during personnel surveillance in early 2006: the P-1 strain (from a pig with pneumonia) was chosen from 4 S. aureus strains collected at the Veterinary Research and Development Center of Upper Northeastern Thailand in 2006.
All isolates were identified by conventional coagulase and DNase tests. All of the MRSA strains from people were taken as part of standard care.
This study was conducted in accordance with the declaration of Helsinki and good clinical practice. It was approved by the Ethics Committee of Khon Kaen University (project number HE551393).
Antimicrobial susceptibility test
The minimum inhibitory concentrations of 7 antimicrobials (vancomycin, cefazolin, oxacillin, cefoxitin, tetracycline, erythromycin and ofloxacin) (Sigma Chemical, St. Louis, USA) were determined using the agar dilution method described by the Clinical and Laboratory Standards Institute [13]. S. aureus ATCC29213 was used as a drug-susceptible standard strain.
Multiplex PCR for SCCmec type
Chromosomal DNA was extracted using the DNeasy tissue Kit (Qiagen, Valencia, USA). Multiplex PCR for detection of the mecA, ccr gene complex type and mec gene complex class were performed using primer sets and conditions as per Kondo et al. [14]. MLEP (mec left extremity polymorphism) typing was performed to estimate the insertion of SCCHg as per Ito et al. [15]. The S. aureus strains used as reference strains for SCCmec typing were: NCTC10442 (type I), N315 (type II), 85/2082 (type III), MR108 (type IV), JCSC3624 (type V), and HDE288 (type VI).
PCR identification for heavy metal resistance genes and potential virulence genes
The mer operon and copA genes and genes encoding for virulent factors such as exfoliative toxin type a, b, d (eta, etb, etd), PVL (lukS-PV, lukF-PV) and TSST (tst) were detected by PCR methods [15-17].
Molecular typing
Coagulase and agr typing was performed as per Sakai et al. [18] and Shopsin et al. [19], respectively. Typing of spa was performed as per Shopsin et al. [20]. The PCR products of the spa genes were then purified using High Pure PCR Product Purification kits (Roche Diagnostics, Indianapolis, USA) and used as DNA templates for sequencing by ABI PRISM 3100 (Applied Biosystems, Foster City, USA). The designation of the spa type was conducted using the RidomStaph Type program (http://www.ridom.de).
Pulsed-field gel electrophoresis (PFGE) was conducted as a method for genotyping. The MSSA strain NCTC8325 and MRSA strain N315 were used as reference strains for PFGE. The analysis of SmaI-digested chromosomal DNA was performed using a contour-clamped homogeneous electric field mapper system as per the manufacturer’s instructions (Bio-Rad, Hercules, USA), with a 48.5-kb ladder (Bio-Rad) as DNA-sized markers. The PFGE patterns for all of the isolates were analyzed using BioNumerics software (version 6.5; Applied Maths, Kortrijk, Belgium) and interpreted as per Tenover et al. [21].
Multilocus sequence typing (MLST) was conducted as per Enright et al. [22]. The alleles of the 7 loci were evaluated by comparing the sequences to those of the corresponding loci in the S. aureus MLST database (http://www.mlst.net). The sequence types (STs) were determined according to the combined patterns of the 7 alleles, while the clonal complexes were defined using the BURST program, based on related sequence types available on the MLST website.
Determination of nucleotide sequences in and around the SCCmec element and coa gene
We determined the entire nucleotide sequences of type IX SCCmec carried by strain O-2 (JCSC6690) and nucleotide sequences of coa genes that could not be identified by PCRs using DNAs of three strains, O-2, N-1, and P-1. The DNA fragments of the SCCmec element from the strain O-2 and its genetics surrounding were amplified using long-range PCRs (list of primers sets in Additional file 1: Table S1). The region from Tn916 to copA gene was amplified by using a constructed DNA template from the Fosmid Library Production Kit (Epicentre Biotechnologies, Madison, WI). The nucleotide sequences were determined by primer-walking. The sequences were analyzed for ORFs with the In Silico Molecular Cloning Program (IMC Load command) and compared with the sequence databases from the National Center for Biotechnology Information using the basic local alignment search tool, BLAST (National Library of Medicine, Bethesda, MD) for annotation and prediction of functions. The nucleotide sequences of the coa gene from strains O-2, N-1 and P-1 were determined as per Watanabe et al. [23].
PCR identification of ORFs at J3 regions of type IX SCCmec elements
The carriage of ORFs at the J3 region of SCCmecIX elements was examined using chromosomal DNAs of strains N-1 and P-1 with three sets of primers (sets a and b as used for the strain O-2 and set c as used for the strain JCSC6943) [12]. The locations and nucleotide sequences of the primers used are indicated in Figure 1 and Additional file 1: Table S1, respectively.
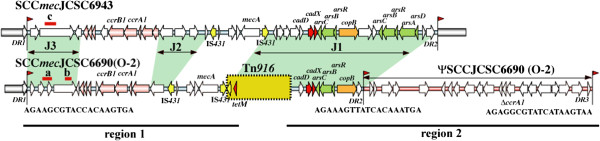
Figure 1.
Structural comparison of type IX SCCmec elements carried by strains O-2 and JCSC6943. Structures of SCCmec are illustrated based on the nucleotide sequences deposited in databases DDBJ/EMBL/GenBank, under accession nos. AB505628 for JCSC6943, AB705452 (region 1, indicated by black bar) and AB705453 (region 2, indicated by black bar) for O-2. The red arrowhead indicates the location of ISS, and the nucleotide sequences are given below. ORFs are colored as follows: yellow, insertion sequences; red, cad operons; lawn green, genes for arsenical resistance operons; orange, copB; salmon, ORFs for ccr gene complexes. The three joining regions (J1, J2, and J3 regions) are indicated in green. The locations of DNA fragments amplified with three pairs of primers (a, b, and c) are indicated in red bars. Nucleotide sequences of primers used for amplification for three loci are listed in Additional file 1: Table S1.
Nucleotide sequence accession numbers
The respective nucleotide sequences of the two chromosomal regions containing the SCCmec element in strain O-2 were deposited in the DDBJ/EMBL/GenBank database under accession numbers AB705452 and AB705453. The nucleotide sequences of three coa genes were deposited in the DDBJ/EMBL/GenBank database under accession numbers AB742446 (strain O-2), AB742447 (strain N-1) and AB787218 (strain P-1).
Results
Characteristics of MRSA isolates
All outpatient and inpatient MRSA strains were multidrug-resistant with high MIC values to cefazolin, oxacillin, cefoxitin, tetracycline, erythromycin and ofloxacin (32 - >64 μg/ml). However, all of them were still susceptible to vancomycin.
None of the MRSA isolates carried the virulence-related genes tested. Thirteen out of 14 outpatient isolates and all 5 inpatient isolates carried a type 3 ccr gene and class A mec gene complex, although 12 out of 13 outpatient isolates and 4 out of 5 inpatient isolates also carried the ccrC. Further PCR experiments showed that these 16 strains contained mercury resistance gene (mer) and the extremity of SCCHg, therefore, they were identified as type III SCCmec strains carrying SCCHg[24]. In conclusion, 18 out of 19 strains carried type III SCCmec. The remaining strain, O-2, carried ccrA1B1 genes and class C2 mec gene complex, which was categorized to type IX SCCmec.
All isolates from both sources were of the agr group I and coagulase type IV except for strain O-2 that belonged to agr group II. Since the coagulase type of strain O-2 could not be determined neither by a genetic method using multiplex PCRs nor by serological typing based on a coagulation inhibition, its coagulase nucleotide sequences were determined, and classified into type XIc, a novel coagulase subtype. The majority of type III SCCmec strains belonged to spa type t037 except for strains O-3 and I-3, which belonged to spa type t233 and nontypeable, respectively. No amplification product of spa-typing was seen when used chromosomal DNA of strain I-3 as DNA templates. MLST genotypes of 5 spa t037 isolates, which were chosen based on their unique PFGE banding patterns, were identified as the ST239, whereas those of strain O-3 and I-3 were ST239 and ST1429 respectively. The strain O-2 belonged to spa type t337 and ST9. The characteristics of strain O-2 were distinct from other 18 strains.
The PFGE profiles revealed that 12 out of 14 outpatient isolates showed similar banding patterns with 1–2 different bands, while strains O-2 and O-3 had a distinct DNA profile (Figure 2). Three of the five inpatient isolates showed closely related PFGE patterns and strain I-3 belonging to ST1429 presented a distinct pattern, whereas the band pattern of strain I-5 was related to the outpatient strains (Figure 2).
Figure 2.
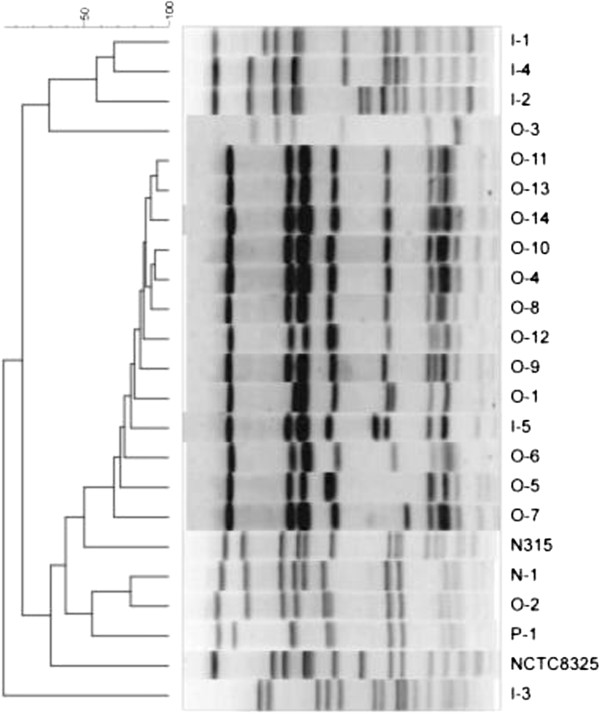
Dendrographic analysis of Pulsed-field Gel Electrophoresis banding patterns of SmaI-digested chromosomal DNAs of MRSA strains from human and a pig. PFGE patterns of 14 MRSA strains from outpatients (O-1 to O-14), 5 MRSA strains from inpatients (I-1 to I-5), an MRSA strain from a healthcare worker (N-1) and an MRSA strain from a pig (P-1) were compared. Curve-based phylogenetic tree using Ranked Pearson Correlation was generated using BioNewmerics.
Structure of type IX SCCmec in strain O-2 (SCCmecO-2)
The nucleotide sequences of the two long regions containing SCCmec element in the strain O-2 were determined. The first 25 kb region contained a class C2 mec gene complex, a type 1 ccr gene complex and the downstream end of a conjugative transposon, Tn916. The second 32 kb region started from the upstream region of Tn916 and ended at the extremity of an SCC. The overall structure of the approximately 70-kb region from the strain O-2 containing the SCCmec element and an SCC without ccr gene (ΨSCC) is illustrated in Figure 1 and the ORFs locating in these regions are listed in Table 1.
Table 1.
ORFs of SCCmec in the S. aureus strain O-2 compared to S. aureus JCSC6943 and S. haemolyticus JCSC1435
| ORFs | Genes | Description of products | % identity to type IX SCC mec in S. aureus JCSC6943a | % identity to S. haemolyticus JCSC1435 |
|---|---|---|---|---|
| KK01 |
orfx |
hypothetical protein |
97 |
80 |
| KK02 |
|
hypothetical protein, similar to modification methylase LlaFI |
<50 |
51.8 |
| KK03 |
|
hypothetical protein, similar to[truncated]modification methylase LlaFI |
<50 |
50.3 |
| KK04 |
|
hypothetical protein, similar to restriction enzyme LlaFI |
<50 |
<50 |
| KK05 |
|
hypothetical protein |
52 |
83 |
| KK06 |
|
hypothetical protein |
65 |
86.3 |
| KK07 |
|
hypothetical protein |
78 |
61.9 |
| KK08 |
|
hypothetical protein |
85 |
83 |
| KK09 |
|
hypothetical protein |
<50 |
51.6 |
| KK10 |
ccrB1 |
cassette chromosome recombinase B1 |
91 |
50 |
| KK11 |
ccrA1 |
cassette chromosome recombinase A1 |
93 |
<50 |
| KK12 |
|
hypothetical protein |
81 |
51.1 |
| KK13 |
|
hypothetical protein |
61 |
<50 |
| KK14 |
|
hypothetical protein |
95 |
<50 |
| KK15 |
tnp |
IS431 transposase |
97.5 |
99.1 |
| KK16 |
|
[truncated]putative HMG-CoAsynthase |
100 |
83 |
| KK17 |
|
Glycerophosphoryl diester phosphodiesterase |
100 |
100 |
| KK18 |
|
hypothetical protein |
100 |
100 |
| KK19 |
mecA |
penicillin-binding protein 2' |
100 |
100 |
| KK20 |
tnp |
transposase for IS431 |
100 |
98 |
| KK21 |
tnp |
Tn916, transposase |
<50 |
<50 |
| KK22 |
|
Tn916, hypothetical protein |
<50 |
<50 |
| KK23 |
|
Tn916, transcriptional regulator |
<50 |
50 |
| KK24 |
tetM |
tetracycline resistance protein TetM |
<50 |
<50 |
| KK25 |
|
hypothetical protein |
<50 |
52.5 |
| KK26 |
cadD |
cadmium binding protein |
96 |
94 |
| KK27 |
cadX |
cadmium resistant accessory protein |
96 |
77 |
| KK28 |
arsC |
arsenate reductase |
93 |
82 |
| KK29 |
arsB |
arsenic efflux pump protein |
88 |
73 |
| KK30 |
arsR |
arsenical resistance operon repressor |
100 |
97 |
| KK31 |
copA |
copper-exporting ATPase |
98 |
67 |
| KK32 | putative lipoprotein | 92 | 98 |
Three integration site sequences (ISS) comprising directly repeated (DR) sequences were identified: the first ISS, at the 3′ end of orfX; the second ISS, at the downstream of the copB gene on the 32-kb fragment; and the third ISS, at the end of the 32 kb fragment (Figure 1). The SCCmecO-2 was estimated to be 43 kb in size, in which the Tn916 was integrated. At the downstream of the SCCmec, an SCC encoding truncated ccrA1 had 83% nucleotide identity with that of type I SCCmec in strain NCTC10442.
We then compared the structure of SCCmecO-2 to a previously reported type IX SCCmec of strain JCSC6943 [12]. The class C2 mec gene complexes of both strains were highly identical, four ORFs around the mecA gene of the strain O-2 had 100% identity with SCCmecJCSC6943. The ccr gene complexes of both strains were located upstream to the mec gene complex. The nucleotide sequences of the ccrA1B1 in SCCmecO-2 showed 91–93% identities with those of SCCmecJCSC6943 and 91–92% identities with those of the type I SCCmec in strain NCTC10442.
The ORFs at J3 region of SCCmecO-2 which encoded for hypothetical proteins related to modification methylase and restriction enzymes were less similar to those of the JCSC6943 (Table 1). At the J1 region of both SCCmec elements, the ORFs encoding resistance genes for heavy metals (e.g., cadmium and copper) were present. But a truncated copy of Tn916 encoding tetracycline resistance gene (tetM) was identified only in the SCCmecO-2 (Table 1).
Comparisons of type IX SCCmec MRSA isolates
The characteristic of type IX SCCmec MRSA strains, N-1 and P-1, were compared with strain O-2. All 3 strains belonged to the same agr type and ST type (Table 2). Nucleotide sequences of coa genes of these three strains were exactly identical, which were classified into type XIc. However, their spa types were not totally identical. Strain P-1 belonged to spa type t044, whereas strains N-1 and O-2 belonged to the same spa type t337. These three isolates showed closely related PFGE patterns with 2–4 different bands (Figure 2).
Table 2.
Characteristics of MRSA strains tested in this study
| Isolate no.a | Origin of specimen | Disease |
Minimum inhibitory concentrationb(μg/mL) |
|
ccr gene type |
mec gene complex class |
mer operon |
copA |
agr type |
Coagulase type |
spa type |
MLST (ST) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Van | Cefa | Oxa | Cefox | Tetra | Erythro | Ofx | |||||||||||
| Isolates from outpatients (n = 14) | |||||||||||||||||
| O-1 |
Pus Left thigh (Human) |
Accident, infected wound |
2 |
> 64 |
> 64 |
> 64 |
32 |
> 64 |
> 64 |
ccrA3B3 |
A |
- |
- |
I |
IV |
t037 |
ST239 |
| O-2 |
Pus Leftfoot (Human) |
Atopic dermatitis |
1 |
32 |
32 |
32 |
> 64 |
> 64 |
> 64 |
ccrA1B1 |
C2 |
- |
+ |
II |
XIc |
t337 |
ST9 |
| O-3 |
Wound (Human) |
Electrical burn |
2 |
> 64 |
> 64 |
> 64 |
64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t233 |
ST239 |
| O-4 |
Pus (Human) |
NA |
2 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
NT |
| O-5 |
Pus (Human) |
Infected wound |
1 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
+ |
I |
IV |
t037 |
NT |
| O-6 |
Tracheal suction (Human) |
NA |
2 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
ST239 |
| O-7 |
Sputum (Human) |
Pulmonary TB |
2 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
NT |
| O-8 |
Wound (Human) |
Accident, head injury |
2 |
> 64 |
> 64 |
> 64 |
64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
NT |
| O-9 |
Wound (Human) |
Accident, infected wound |
2 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
NT |
| O-10 |
Pus Left cheek (Human) |
NA |
1 |
> 64 |
> 64 |
> 64 |
64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
NT |
| O-11 |
Pus scalp (Human) |
Chronic scalp impetigo |
2 |
> 64 |
> 64 |
> 64 |
64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
NT |
| O-12 |
Tissue (Human) |
Gas gangrene |
2 |
> 64 |
> 64 |
> 64 |
32 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
NT |
| O-13 |
Sputum (Human) |
NA |
2 |
> 64 |
> 64 |
> 64 |
64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
NT |
| O-14 |
Pus Lt thigh (Human) |
Accident, infected wound |
2 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
NT |
| Isolates from in patients (n = 5) | |||||||||||||||||
| I-1 |
Blood (Human) |
NA |
2 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
ccrA3B3 |
A |
+ |
- |
I |
IV |
t037 |
NT |
| I-2 |
Blood (Human) |
NA |
1 |
> 64 |
> 64 |
> 64 |
> 64 |
16 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
NT |
| I-3 |
Blood (Human) |
NA |
1 |
> 64 |
> 64 |
> 64 |
8 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
+ |
I |
IV |
UDc |
ST1429 |
| I-4 |
Sputum (Human) |
NA |
2 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
+ |
- |
I |
IV |
t037 |
ST239 |
| I-5 |
Sputum (Human) |
NA |
2 |
> 64 |
> 64 |
> 64 |
32 |
> 64 |
> 64 |
ccrA3B3ccrC |
A |
- |
- |
I |
IV |
t037 |
ST239 |
| An isolate from healthy worker (n = 1) | |||||||||||||||||
| N-1 |
Nasal swab (Human) |
colonize |
1 |
> 64 |
64 |
32 |
64 |
1 |
> 64 |
ccrA1B1 |
C2 |
- |
+ |
II |
XIc |
t337 |
ST9 |
| An isolate from pig (n = 1) | |||||||||||||||||
| P-1 | Lung (pig) | pneumonia | 1 | > 64 | > 64 | 64 | > 64 | 64 | > 64 | ccrA1B1 | C2 | - | + | II | XIc | t044 | ST9 |
a O, outpatient; I, inpatient; N, nurse; P, pig; NA, information was not available.
b Van, vancomycin; Cefa, cefazolin; Oxa, oxacillin; Cefox, cefoxitin; Tetra, tetracycline; Erythro, erythromycin; Ofx, ofloxacin.
C ND,Undetermined; because no PCR product was amplified.
The carriage of genes in strains N-1 and P-1 examined by using primer’s pairs (a, b and c) showed that strains O-2 and N-1 were positive in PCRs using primer pairs a and b, while strain P-1 was negative with all three primer’s pairs. Although strains N-1 and P-1 were positive in PCRs identifying tetM and copB genes, the locations of these genes were not examined yet.
Discussion
Characteristics of type IX SCCmec in strain O-2
The SCCmecO-2, which was located on the downstream region of the orfX, was classified into type IX. It had type I ccr gene complex carrying ccrA and ccrB genes that were classified into ccrA1 and ccrB1 based on the criteria suggested by IWG-SCC [2]. The class C2 mec gene complex of SCCmecO-2 was very similar to that of SCCmecJCSC6943 [12], the ccr gene complex was located between orfX and mec gene complex. Furthermore, a number of ORFs encoding genes associated with heavy metal resistance (i.e., cadD, cadX, arsC, arsB, arsR and copB) were also found and showed 92-100% nucleotide identities to those of the SCCmecJCSC6943. However, a number of ORFs were absent in the SCCmecJCSC6943, e.g., hypothetical proteins related to restriction-modification system at J3 region and a conjugative transposon Tn916 encoding tetM at J1 region. The transposon Tn916 was located just downstream of the class C2 mec gene complex and the extremity of Tn916 including int gene was truncated. The transposition of Tn916 was not site-specific. We presume that Tn916 transposed firstly into the SCC element, then the class C2 mec gene complex was integrated to the SCC harboring Tn916 by disrupting its extremity.
The class C2 mec gene complex was formerly reported predominantly in S. haemolyticus[1]. Comparison of the ORFs of SCCmecO-2 and those of S. haemolyticus strain JCSC1435 revealed that their heavy metal-resistant genes (e.g., cadD, cadX, arsC, arsB, arsR, copB) had 67-97% identities. Three ORFs at the J3 region had more than 80% identities with those of the S. haemolyticus strain JCSC1435 [25]. These data suggested that although the SCCmecO-2 and SCCmecJCSC6943 were classified into type IX, they might have evolved independently. Further study will clarify whether any staphylococcal strains such as the S. haemolyticus, might serve as reservoirs for SCC or SCCmec elements (Table 1).
MRSA clones in Thailand
MRSA infections in the hospitals have been a big concern in Thailand. So far the HA-MRSA strains have been analyzed, ST239-SCCmecIII clone was the major, although ST5-SCCmecII clone was identified as the minor one [10,11]. On the other hand, CA-MRSA in Thailand was reported to be very rare [9]. The incidence of CA-MRSA was undetermined because most outpatients with non severe infections were treated empirically without collecting samples for culture [26]. Most of the MRSA isolates from outpatients in the present study were ST239-SCCmecIII-spa type t037, which was the dominant clone reported in Asian countries except for Korea and Japan [10]. CA-MRSA has been reported to be susceptible to a wide range of non-β-lactam antibiotics, low oxacillin MIC, but our tested strains were multidrug-resistant to non-β-lactam antimicrobials similar to HA-MRSA strains [27].
Our results were consistent with the report from ANSORP that ST239-SCCmecIII and ST5-SCCmecII MRSA clones were found among CA-MRSA isolates from patients in Asian countries (e.g., Vietnam, Thailand, Sri Lanka, Taiwan, and Hong Kong), without any risk factors for HA-MRSA infection [5]. The results of our PFGE experiment suggested that ST239–SCCmecIII strains disseminated in the community were rather clonal and distinct from those in the hospital. Further detailed study will be needed to clarify this hypothesis.
A novel MRSA clone in the Thai community
It has been reported that CA-MRSA strains tend to carry types IV or V SCCmec element and belonged to particular genotypes. The distribution of CA-MRSA clones varied by geographical area: the ST1-SCCmecIV and ST8-SCCmecIV strains (called as USA400 and USA300 based on PFGE patterns) in the USA; the CC80-SCCmecIV strains in Europe and Africa; ST22-SCCmecIV and SCCmecV- ST772 strains in India; ST30-SCCmecIV and ST93-SCCmecIV strains in Australia; and CC59-SCCmecV strains in Asia [28,29]. Interestingly, most of these CA-MRSA strains carried the Panton Valentine Leukocidin (PVL) gene that encodes a necrotizing cytotoxin, which may be responsible for the invasiveness and virulence of the organism [30]. Recently livestock associated MRSA (LA-MRSA) have emerged in the community and spread into hospitals [31-34]. The majority of LA-MRSA strains belonged to ST398, while strains belonged to other STs, e.g. ST9, has also been reported.
In this study, we analyzed three ST9 MRSA strains carrying SCCmecIX, the same SCCmec type identified in an ST398-MRSA isolated from a Thai veterinarian (strain JCSC6943) [12].
The strain O-2 was isolated in March 2006, from a young boy with underlying atopic dermatitis and suffering from chronic impetigo of the left foot. He had not been admitted to hospital in the prior year. Therefore, we regarded the strains as CA-MRSA and as a novel MRSA clone emerged in the Thai community. We explored the clone and identified 2 strains, one (strain P-1) from a diseased pig with pneumonia in the same province and the other (strain N-1) from a nasal swab of a nurse isolated during hospital-personnel surveillance in the same year as the first case. The ST9-MSSA has occasionally been isolated from healthy human carriers [35] and has been able to colonize and be transmitted between humans and pigs as it has also been isolated from pigs and workers on pig farms [36,37]. Recently ST9-SCCmecIX strains have been isolated from swine in Thailand [38,39]. In the present study, 2 isolates of ST9-SCCmecIX were from humans. To our knowledge, this is the first report of ST9-SCCmecIX strain from humans (a patient and a healthy healthcare worker) and a diseased pig in Thailand. Unfortunately, information regarding any animal contact by these persons or their families was not available. The ST9-SCCmecIX strain would belong to a newly identified CA-MRSA clone disseminating in the Thai community, although no isolate harbored any potential virulence genes including the pvl gene, which is usually found in CA-MRSA strains.
The detailed comparison of 3 ST9-SCCmecIX strains: O-2, N-1, and P-1, suggested that strains isolated from human and pig were not exactly identical. Their PFGE patterns were not totally identical but they were closely related, in contrast, that of ST398 strain JCSC6943 showed a non-typeable pattern (absence of DNA fragment). The spa type of strains O-2 and N-1 belonged to t337, whereas strain P-1 belonged to t044. Recently, MRSA isolates from pigs and pork in Thailand carrying the SCCmecIX-spa type t337 have been reported [38,39]. The ST9-spa type t899 MRSA and MSSA were recently isolated from dust samples taken from Chinese pig farms [40]. Most of these isolates were resistant to various classes of antimicrobials such as ß-lactams, macrolides and tetracycline, which is consistent with the strains O-2, N-1 and P-1. However, those strains belonged to the SCCmec type III.
PCR identification of J regions in strains O-2 and N-1 suggested that they carried the same subtype of type IX SCCmec, which was distinct from that of strain P-1 and JCSC 6943. On the other hand, strain P-1 was assumed to carry a novel subtype of type IX SCCmec, since it was distinct from that of strains O-2 and JCSC6943. We presume that livestock, e.g., pigs, might be the reservoir of ST9-S. aureus in Asian countries. The ST9-S. aureus strain might become an MRSA by acquiring SCCmec elements from different sources, since ST9-SCCmecIII strains were identified in China [40]. Identification of several subtypes of type IX SCCmec elements suggested that such elements might come from elsewhere and integrated into ST9 strains. In this study, it could not be concluded that SCCmecIX strains from human were derived from pig, since strain P-1 was not identical to strains O-2 and N-1. However, SCCmecIX strains with the same spa type (t337) have been identified from pigs (38, 39). Further study of these isolates will clarify whether ST9 MRSA strains isolated from human might be derived from livestock and disseminated in the Thai community.
Conclusions
ST9-SCCmecIX MRSA strains from humans (a patient and a healthy healthcare worker) and a diseased pig in Thailand were reported as a newly identified CA-MRSA clone disseminating in the Thai community.
Competing interests
All authors declare that they have no competing interests.
Authors’ contributions
AL participated in study design, data collection and interpretation, drafted and revised the manuscript. TI contributed to study design and revised the manuscript. SSL, HX and MXX completed all data analyses. CE, AC, CW, and NJ participated in data collection and analyses. KH contributed to study design and revised the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Sequences of oligonucleotide primers for long range PCR.
Contributor Information
Aroonlug Lulitanond, Email: arolul@kku.ac.th.
Teruyo Ito, Email: teruybac@juntendo.ac.jp.
Shanshuang Li, Email: teruybac@juntendo.ac.jp.
Xiao Han, Email: hx2005@juntendo.ac.jp.
Xiao Xue Ma, Email: zjfcn@msn.com.
Chulapan Engchanil, Email: chulapan.engchanil@gmail.com.
Aroonwadee Chanawong, Email: aroonwad@kku.ac.th.
Chotechana Wilailuckana, Email: wchote@kku.ac.th.
Netchanok Jiwakanon, Email: nokjiw@hotmail.com.
Keiichi Hiramatsu, Email: khiram06@juntendo.ac.jp.
Acknowledgements
The authors thank (a) the CMDL, Faculty of Associated Medical Sciences, Khon Kaen University, for support (b) the staff of the Clinical Microbiology Laboratory at Srinagarind Hospital, Faculty of Medicine, Khon Kaen University for collecting the clinical isolates (c) Dr. Niyomsak Uppatum, Director of Veterinary Research and Development (Upper Northeastern) Center, Khon Kaen for providing the isolates from pigs and (d) Mr. Bryan Roderick Hamman and Mrs. Janice Loewen-Hamman for assistance with the English-language presentation.
Financial support
This work was supported by a Grant-in-Aid for 21st Century COE Research and Japan Society for the Promotion of Science. This study was supported in part by a Grant-in-Aid (C19590456) and Supported Program for the Strategic Research Foundation at Private Universities, 2012 – 2017 from MEXT (Ministry of Education, Culture, Sports, Science and Technology of Japan).
References
- Katayama Y, Ito T, Hiramatsu K. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob Agents Chemother. 2001;45(7):1955–1963. doi: 10.1128/AAC.45.7.1955-1963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53(12):4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlej A, Hryniewicz W, Empel J. Staphylococcal Cassette Chromosome mec (SCCmec) classification and typing methods: an overview. Pol J Microbiol. 2011;60(2):95–103. [PubMed] [Google Scholar]
- Deresinski S. Methicillin-resistant Staphylococcus aureus: an evolutionary, epidemiologic, and therapeutic odyssey. Clin Infect Dis. 2005;40(4):562–573. doi: 10.1086/427701. [DOI] [PubMed] [Google Scholar]
- Song JH, Hsueh PR, Chung DR, Ko KS, Kang CI, Peck KR, Yeom JS, Kim SW, Chang HH, Kim YS, Jung SI, Son JS, So TM, Lalitha MK, Yang Y, Huang SG, Wang H, Lu Q, Carlos CC, Pepera JA, Chiu CH, Liu JW, Chongthaleong A, Thamlikikul V, Van PH. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J Antimicrob Chemother. 2011;66(5):1061–1069. doi: 10.1093/jac/dkr024. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Methicillin-resistant Staphylococcus aureus (MRSA) infections. CDC 24/7: Saving Lives. 2011. serial online] [cited 2013 April 13]. Available from: URL: http://www.cdc.gov/mrsa/diagnosis/index.html.
- Wulf MWH, Markestein A, van der Linden FT, Voss A, Klaassen C, Verduin CM. First outbreak of methicillin-resistant Staphylococcus aureus ST398 in a Dutch Hospital, June 2007. Euro surveillance. 2008;13(9):1–2. [PubMed] [Google Scholar]
- Köck R, Schaumburg F, Mellmann A, Köksal M, Jurke A, Becker K, Friedrich AW. Livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) as causes of human infection and colonization in Germany. PLoS One. 2013;8(2):e55040. doi: 10.1371/journal.pone.0055040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekviwattanawong S, Srifuengfung S, Chokepaibulkit K, Lohsiriwat D, Thamlikitkul V. Epidemiology of Staphylococcus aureus infections and the prevalence of infection caused by community-acquired methicillin-resistant Staphylococcus aureus in hospitalized patients at Siriraj Hospital. J Med Assoc Thai. 2006;89(Suppl 5):S106–S117. [PubMed] [Google Scholar]
- Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, Jamklang M, Chavalit T, Song JH, Hiramatsu K. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother. 2006;50(3):1001–1012. doi: 10.1128/AAC.50.3.1001-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulitanond A, Chanawong A, Sribenjalux P, Wilailuckana C, Kaewkes W, Vorachit M, Ito T, Hiramatsu K. Preliminary report of SCCmec types and antimicrobial susceptibilities of methicillin-resistant Staphylococcus aureus isolates from a university hospital in Thailand. Southeast Asian J Trop Med Public Health. 2010;41(4):920–927. [PubMed] [Google Scholar]
- Li S, Skov RL, Han X, Larsen AR, Larsen J, Sørum M, Wulf M, Voss A, Hiramatsu K, Ito T. Novel types of staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2011;55(6):3046–3050. doi: 10.1128/AAC.01475-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 21st informational supplement M100-S21. Wayne, PA, USA: CLSI; 2011. [Google Scholar]
- Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51(1):264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45(5):1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XX, Ito T, Chongtrakool P, Hiramatsu K. Predominance of clones carrying Panton-Valentine leukocidin genes among methicillin-resistant Staphylococcus aureus strains isolated in Japanese hospitals from 1979 to 1985. J Clin Microbiol. 2006;44(12):4515–4527. doi: 10.1128/JCM.00985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Nishifuji K, Sasaki M, Fudaba Y, Aepfelbacher M, Takata T, Ohara M, Komatsuzawa H, Amagai M, Sugai M. Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect Immun. 2002;70(10):5835–5845. doi: 10.1128/IAI.70.10.5835-5845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai F, Takemoto A, Watanabe S, Aoyama K, Ohkubo T, Yanahira S, Igarashi H, Kozaki S, Hiramatsu K, Ito T. Multiplex PCRs for assignment of staphylocoagulase types and subtypes of type VI staphylocoagulase. J Microbiol Methods. 2008;75(2):312–317. doi: 10.1016/j.mimet.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Shopsin B, Mathema B, Alcabes P, Said-Salim B, Lina G, Matsuka A, Martinez J, Kreiswirth BN. Prevalence of agr specificity groups among Staphylococcus aureus strains colonizing children and their guardians. J Clin Microbiol. 2003;41(1):456–459. doi: 10.1128/JCM.41.1.456-459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopsin B, Gomez M, Montgomery SO, Smith DH, Waddington M, Dodge DE. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37(11):3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murry BE, Persing DH, Swaminathan B. Interpretating chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Ito T, Sasaki T, Li S, Uchiyama I, Kishii K, Kikuchi K, Skov RL, Hiramatsu K. Genetic diversity of staphylocoagulase genes (coa): insight into the evolution of variable chromosomal virulence factors in Staphylococcus aureus. PLoS One. 2009;4(5):e5714. doi: 10.1371/journal.pone.0005714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Mediavilla JR, Smyth DS, Chavda KD, Ionescu R, Roberts RB, Robinson DA, Kreiswirth BN. Identification of a novel transposon (Tn6072) and a truncated staphylococcal cassette chromosome mec element in methicillin-resistant Staphylococcus aureus ST239. Antimicrob Agents Chemother. 2010;54(8):3347–3354. doi: 10.1128/AAC.00001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui L. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol. 2005;187(21):7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangsathapornpong A. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging problem in children. J Infect Dis Antimicrob Agents. 2006;23:141–148. [Google Scholar]
- Chambers HF. The changing epidemiology of Staphylococcus aureus. Emerg Infect Dis. 2001;7(2):178–182. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybold U, Kourbatova EV, Johnson JG, Halvosa SJ, Wang YF, King MD, Ray SM, Blumberg HM. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of healthcare-associated blood stream infections. Clin Infect Dis. 2006;42(5):647–656. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- Udo EE, O'Brien FG, Al-Sweih N, Noronha B, Matthew B, Grubb WB. Genetic lineages of community-associated methicillin-resistant Staphylococcus aureus in Kuwait hospitals. J Clin Microbiol. 2008;46(10):3514–3516. doi: 10.1128/JCM.00966-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte W, Braulke C, Cuny C, Strommenge B, Werner G, Heuck D, Jappe U, Wendt C, Linde HJ, Harmsen D. Emergence of methicillin-resistant Staphylococcus aureus with Panton-Valentine leukocidin in central Europe. Eur J Clin Microbiol Infect Dis. 2005;24(1):1–5. doi: 10.1007/s10096-004-1262-x. [DOI] [PubMed] [Google Scholar]
- Weese JS, Van Duijkeren E. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet Microbiol. 2010;140(3–4):418–429. doi: 10.1016/j.vetmic.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Graveland H, Duim B, van Duijkeren E, Heederik D, Wagenaar JA. Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans. Inter J Med Microbiol. 2011;301(8):630–634. doi: 10.1016/j.ijmm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Lewis HC, Mølbak K, Reese C, Aarestrup FM, Selchau M, Sørum M, Skov RL. Pigs as source of methicillin-resistant Staphylococcus aureus CC398 infections in humans, Denmark. Emerg Infect Dis. 2008;14(9):1383–1389. doi: 10.3201/eid1409.071576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köck R, Harlizius J, Bressan N, Laerberg R, Wieler LH, Witte W, Deurenberg RH, Voss A, Becker K, Friedrich AW. Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus (MRSA) among pigs on German farms and import of livestock-related MRSA into hospitals. Eur J Clin Microbiol Infect Dis. 2009;28(11):1375–1382. doi: 10.1007/s10096-009-0795-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano C, Aspiroz C, Saenz Y, Ruiz-Garcia M, Royo-Garcia G, Gomez-Sanz E, Ruiz-Larrea F, Zarazaga M, Torres C. Genetic environment and location of the Inu(A) and Inu(B) genes in methicillin-resistant Staphylococcus aureus and other staphylococci of animal and human origin. J Antimicrob Chemother. 2012;67(12):2804–2808. doi: 10.1093/jac/dks320. [DOI] [PubMed] [Google Scholar]
- Bridge Kuehn MSJ. MRSA may move from livestock to humans. JAMA. 2012;308(17):1726. doi: 10.1001/jama.2012.14814. [DOI] [PubMed] [Google Scholar]
- Armand-Lefevre L, Ruimy R, Andremont A. Clonal comparison of Staphylococcus aureus isolated from healthy pig farmers, human controls, and pigs. Emerg Infect Dis. 2005;11(5):711–714. doi: 10.3201/eid1105.040866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard M, Cavaco LM, Sirichote P, Unahalekhaka A, Dangsakul W, Svendsen CA, Aarestrup FM, Hendriksen RS. SCCmec type IX element in methicillin resistant Staphylococcus aureus spa type t337 (CC9) isolated from pigs and pork in Thailand. Front Microbiol. 2012;3(103):1–4. doi: 10.3389/fmicb.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen J, Imanishi M, Hinjoy S, Tharavichitkul P, Duangsong K, Davis MF, Nelson KE, Larsen AR, Skov RL. Methicillin-resistant Staphylococcus aureus ST9 in pigs in Thailand. PLoS One. 2012;7(2):e31245. doi: 10.1371/journal.pone.0031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Li J, Hu C, Jin S, Li F, Guo Y, Ran L, Ma Y. Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J Antimicrob Chemother. 2009;64(4):680–683. doi: 10.1093/jac/dkp275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of oligonucleotide primers for long range PCR.