Summary
Shank family proteins (Shank1, Shank2, and Shank3) are synaptic scaffolding proteins that organize an extensive protein complex at the postsynaptic density (PSD) of excitatory glutamatergic synapses. Recent human genetic studies indicate that SHANK family genes (SHANK1, SHANK2, and SHANK3) are causative genes for idiopathic autism spectrum disorders (ASD). Neurobiological studies of Shank mutations in mice support a general hypothesis of synaptic dysfunction in the pathophysiology of ASD. However, the molecular diversity of SHANK family gene products, as well as the heterogeneity in human and mouse phenotypes, pose challenges to modeling human SHANK mutations. Here, we review the molecular genetics of SHANK mutations in human ASD and discuss recent findings where such mutations have been modeled in mice. Conserved features of synaptic dysfunction and corresponding behaviors in Shank mouse mutants may help dissect the pathophysiology of ASD, but also highlight divergent phenotypes that arise from different mutations in the same gene.
Introduction
In recent years, tremendous progress has been made in recognizing and diagnosing autism, a condition that was first described by Kanner and Asperger nearly 70 years ago (Asperger, 1944; Kanner, 1943; Volkmar et al., 2009). Clinically, autistic phenotypes are present in a group of heterogeneous conditions, termed autism spectrum disorders (ASD) (Lord et al., 2000a). Genetic risk contributes significantly to idiopathic ASD, but the specific genetic alterations remain elusive in the majority of cases (Abrahams and Geschwind, 2008; Folstein and Rosen-Sheidley, 2001; State, 2010b). Remarkably little is known about the underlying pathophysiology or neurological basis of ASD (Amaral et al., 2008; Courchesne et al., 2007; Geschwind and Levitt, 2007; Rubenstein, 2010; Zoghbi, 2003). The development of animal models is an important step in bridging the human genetics of ASD to circuit-based deficits underlying the clinical presentation, and ultimately to discovering, designing, and deploying effective therapeutic strategies.
SHANK/ProSAP family proteins (SHANK1, SHANK2, SHANK3) have emerged as promising candidates for modeling ASD in mice due to strong genetic evidence showing molecular defects of SHANK in patients with ASD (Berkel et al., 2010; Berkel et al., 2012; Durand et al., 2007; Gauthier et al., 2010; Marshall et al., 2008; Pinto et al., 2010; Sato et al., 2012). Mutations of SHANK3 were the first (Durand et al., 2007) and remain the best characterized SHANK mutations in human ASD (Boccuto et al., 2012; Moessner et al., 2007). Recently, mutations in SHANK1 and SHANK2 have also been associated with ASD (Berkel et al., 2010; Berkel et al., 2012; Sato et al., 2012), supporting a general function for this gene family in common molecular pathways associated with ASD. Shank family proteins are scaffolding proteins that organize a cytoskeleton-associated signaling complex at the postsynaptic density (PSD) of nearly all excitatory glutamatergic synapses in the mammalian brain (Grabrucker et al., 2011b; Gundelfinger et al., 2006; Kreienkamp, 2008; Sheng and Kim, 2000). The genetic association of ASD with SHANK family genes provided an immediate link between synaptic dysfunction and the pathophysiology of ASD. Shank1 mutant mice were first reported in 2008 (Hung et al., 2008). Recently, two Shank2 (Schmeisser et al., 2012; Won et al., 2012) and five Shank3 mutant mice have been reported (Bozdagi et al., 2010; Peca et al., 2011; Schmeisser et al., 2012; Wang et al., 2011). Analysis of these mutant mice has yielded a wealth of new information and raised numerous questions. Here we compare and contrast the various Shank mouse models with a focus on Shank3, discuss the potential relevance to human ASD, and highlight key challenges and opportunities in studying the role of SHANK genes in ASD.
Genetic Linkage of SHANK Genes to Autism Spectrum Disorders
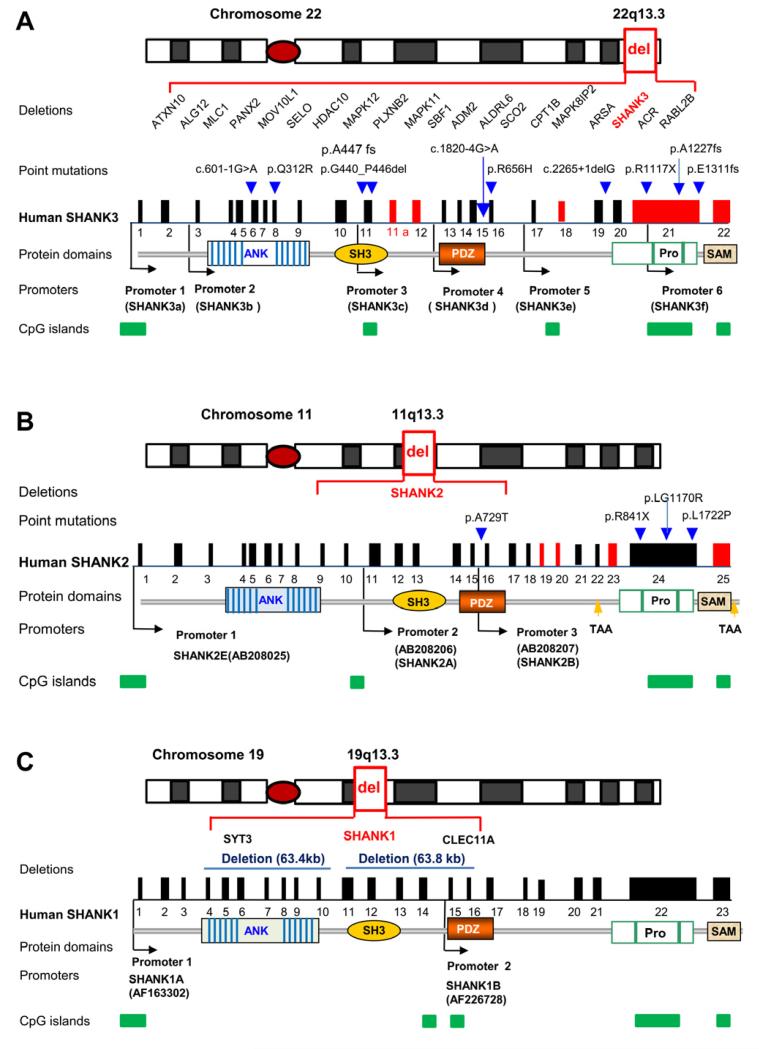
In humans, SHANK3 is one of the best characterized genes implicated in ASD. SHANK3 maps to the critical region of 22q13.3 deletion syndrome (Phelan-McDermid syndrome, PMS) (Figure 1A) (Wilson et al., 2003). The key clinical features associated with PMS are global developmental delay, hypotonia, absent or severely delayed language, autistic behaviors, and intellectual disability (Phelan, 2007). Atypical bipolar disorder has also been associated with 22q13.3 deletions in recent case reports (Denayer et al., 2012; Verhoeven et al., 2013; Verhoeven et al., 2012). The size of the deletions in PMS is extremely variable (0.1-10 Mb) (Dhar et al., 2010; Wilson et al., 2003), but deletions of SHANK3 have been reported in all cases except in one report of two children who have deletions proximal to SHANK3 (Wilson et al., 2008), suggesting that other genes in 22q13.3 may also be important for brain function. Much smaller deletions specific to SHANK3 or balanced translocations within the SHANK3 gene have been reported in patients with neurobehavioral features indistinguishable from patients with large deletions including SHANK3 (Anderlid et al., 2002; Bonaglia et al., 2005, 2006; Wong et al., 1997). These observations have led to the conclusion that haploinsufficiency of SHANK3 is a major contributor to the neurobehavioral features in 22q13.3 deletion patients. Subsequently, point mutations and microdeletions of SHANK3 have been identified in idiopathic ASD cases (Boccuto et al., 2012; Dhar et al., 2010; Durand et al., 2007; Gauthier et al., 2010; Gong et al., 2012; Marshall et al., 2008; Moessner et al., 2007; Waga et al., 2011). In all, six types of molecular defects have been identified in SHANK3 in more than 1000 human patients. These include: (1) cytogenetically visible terminal deletion of 22q13.3 or ring chromosome of 22 (Jeffries et al., 2005; Wilson et al., 2003), (2) a microdeletion detected by array-based methods (Boccuto et al., 2013; Dhar et al., 2010), (3) microduplication (Okamoto et al., 2007), (4) translocations with breakpoints within the SHANK3 gene (Bonaglia et al., 2005, 2006), (5) small intragenic deletions (Bonaglia et al., 2011), and (6) point mutations (Boccuto et al., 2012; Durand et al., 2007; Moessner et al., 2007).
Figure 1.
SHANK Mutations Causing Autism-Related Phenotypes
(A) SHANK3 is part of a large gene cluster associated with deletions in chromosome 22q13.3 deletion syndrome, or Phelan-McDermid syndrome, associated with autistic behaviors and intellectual disability (select genes are shown). The deletion sizes vary from 17 kb within SHANK3 to 10 Mb in 22q13.3. SHANK3 gene structure, mutations, and protein domains are shown. Human SHANK3 has 23 exons as deduced from cDNA AB569469 deposited in GenBank. Exon 11a is a newly identified exon. The positions of six identified promoters are indicated as black arrows. The exons in red are alternatively spliced. The positions of point mutations are indicated as blue arrows and the nature of point mutations are as described above the arrow. c.601-1G>A splicing mutation in intron 5 (Hamdan et al., 2011), p.Q312R in exon 8 (Moessner et al., 2007), p.G440_P446del and p.A447fs in exon 11 (Boccuto et al., 2012; Waga et al., 2011), c.1820-4 G>A splicing mutation in intron 15 (Boccuto et al., 2012), p.R656H in exon 16 (Waga et al., 2011), c.2265+1delG splicing mutation in intron 19 (Gauthier et al., 2009), and p.R1117X, p.A1227fs, p.E1311fs in exon 21 (Boccuto et al., 2012; Durand et al., 2007; Gauthier et al., 2010). Protein domains are shown and aligned to corresponding exons (ANK, ankyrin repeat domain; SH3, Src homology 3 domain; PDZ, PSD-95/Discs large/ZO-1 domain; Pro, a proline-rich region containing homer- and cortactin-binding sites; SAM, sterile alpha motif domain). CpG islands are sites of differential methylation and are indicated by green bars.
(B) SHANK2 gene structure, protein domains, isoforms, microdeletions and mutations in ASD. Exons in red are alternatively spliced exons. Microdeletions found in ASD are intragenic and within SHANK2 genomic region. Point mutations found in ASD are indicated as blue arrows and the nature of mutations are described above. Three identifiable promoters corresponding to SHANK2E, 2A, 2B are indicated as black arrow. Two alternative stop codons (TAA) are indicated as yellow arrows.
(C) SHANK1 gene structure, protein domains, isoforms, and microdeletions in ASD. Two small deletions including SHANK1 and two other genes are shown. No pathological point mutations in SHANK1 have been reported in ASD. The positions for two promoters are shown (Lim et al., 1999). The alternative splicing and isoforms have not been fully characterized. In (A)-(C), genomic distance and exons are not drawn to scale.
De novo sequence changes in SHANK3 including missense, frame shift, and splice site mutations have been reported in ASD patients (Boccuto et al., 2012; Durand et al., 2007; Gauthier et al., 2010; Gauthier et al., 2009; Gong et al., 2012; Hamdan et al., 2011; Moessner et al., 2007; Schaaf et al., 2011; Waga et al., 2011). The positions of these point mutations that are likely pathological are depicted in Figure 1A and the major clinical features extracted from case reports are summarized in Table 1. Point mutations affecting the splice acceptor site in intron 5 (Hamdan et al., 2011) and splice donor site of intron 19 (Gauthier et al., 2009), as well as a one base pair insertional mutation in exon 21 causing a frame shift (p.A1227fs) (Durand et al., 2007), were found in children with ASD and severe speech delay. The p.A447fs mutation was found in a child with atypical autism disorder and speech delay. This mutation was inherited from his father who also exhibits learning disability and attention deficit hyperactivity disorder (Boccuto et al., 2012). In contrast, a splice mutation of c1820-4G>A is associated with a child with mild ASD (Asperger syndrome) (Boccuto et al., 2012). Intellectual disability was mild in the patient with the intron 5 splicing mutation (Hamdan et al., 2011), severe in the patient with the p.A1227fs exon 21 mutation (Durand et al., 2007), and was not described in the patient with the intron 19 splice mutation (Gauthier et al., 2009). The p.E1331fs mutation, which is in the last coding exon close to stop codon, was found in children with pervasive developmental disorder not otherwise specified (PDD-NOS) and severe intellectual disability (Boccuto et al., 2012). Several small intragenic or interstitial deletions have also been reported (Bonaglia et al., 2011). Intragenic deletions of exons 1-9 or exons 1-17 of SHANK3 have been found in patients exhibiting severe language delay and significant intellectual disability, but a formal evaluation for ASD was not performed in these two cases (Bonaglia et al., 2011). Together, these data strongly support a conclusion that molecular defects of SHANK3 can cause ASD but with variable presentations. However, the frequency of putatively pathological mutations in SHANK3 appears to be rare in ASD (< 0.75%) (Moessner et al., 2007), and the degree to which SHANK3 mutations contribute to the population attributable risk of ASD is small (Betancur, 2011; Buxbaum, 2009; State, 2010b). Many missense mutations have been identified in SHANK3 (Gauthier et al., 2009; Moessner et al., 2007; Schaaf et al., 2011), but their clinical relevance has not been determined. Although the number of cases with point mutations that are clearly pathological in nature is still small, the clinical features from these cases reports suggest a genotype-phenotype correlation related to the autism diagnosis. Interestingly, point mutations, including a nonsense mutation in exon 21 (p.R1117X) of SHANK3 have been reported in families with schizophrenia and mild intellectual disability (Gauthier et al., 2010). This observation is consistent with recent reports that similar copy number variants (CNVs) are found across many genomic loci in both ASD and schizophrenia (Cook and Scherer, 2008; Gejman et al., 2011; Moreno-De-Luca et al., 2010; Sanders et al., 2011). Microduplications of SHANK3 have also been reported in children with developmental delay and dysmorphic features (Okamoto et al., 2007), suggesting that SHANK3 gene dosage affects brain function.
Table 1. Genotype/Phenotype Correlations of Human SHANK3 Mutations.
| Genetic defect | No. of case |
SHANK3 isoforms affected |
Other genes disrupted |
ASD related diagnosis |
Intellectual disability (ID) |
Other clinical features |
||
|---|---|---|---|---|---|---|---|---|
|
22q13.3 deletion (including SHANK3) (0.1-10Mb) |
>1000 | All isoforms disrupted |
From 2- 30 other genes |
ASD diagnosis in >75% of cases |
>95% cases with developmental delay, moderate to severe ID, absent speech or severe speech delay |
Hypotonia, seizure, motor development delay, facial dysmorphism, increased pain threshold, bipolar disorder, mild congenital anomaly |
||
| Microdeletion of SHANK3 | 3 | All isoforms disrupted |
None or /ACR |
ASD | Speech delay and mild ID |
Hyperactivity, hypospadias, Behavioral issues Seizure/regression |
||
|
Intragenic deletion |
Deletion size |
Exons/ domain deleted |
||||||
| 38kb | exon 1-9 /ANK |
1 | SHANK3a/b | none | Not mentioned | Moderate ID, Profound delayed in language acquisition |
Microcephaly. Astigmatism |
|
| 74kb | exon 1-17 ANK/SH3/ PDZ |
1 | SHANK3a-e | none | Not evaluated | Profound ID | Mild congenital anomalies |
|
| 44kb | Exon 19-23 Homer binding/ SAM |
1 | SHANK3f | ACR | ASD | Moderate ID/hyperactivity disorder |
Short stature/facial dysmorphism/astigma tism |
|
| 27kb | exon 20-23 Homer binding/ SAM |
1 | SHANK3f | ACR# | Classical autism |
Not mentioned | ADHD, no facial dysmorphism |
|
| 17kb | Exon 23 SAM |
1 | SHANK3f | ACR | No ASD by CARS and ABC |
Mild ID, severe delayed in language acquisition |
Mild facial dysmorphism, mild motor delay |
|
| Point mutation/ indel |
Exon/prote
in domain |
1 | ||||||
| c.601 - 1 G>A |
ANK | 1 | SHANK3a-b | None | No ASD | Mild ID, severe language impairment |
||
| p.Q312R* | Exon 8 ANK |
1 | SHANK3a-b | None | ASD by ADI-R and ADOS |
language delay | Abnormal EEG but no seizures. Has self injurious behavior |
|
| p.A447fs^ | Exon 11 SH3 |
1 | SHANK3a-c | None | Borderline score for ASD evaluation |
Language delay | No facial dysmorphism |
|
| p.G440 P 446 del |
Exon 11 SH3 |
1 | SHNK3a-c | None | ASD | Severe ID | Delayed psychomotor development |
|
| c.1820- 4G>A |
PDZ | 1 | SHANK3a-d | None | Asperger’s syndrome |
Normal speech and some behavioral problems |
Facial dysmorphism/mild congenital anomaly |
|
| p.R656H& | Exon 16 PDZ |
1 | SHANK3a-d | None | ASD | Mild ID, development delay |
||
| c.2265+1 del G |
1 | SHANK3a-e | None | ASD | Not mentioned | |||
| p.R1117X | Exon 21 Homer binding | 1 | SHANK3f | None | No evidence for ASD |
Mild to moderate ID | Schizophrenia Hyperactivity/no facial dysmorphism |
|
| p.A1227 fs |
Exon 21 Homer- binding |
1 | SHANK3f | None | ASD | Severe ID and impaired speech |
||
| p.E1311 fs | Exon 21 Homer- binding |
1 | SHANK3f | None | PDD-NOS | Severe ID and absent speech |
Seizure, facial dysmorphism, motor development delay |
|
parents are first cousin.
proband has a chromosome 9p24.3 copy number gain that is inherited from mother and considered a benign variant.
variant is also found in healthy and normal father.
the variant is inherited from father who has learning disability and attention deficit disorder.
The information in the Table is extracted from following reports (Boccuto et al., 2012; Bonaglia et al., 2011; Dhar et al., 2010; Durand et al., 2007; Gauthier et al., 2010; Gauthier et al., 2009; Moessner et al., 2007; Phelan, 2007; Sarasua et al., 2011; Waga et al., 2011; Wilson et al., 2003; Wong et al., 1997). ACR: Acrosin, a sperm specific proteinase which has no known function in brain; ABC, autism behavioral checklist; ADHD, attention deficithyperactivity disorder; ADI-R, Autism diagnosis interview-revised; ADOS, autism diagnosis observation scale; CARS, Child autism rating scale; Del, deletion; EEG, Electroencephalography; ID, intellectual disability. PDD-NOS, pervasive developmental disorder-otherwise not specified. N/A not
More recently, point mutations of SHANK2 and microdeletions of SHANK1 and SHANK2 have been found in patients with ASD and intellectual disability (Figures 1B and 1C) (Berkel et al., 2010; Leblond et al., 2012; O’Roak et al., 2012b; Pinto et al., 2010; Sato et al., 2012). Compared to SHANK3, the number of cases with SHANK2 mutations is small but convincing. All microdeletions found in ASD are intragenic deletions that disrupt the SHANK2 protein. A nonsense mutation in SHANK2 exon 24 encoding the proline-rich homer-binding domain has also been found in an ASD proband (Figure 1B) (Berkel et al., 2010). Mutations of SHANK3 at similar location were also found in ASD (Boccuto et al., 2012; Durand et al., 2007). In the case of SHANK1, microdeletions including SHANK1 and two other adjacent genes were reported in five ASD individuals in two families with mild ASD (Figure 1C). Pathological point mutations have not yet been reported in ASD so far (Sato et al., 2012)
To date, correlation between genotype and phenotype has been described in patients with 22q13.3 deletions including SHANK3 (Sarasua et al., 2011). In most reports, clinical features were described by self-reporting or extracted from existing medical records. A summary of the molecular and clinical finding related to SHANK3 variants is provided in Table 1 based on the available data from individual reports in the literature (Boccuto et al., 2012; Bonaglia et al., 2011; Dhar et al., 2010; Gauthier et al., 2010; Gauthier et al., 2009; Moessner et al., 2007; Philippe et al., 2008; Sarasua et al., 2011; Waga et al., 2011; Wilson et al., 2003). Importantly, the quality of clinical data varies and is often incomplete in these reports and thus direct comparisons should be made with caution. Diagnostic evaluations for ASD were not conducted in the majority of cases or were carried out using different evaluation protocols. Because the clinical data are not complete in most cases, it is difficult to draw firm conclusions about the relationship between specific SHANK3 variants and clinical features related to ASD. This challenge is best illustrated in three cases with very similar mutations (p.A1227fs, p.E1311fs, and p.R1117X) in exon 21 encoding Homer binding site of SHANK3 (Figure 1A). Mutations p.A1227fs and p.E1331fs were found in patients with ASD or PDD-NOS, severe language delay, and significant intellectual disability (Boccuto et al., 2012; Durand et al., 2007), while p.R1117X was found in patients with schizophrenia and mild intellectual disability (Gauthier et al., 2010). Similarly, in other cases with almost identical small microdeletions (<100 kb) including SHANK3, neurobehavioral phenotypes were quite variable (Boccuto et al., 2013; Bonaglia et al., 2011; Dhar et al., 2010). One hypothesis to explain these differences is the presence of a genetic or epigenetic variant in the other allele of SHANK3, or haploinsufficiency and positional effects of deletions on other genes known to cause autosomal recessive neurological disorders in the 22q13.3 region. For example, genes implicated in metachromatic leukodystrophy (ARSA), congenital disorders of glycosylation (ALG12), and spinocerebellar ataxia type 10 (ATXN10) are mapped within the 22q13.3 region. In addition, mutations or allelic variation in as yet unidentified genes that function as epistatic modifiers for SHANK3 could influence the phenotypes associated with SHANK3 defects.
In the cases of SHANK1 and SHANK2 mutations associated with ASD, no studies relating genotype and phenotype have been reported. The penetrance of SHANK2 mutations in ASD is not complete in some cases (Leblond et al., 2012). This observation has led to the proposal of a multiple hit model to explain the clinical relevance of SHANK2 mutations. Because the number of ASD cases with two genetic hits including SHANK2 is small, the validity of the model remains to be tested in additional patient cohorts or by functional studies. Interestingly, microdeletion of SHANK1 is only penetrant in males with mild ASD in families studied (Sato et al., 2012). The molecular basis for gender-specific penetrance related to SHANK1 mutations is not immediately clear but may provide an opportunity to investigate the mechanism underlying higher male gender-specific risk in ASD.
Deletions involving entire SHANK family genes in ASD predict that haploinsufficiency is the primary mechanism underlying ASD pathogenesis (Wilson et al., 2003). By comparison, for point mutations such as missense mutation and small intragenic deletions identified in SHANK2 and SHANK3, the pathogenic mechanism is less clear (Bonaglia et al., 2011; Durand et al., 2011; Moessner et al., 2007). The possibility of gain-of-function or dominant negative effects in addition to loss-of-function should be considered if mutated mRNA transcripts produce stable mutated or truncated proteins. Different SHANK mutations may thus act through different mechanisms to alter protein-protein interactions at the PSD and cause synaptic dysfunction that may underlie clinical presentations of disorder. However, to date, we have little information on the molecular mechanisms by which more subtle mutations in SHANK3 alter protein function at synapses (Durand et al., 2007; Durand et al., 2011).
SHANK Encodes a Multidomain Scaffolding Protein Present at Glutamatergic Synapses
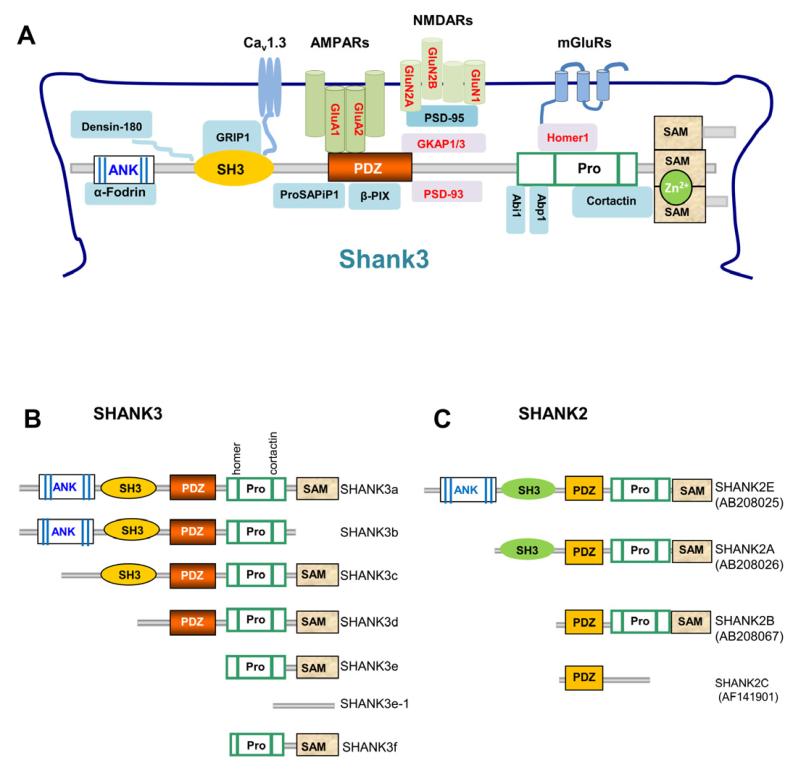
Shank/ProSAP family members including Shank3 have five conserved protein domains – an ankyrin repeat domain (ANK), Src homology 3 (SH3) domain, PSD-95/Discs large/ZO-1 (PDZ) domain, a proline-rich region containing homer- and cortactin-binding sites, and a sterile alpha motif (SAM) domain (Figure 2A). Shanks are scaffolding proteins that interact with many synaptic proteins in the PSD (Ehlers, 1999; Gundelfinger et al., 2006; Kreienkamp, 2008; Sheng and Kim, 2000). More than 30 synaptic proteins have been reported to interact with Shank family proteins (Figure 2 and Table 2). Due to the similarity of protein domains among Shank family proteins, in vitro binding experiments have shown a significant overlap in protein-protein interactions involving Shank1-3. Shank3-interacting proteins include receptors, ion channels, cytoskeletal proteins, scaffolding proteins, enzymes, and signaling molecules (Grabrucker et al., 2011b; Kreienkamp, 2008). The large protein complex organized by Shanks performs a variety of functions at the postsynaptic membrane including actin-based cytoskeletal remodeling, synapse formation, AMPA receptor endocytosis, and regulation of synaptic transmission and plasticity (Table 2). Whether all these protein-protein interactions occur in vivo are unknown and the precise function for these interactions remains to be fully elucidated.
Figure 2.
Shank Protein Interactions and Isoform-Specific Domain Structure
(A) Schematic of the partial Shank protein interactome at the PSD with Shank3 as a model. A more complete list of Shank family interacting proteins is shown in Table 2. Protein domains in Shank family members are similar. Many interacting proteins interact with all three Shank family proteins (Shank1, Shank2, and Shank3) in in vitro assays. The proteins in red font are altered in Shank3 mutant mice.
(B) Diagram of SHANK3 protein isoforms SHANK3a-f. Protein domain structure was deduced from confirmed mRNAs expressed from different promoters in human and mouse brains. Polypeptides have not been validated due to the lack of isoform-specific antibodies. Pro, proline rich region.
(C) Diagram of SHANK2 protein isoforms of SHANK2A, 2B, 2C, and 2E [modified from (Leblond et al., 2012)]. SHANK2C has an alternative stop codon due to the alternative splicing of exons 19 and 20 as shown in Figure 1B.
Table 2. SHANK Family Interacting Proteins.
| Domain | Direct Partner | Function | Select References |
|---|---|---|---|
| ANK | α-fodrin, sharpin | Actin-based cytoskeleton, dendritic spine development |
Bockers et al., 2001; Lim et al., 2001 |
| SH3 | GRIP1, Cav1.3, Densin-180 | AMPA receptor trafficking, CREB, Wnt signaling pathways, dendritic spine remodeling, interaction with CaMKII, |
Quitsch et al., 2005; Sala et al., 2001; Sheng and Kim, 2000; Uemura et al., 2004; Zhang et al., 2005 |
| PDZ | GKAP1(SAPAP1), GKAP3(SAPAP3), PSD-93, mGluR1/5, AMPA receptors, β-PIX, ProSAPiP1, LASPER1, ProSAPiP2, PLC-β3 |
Cytoskeleton, dendritic spine formation and remodeling, synaptic transmission and plasticity, self- multimeirzation |
Gundelfinger et al., 2006; Kreienkamp, 2008; Liebau et al., 2009; Wendholt et al., 2006 |
| Proline- rich/Homer and Cortactin binding |
Homer1, Dynamin-2, Abp1, Abi1, Cortactin, IRSp53 |
Arp 2/3 complex, regulation of actin-based cytoskeleton, dendritic spine formation and remodeling , synaptic transmission and plasticity, AMPA receptor endocytosis, |
Haeckel et al., 2008; Lu et al., 2007; Okamoto et al., 2001; Proepper et al., 2007; Qualmann et al., 2004; Soltau et al., 2004; Soltau et al., 2002; Tu et al., 1999 |
| SAM | Shank3 | Synaptic targeting, self- multimerization, Zn2+ binding |
Baron et al., 2006; Boeckers et al., 2005; Naisbitt et al., 1999 |
| SHANK3 full length protein |
ACTN2, CLU, GKAP1, GKAP3, HNRNPC, LZTS3, PICK1, SYNGAP1 |
Shared interactions with PSD- 95. No functional studies described |
Sakai et al., 2011 |
ANK, ankyrin repeat domain; SH3, Src homology 3 domain; PDZ, PSD-95/Discs large/ZO-1 domain; Pro, a proline-rich region containing homer- and cortactin-binding sites; SAM, sterile alpha motif domain; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
Concentrated at glutamatergic synapses, Shanks interact directly or indirectly with all major types of glutamate receptors – NMDA receptors, AMPA receptors, and mGluRs – via different domains (Ehlers, 1999; Naisbitt et al., 1999; Tu et al., 1999; Uchino et al., 2006; Verpelli et al., 2011). When overexpressed in cultured neurons from mice, Shanks recruit GluA1 AMPA receptors and increases the formation of new synapses (Roussignol et al., 2005). Expression of Shank3 with deletions of various domains in cultured mouse neurons has demonstrated distinct roles for each domain in dendritic spine development (Roussignol et al., 2005). For example, mutation of the PDZ domain of Shank3 results in a reduction in dendritic spine formation while mutation of ANK-SH3 domains leads to spines with normal length but reduced spine head area. In contrast, mutation in the cortactin binding site results in longer spines with reduced spine head area (Roussignol et al., 2005).
At present, it is unclear how the interactions of Shanks with various glutamate receptor subtypes are coordinated and regulated at a given synapse. In cultured rat neurons, knock-down of Shank3 selectively reduces synaptic mGluR5 receptor and impairs mGluR5-dependent signaling and plasticity (Verpelli et al., 2011). Whether similar deficits are present in vivo is not yet clear, although excessive mGluR5 signaling has been implicated in fragile X syndrome (Krueger and Bear, 2011), which has clinical overlap with 22q13.3 deletions (Phelan, 2008). Among the various Shank binding proteins, Homer family members have been shown to regulate diverse synaptic functions (Hayashi et al., 2009; Sala et al., 2001; Tu et al., 1999). Homer1 and Shank1 form a mesh-like matrix that is thought to function as an organizing lattice for PSD proteins (Hayashi et al., 2009). Shank3 shares very similar protein domain structure to Shank1, suggesting that Shank3 participates in a similar protein network with Homer1. As with interactions involving glutamate receptors, it is not yet known how the multitude of Shank interactions with other scaffolding and signaling proteins at a given synapse are coordinated and regulated.
Shank3 shares a similar protein domain structure but has a different expression pattern and subcellular localization than Shank1 and Shank2 (Bockers et al., 2004; Peca et al., 2011; Tao-Cheng et al., 2010). Shank3 forms multimers via its C terminal SAM domain (Boeckers et al., 2005; Hayashi et al., 2009; Naisbitt et al., 1999) as well as its PDZ domain (Iskenderian-Epps and Imperiali, 2010). The SAM domain of Shank3 has a Zn2+ binding site that is important for Shank3 protein folding at the PSD as well as for synaptogenesis and synapse maturation in vitro (Baron et al., 2006; Grabrucker et al., 2011a). Biochemically, Shank family proteins are ubiquitinated in an activity-dependent manner in neurons (Ehlers, 2003). The exact biochemical mechanism responsible the ubiquitination of Shank family protein remains to be determined. Many interesting questions related to the molecular function of Shank3 await further investigation. Does Shank3 interact with different proteins in a synapse-specific manner? Is the interaction of Shank3 with synaptic proteins regulated by activity? How do these interactions and post-translational modifications contribute to the synaptic defects in human ASD and intellectual disability associated with the SHANK3 defects? Because point mutations and microdeletions in similar domains of SHANK1 and SHANK2 have been reported in ASD (Berkel et al., 2010; Pinto et al., 2010; Sato et al., 2012), an interesting question is do various SHANK mutations cause ASD by disrupting similar mechanisms at the synapse (State, 2010a)?
Molecular Diversity of SHANK Gene Products
SHANK genes display a complex transcriptional regulation with multiple intragenic promoters and extensive alternatively spliced exons both in humans and mice (Leblond et al., 2012; Lim et al., 1999; Maunakea et al., 2010; McWilliams et al., 2004; Redecker et al., 2006; Wang et al., 2011; Wilson et al., 2003). Transcripts of SHANK3 the best characterized among the three family members in human and mice (Durand et al., 2007; Maunakea et al., 2010). The alternatively spliced exons of Shank3 encode the SH3, proline-rich, and SAM domains (Figure 1A). Combinations of multiple intragenic promoters and alterative splicing result in an extensive array of mRNA and deduced protein isoforms. The exact number of protein isoforms has not been determined, but selected mRNA isoforms analyzed in silico indicate that each Shank3 isoform (Shank3a-f) has a unique combination of different protein domains as illustrated in Figure 2B (Wang et al., 2011). For example, Shank3e and Shank3f mRNAs lack exons encoding the PDZ domain that is responsible for the interaction with NMDA and AMPA receptors (Naisbitt et al., 1999). Shank3b lacks the proline-rich and SAM domains that are critical for Homer binding and multimerization (Tu et al., 1999). Because each protein domain mediates a unique complement of protein-protein interactions (Hayashi et al., 2009; Roussignol et al., 2005), it is likely that each Shank3 isoform has a distinct set of functions. An important area for future research will be to determine the function for each isoform in vivo and its relevance to synaptic and behavioral phenotypes. The diversity of SHANK3 isoforms may contribute to synaptic signaling and postsynaptic protein composition. In addition, since Shank3 mRNA has been found in dendrites (Bockers et al., 2004), the specific Shank3 mRNAs targeted to dendrites or perhaps individual synapses may also be isoform-specific.
The complexity of SHANK3 transcript structure indicates that point mutations, translocations, and intragenic deletions of SHANK3 found in ASD patients are isoform-specific. For example, the intron 5 splicing mutations in the exon encoding the ANK domain is only predicted to affect two long isoforms of SHANK3 initiated from promoters 1 and 2 (SHANK3a and SHANK3b) (Figures 1A and 2B). The intron 19 splicing mutation will disrupt most isoforms but leave SHANK3f and other short isoforms intact. Mutations in exon 21 are expected to have no effect on mRNAs lacking exon 21 or other short SHANK3 mRNAs truncated before exon 21. Similarly, deletions within exons 1-9 and exons 1-17, and translocation breakpoints within intron 8 and exon 21, will affect different isoforms predicted from the SHANK3 gene structure. In contrast, microdeletions or large cytogenetic deletions will disrupt all SHANK3 isoforms. Therefore, the molecular consequences at the RNA and protein levels for each SHANK3 mutation are almost certainly different. A determination of how different mutations and genetic variants influence the array of potential Shank3 proteins awaits the generation of isoform-specific antibodies.
If each Shank3 isoform has distinct functions at the synapse, one attractive hypothesis is that isoform-specific disruption of SHANK3 will result in different phenotypic consequences. This may offer an explanation for the clinical heterogeneity in ASD caused by SHANK3 mutations. A similar principle may be applicable to ASD caused by defects in other genes. Consistent with this notion, many known ASD genes, such as neuroligins and neurexins, display a complex pattern of isoform-specific expression in brain (Boucard et al., 2005; Sudhof, 2008), with different isoforms having very distinct functions (Chih et al., 2006). In the cases of neurexins, more than 1000 isoforms have been reported (Missler and Sudhof, 1998).
The expression of Shank3 isoforms is cell type-specific and developmentally regulated (Lim et al., 1999; Maunakea et al., 2010). RNA in situ hybridization in rat brain using a single probe from exon 21 encoding the proline-rich domain showed that Shank3 is widely expressed in all brain regions at a low level at birth but increases after 2 weeks of age in the striatum, hippocampus, cerebellum, and in layers 1 and 2 of the neocortex (Bockers et al., 2001; Bockers et al., 2004). Similar findings were reported in mouse brain using a probe from exon 21 encoding the proline-rich domain of mouse Shank3 (Peca et al., 2011). Peak expression of Shank3 occurs at an important developmental stage of synaptic plasticity and experience-dependent circuit maturation (Bockers et al., 2004). These studies, however, have not defined the isoform-specific expression of Shank3, and thus the expression profile for different Shank3 isoforms and regulation of isoform-specific expression remain to be elucidated. To add further complexity, SHANK3 has five CpG islands across the gene and these CpG islands display brain-specific and cell type-specific DNA methylation (Figure 1A) (Beri et al., 2007; Ching et al., 2005; Maunakea et al., 2010). Both DNA methylation and histone deacetylase inhibitors have been shown to modulate the isoform specific gene expression of Shank3 in cultured neurons (Beri et al., 2007; Maunakea et al., 2010). Thus, in addition to alternate promoter use and mRNA splicing, epigenetic mechanisms such as DNA methylation and histone acetylation regulate the expression of the Shank3 gene in an isoform-specific manner. Multiple intragenic CpG islands are also associated with SHANK1 and SHANK2 (Figures 1B and 1C), but the role of these CpG islands in transcriptional regulation remains to be investigated.
SHANK2 exhibits transcriptional regulation similar to SHANK3 (Leblond et al., 2012). Specifically, SHANK2 has several isoforms driven by multiple promoters and alternative splicing of coding exons (Figure 1B). The longest Shank2e isoform containing all five protein domains was initially reported as an epithelia-specific isoform in rat (McWilliams et al., 2004). However, a recent report indicates that SHANK2E is also expressed in brain tissues in humans (Leblond et al., 2012). Several short isoforms (SHANK2A, SHANK2B, and SHANK2C) are transcribed from downstream promoters (SHANK2A, SHANK2B) or result from alternative splicing (SHANK2C) that contain distinct combinations of protein domains (Figure 2C). Tissue-specific alternative splicing of exons 19-20, exon 23, and an alternative stop codon in exon 22 of SHANK2C have also been identified in humans (Figure 1B) (Leblond et al., 2012). The alternative splicing of exons 19-20 and isoforms of SHANK2E (Jiang, unpublished data), SHANK2A, and SHANK2B (Lim et al., 1999) were conserved in mice but the status of SHANK2C and other spliced exons has not been confirmed. Similar to SHANK3, the combination of different promoters and splicing is expected to produce substantial protein diversity of SHANK2 that may carry out distinct functions at synapses. Although similar complexity of transcriptional structure has been suggested for SHANK1 (Figure 1C), detailed transcript profiles related to alternative promoters and exons remain to be delineated (Lim et al., 1999). Together, the available evidence indicates that the overall transcriptional structure of SHANK family genes is conserved in mice (Wang et al., 2011). However, the complexity of transcriptional regulation poses a significant technical challenge to adequately model human SHANK mutations in mice.
Modeling SHANKs Mutations in Mice
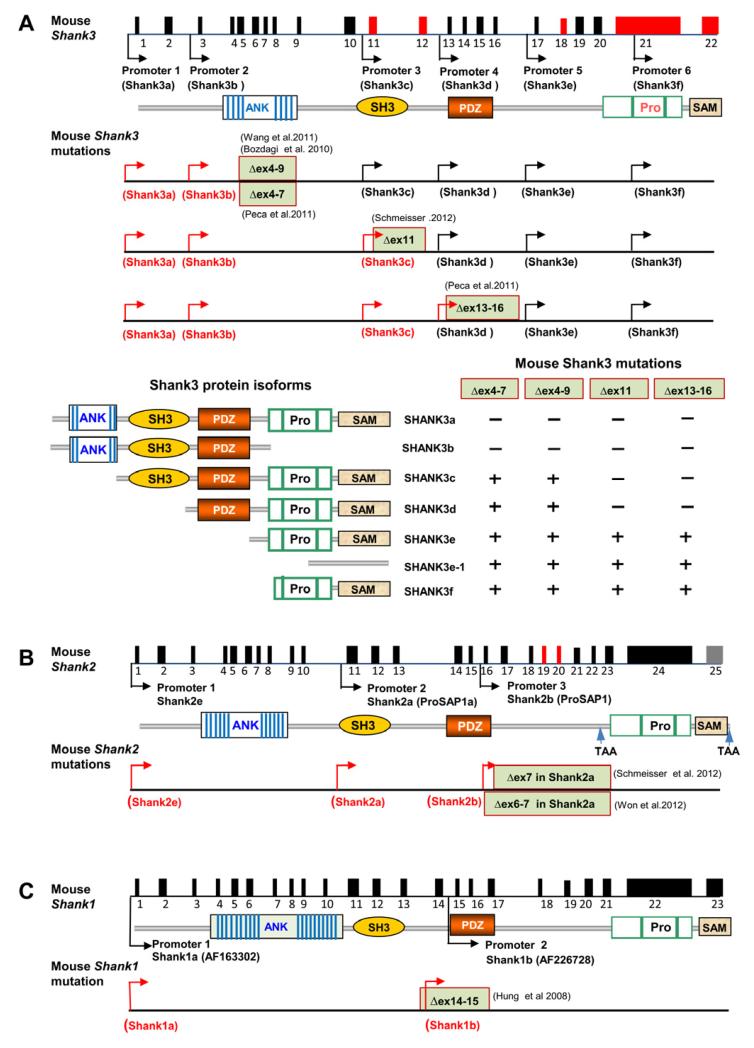
Mutant mice for all Shank family genes have now been produced and characterized (Figure 3). Shank1 mutant mice with a deletion of exons 14-15 encoding the PDZ domain were first reported in 2007 (Hung et al., 2008) (Figure 3C) and more extensive behavioral analyses were conducted subsequently (Silverman et al., 2011; Wohr et al., 2011). The targeted deletion of exons 14-15 is believed to produce a null allele of Shank1. Because the transcript structure has not been fully worked out, the possibility that this is not a complete Shank1 knockout cannot be ruled out. The major molecular and behavioral phenotypes of Shank1 mutant mice are summarized in Table 3. The synaptic proteins GKAP/SAPAP and Homer are reduced in the PSD of Shank1−/− mouse brain. Smaller dendritic spines were observed, but the ultrastructure of the PSD is unaffected at CA1 synapses of Shank1−/− mice. Basal synaptic transmission was reduced but long-term potentiation (LTP) and long-term depression (LTD) in CA1 hippocampus were unaffected (Hung et al., 2008). Behavioral analyses revealed subtle impairments in social interaction and communication as well as increased repetitive behaviors (Silverman et al., 2011; Wohr et al., 2011). Intriguingly, spatial learning and memory was enhanced in Shank1−/− mice (Hung et al., 2010).
Figure 3.
Targeted Mutations in Shank Genes in Mice
(A) Schematic of the mouse Shank3 gene structure deduced from cDNA AB230103 deposited in GenBank. The promoters are shown by arrows and alternatively spliced exons are indicated in red. The positions of targeted mutations in five different lines of Shank3 mutant mice are shown. The transcripts from promoters upstream of deleted exons are predicted to be truncated or disrupted (red arrows) and the transcripts from promoter downstream of deleted exons are predicted to be intact in each mutant line of mice (black arrows). Bottom panels depict predicted isoform-specific expression of Shank3 mRNA and proteins in Shank3 mutant mice. The “−” indicates that the isoform is disrupted and “+” indicates the isoform remains intact. The full complement of Shank3 mRNA and protein isoforms that derive from combinations of alternative promoters and mRNA splicing remains unknown. Therefore, the pattern of isoform-specific expression and disruption by specific mutations is likely more complex than indicated.
(B) Schematic of mouse Shank2 gene structure and mouse mutations of Shank2. The sequence of Shank2 full length mRNA is not available and the intron-exon structure is deduced from a longest rat Shank2 cDNA deposited in GenBank (NM_201350). Exons in red are alternatively spliced. The human isoforms of SHANK2E, SHANK2A, and SHANK2B are conserved in mice (Shank2e, Shank2a, and Shank2b) but the status of SHANK2C is unknown. The Δex7 and Δex6-7 mutations (exon numbering based on the sequences of Shank2a/ProSAP1A cDNAs NM_001113373 or AB099695) described in the Shank2 mutant mice are shown (Schmeisser et al., 2012; Won et al., 2012). Exons 6 and 7 in Shank2a are deduced to correspond to exons 16 and 17 in the full length Shank2 gene structure diagram. The Shank2 Δex7 or Δex6-7 is predicted to cause truncation of all transcripts of Shank2 from the promoters upstream of exons 6-7 (red arrows).
(C). Schematic of mouse Shank1 gene structure and mouse mutation of Shank1. The deletion of exons 14-5 is predicted to disrupt all isoforms of Shank1 (Shank1a and 1b in red arrows) (Hung et al., 2008). In (A)-(C), genomic distance and exons are not drawn to scale.
Table 3. Molecular, Biochemical, Synaptic, and Behavioral Phenotypes of Shankl and Shank2 Mutant Mice.
| Shankl (Hung et al., 2008) (Silverman et al. 2011) (Wohr et al. 2011) |
Shank2 (Won et al., 2012) | Shank2 (Schmeisser et al., 2012) | |
|
Exons/domain targeted |
Exons 14-15/PDZ (Δex14-15) | Exons 6-7/PDZ (Δex6-7) | Exon 7/PDZ (Δex7) |
| Strain/Background | 129SvJ ES cell and backcrossing to C57BL/6 for 3 generations |
129SvJ ES cells and backcrossing to C57BL/6 for > 5 generations |
129SV R1 ES cells and backcrossing to C57BL/6J for 10 generations |
| Age of mice tested | Biochemistry:3 months Morphology:E18 day for neuron culture and adult mice Electrophysiology:3-5 weeks Behavior:3-5 months |
Biochemical:3-4 weeks Morphology:8-9 weeks Electrophysiology:3-9 weeks Behaviors:1-5 months |
Biochemical:p25 and p70 days Morphology:E18 neuron culture and adult mice Electrophysiology:p21, p27, and 90 days Behaviors:6-8 months |
|
Transcripts not disrupted |
Predicted to be none | Predicted to be none | Predicted to be none |
|
Genotype of mice analyzed |
Homozygous, some heterozygous | Homozygous, some heterozygous | Homozygous, some heterozygous |
|
Altered synaptic proteins |
Reduced GKAP and Homer | Whole brain: Reduction of p- CaMKIIα/β (Thr286), p-ERK1/2, p-p38, p-GluA1 (Ser831 /S845), Increased GluN1 |
Hippocampus: increased GluN2B Striatum: increased GluN1, GluN2A, GluA2, SHANK3 |
|
Brain and synaptic morphology |
CA1 hippocampus Reduced spine density, smaller and thinner PSD |
CA1 Hippocampus Normal spine density No change in length and thickness of PSD |
CA1 Hippocampus Reduced spine density, PSD structure normal |
| Synaptic physiology |
CA1 Hippocampus Reduced AMPAR-mediated basal transmission. Normal mEPSC amplitude. Decreased mEPSC frequency, Normal paired-pulse ratio. Normal NMDA/AMPA ratio Normal LTP and LTD |
CA1 Hippocampus No change in basal synaptic transmission and normal mEPSC Reduced NMDA/AMPAR ratio Reduced NMDAR dependent LTP and LTD. mGluR dependent LTD is normal Medial prefrontal cortex Normal NMDA/AMPA ratio |
CA1 Hippocampus Reduced synaptic transmission Reduced mEPSC frequency. No difference for mEPSC amplitudes; Normal mIPSC frequency but slightly reduced mIPSC amplitudes, Normal sEPSC. The ratio of NMDA/AMPA is increased. Decreased I/O ratio. LTP is slightly enhanced but LTD is normal |
| Social behaviors | Reduced social sniffing by males in male-female interactions |
Reduced interest in novel mice in non- social vs novel social pairing in 3 chamber test. Normal social recognition and normal olfaction. |
Normal initiation for social contact but Impairment in maintaining social intact in resident-intruder test. Impaired social interaction in three chambers test. Normal olfaction |
|
Ultrasonic vocalizations |
Reduced calls by males in male- female reciprocal social interaction. |
Male mice made less USV calls while females were presented and had longer latency for the first call |
Female pup made more USV calls. No difference for USVs in adult during male-male interaction but reduced call and longer latency during female- female interaction |
| Repetitive behaviors | increased self-grooming and repetitive behaviors |
Increased locomotor activity, No increased self-grooming in home cage but increased in novel objective recognition task. Females show repetitive jumping and reduced digging |
Increased self-grooming/increased locomotor activity |
| Learning & memory | Enhanced spatial learning and memory task Impaired fear conditioning |
Impaired spatial and learning and memory in Morris water maze. Novel objection recognition memory is normal. |
normal working memory, novel objective recognition memory |
| Other behaviors | partial anxiety-like behavior; reduced scent marking behaviors |
Increased anxiety-like behavior, impaired nesting behavior. |
Increased anxiety-like behaviors, |
AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; LTP, long-term potentiation; LTD, long-term depression; mEPSC, miniature excitatory postsynaptic current; mIPSC, miniature inhibitory postsynaptic current; NMDA, N-Methyl-D-aspartate; I/O, input/output; mGluR5, metabotropic glutamate receptor 5; N/A, not analyzed; USV, ultrasonic vocalization.
Two different lines of Shank2 mutant mice have been recently reported (Schmeisser et al., 2012; Won et al., 2012). Schmeisser et al. reported Shank2 exon 7 deletion mutant mice (Shank2 Δex7) while Won et al. described mice with both exons 6 and 7 deleted (Shank2 Δex6-7). Exons 6-7 encode the PDZ domain of Shank2. Importantly, the exon numbering is defined based on mouse Shank2a/ProSAP1a isoform cDNA (AB099695 or NM_00111373). Full length mouse Shank2 mRNA has not been reported or deposited in a public database. Using the longest rat cDNA of Shank2 deposited in GenBank (NM_201350) as a reference, one can deduce that exons 6 and 7 in mouse Shank2a isoform correspond to exons 16 and 17 of predicted full length mouse Shank2 deduced from rat cDNA (NM_201350) (Figure 3B). The difference in exon numbering for SHANK2 in different organisms in other reports is likely due to the pattern of uncharacterized exons or alternative splicing (Leblond et al., 2012; Lim et al., 1999; McWilliams et al., 2004). The deletion of exon 7 and exon 6-7 of Shank2a (exons 17 and exons 16-17 of predicted full length Shank2) resulted in a frame shift of the open reading frame immediately after exon 7. Therefore, the molecular nature of these two targeted mutations is predicted to be very similar at protein level. Analyses of protein composition, synaptic development and function, and behaviors have revealed similarity but also significant differences between these two lines of Shank2 mutant mice (Table 3). Below, we utilize the exon 6-7 nomenclature based on numbering from promoter 2 of Shank2a/ProSAP1a. Full-length exon numbering is depicted in Figure 3B.
Biochemically, protein composition at synapses was altered in both Shank2 Δex7 and Δex6-7 mice but with slight differences. In Shank2 Δex7−/− mice, GluN1 and GluN2B NMDA-type glutamate receptors in hippocampus and GluN1, GluN2A, and GluA1 in striatum are increased (Schmeisser et al., 2012). Interestingly, Shank3 was up-regulated in striatum of Shank2 Δex7−/− mice. In Shank2 Δex 6-7−/− mice, reduction of phosphorylated CaMKIIα/β (T286), ERK1/2, p38, and GluA1 (Ser831/S845) was observed in hippocampus (Won et al., 2012). Similar to Shank2 Δex7−/− mice (Schmeisser et al., 2012), GluN1 is increased in the hippocampus of Shank2 Δex6-7−/− mice (Won et al., 2012). Whereas baseline synaptic transmission was reduced in Shank2 Δex7−/− mice (Schmeisser et al., 2012), normal synaptic transmission was observed in Shank2 Δex6-7−/− mice (Won et al., 2012). mEPSCs recorded from CA1 hippocampal neurons were unaltered in Shank2 Δex6-7−/− mice, but reduced in Shank2 Δex7−/−. Interestingly, the ratio of NMDA/AMPA currents was reduced at CA1 synapses in Shank2 Δex6-7−/− mice but increased at the same synapses of Shank2 Δex7−/− mice. NMDA receptor-dependent LTP in hippocampal CA1 synapses was increased and LTD was unaffected in Shank2 Δex7−/− mice. In contrast, both NMDA receptor-dependent LTP and LTD at CA1 synapses were reduced in Shank2 Δex6-7−/− mice. Behaviorally, hyperactivity, impaired social interaction, altered ultrasonic vocalizations, and increased self-grooming were observed in both Shank2 Δex6-7−/− and Shank2 Δex7−/− mice. Spatial learning and memory was impaired in Shank2 Δex6-7−/− but normal in Shank2 Δex7−/− mice. The basis for apparent discrepancies in synaptic physiology but similar behavioral profiles between Shank2 Δex6-7−/− and Shank2 Δex7−/− mice is not immediately clear and further investigation is warranted. Interestingly, treatment with D-cycloserine, a NMDA receptor agonist, restored the NMDA/AMPA ratio and improved social interaction in the three-chamber social interaction assay in Shank2 Δex6-7−/− mice (Won et al., 2012). In addition, treatment with CDPPB, an mGluR5-selective positive allosteric modulator, not only rescued the reduced NMDA/AMPA ratio but also recovered the defective LTP and LTD in hippocampus as well as biochemical changes in Shank2 Δex6-7−/− mice. CDPPB also reversed the impaired social interaction impairment in Shank2 Δex6-7−/− mice without affecting other behavioral impairments in these mice (Won et al., 2012).
Five lines of Shank3 mutant mice carrying different mutations in Shank3 have been reported (Bozdagi et al., 2010; Peca et al., 2011; Schmeisser et al., 2012; Wang et al., 2011) (Figures 3A). The mutations in these mice include deletions of exons 4-9 by two groups with slightly different design [(Δex4-9Buxbaum(B)) (Bozdagi et al., 2010) and Δex4-9Jiang (J) (Wang et al., 2011)], deletion of exons 4-7(Δex4-7) (Peca et al., 2011) encoding the ANK repeat domain, deletion of exon 11(Δex11) encoding the SH3 domain (Schmeisser et al., 2012), and deletion of exons 13-16 (Δex13-16) encoding the PDZ domain (Peca et al., 2011). Because all these deletions cause a frame shift for targeted transcripts, they all resulted in either a truncated Shank3 protein or possible disruption of full length RNA or protein isoforms due to the stability of encoded mRNA or protein. Based on current knowledge of Shank3 promoters and alternative splicing, each of these mice is expected to have disruption of different Shank3 isoforms (Wang et al., 2011) (Figure 3A). Isoform-specific disruption of Shank3 was evident in Δex4-7, Δex4-9J, Δex11, and Δex13-16 mice (Peca et al., 2011; Schmeisser et al., 2012; Wang et al., 2011). The Δex4-9J deletion disrupted mRNA transcripts from promoters 1 and 2 (Shank3a and Shank3b) but not Shank3c-f as confirmed by isoform-specific RT-PCR analysis (Wang et al., 2011). One unexpected finding from RNA expression analysis of Δex4-9J mice was the presence of an mRNA splice isoform from exon 2 to exon 10, in addition to the expected splicing isoform from exon 3 to exon 10 due to the deletion of exons 4-9 (Wang et al., 2011). Intriguingly, this cryptic splicing from exon 2 to 10 occurred only in brain but not in kidney of Δex4-9J−/− mice. The mRNAs with joining of exons 2-10 and exons 3-10 were stable and were predicted to result in a frame shift in protein sequence shortly after exon 10. Whether the same cryptic splicing occurs in the Δex4-9B mutant mice has not been investigated (Bozdagi et al., 2010). Although targeted deletion may interfere with pre-mRNA splicing mechanisms, the basis for tissue specificity of cryptic splicing is unknown. This observation adds to the complexity of predicting the molecular consequence of different mutations in Shank3 mutant mice, and perhaps in human SHANK3 mutations, and suggests that otherwise similar mutations such as Δex4-9B, Δex4-9J, and Δex4-7 may have different molecular consequences for Shank3 at the mRNA and protein level. The Δex11 mutation (Schmeisser et al., 2012) is predicted to disrupt promoters 1 to 3 for Shank3a-c but not promoters 4 to 6 for Shank3d-f, while the Δex13-16 mutation (Peca et al., 2011) is predicted to disrupt transcripts from promoter 1 to 4 (Shank3a-d) but not from promoter 5 to 6 (Shank3e-f) (Figure 3A) (Peca et al., 2011), although this prediction requires molecular confirmation. The effect on alternative splicing of these targeted mutations has not been determined and, as of yet, the full complement of Shank3 mRNA transcripts and splice variants is not known and awaits characterization at the mRNA and protein level. Beyond the mouse models, it will be important to know the isoform expression of SHANK3 protein in patients carrying various mutations if postmortem brain tissue becomes available.
Phenotypic analyses at the biochemical, synaptic, and behavioral levels were performed extensively on either heterozygotes or homozygotes at different ages for Shank3 Δex4-7, Δex4-9J, Δex4-9B, and Δex13-16 but to a lesser degree in Δex11 mutant mice (Bozdagi et al., 2010; Peca et al., 2011; Schmeisser et al., 2012; Wang et al., 2011; Yang et al., 2012). The methods and techniques used in these analyses were similar but not identical. Different brain regions including hippocampus, striatum, and neocortex were analyzed in different lines of mutant mice. Overall, the data obtained from these studies support a general conclusion that synaptic function is impaired and social behaviors are abnormal in mice with Shank3 mutations. In the following sections, we compare and contrast phenotypes observed with the various Shank3 mutant mice which are also summarized in Table 4.
Table 4. Molecular, Biochemical, Synaptic, and Behavioral Phenotypes of Shank3 Mutant Mice.
|
Bozdagi et al., 2010), Yang et al., 2012) |
Wang et al., 2011 | Peca et al., 2011 | Schmeisser et al., 2012 | ||
|---|---|---|---|---|---|
|
Exons/domain targeted |
Exons 4-9B/ANK repeat (Δex4-9B) |
Exons 4-9J/ANK repeat (Δex4-9J) |
Exons 4-7/ANK repeat (Δex4-7) |
Exons 13-16/PDZ (Δex13-16) |
Exon 11/PDZ (Δex11) |
|
Strain/ Background |
Bruce4 C57BL/6 ES cell and maintain on C57BL/6 |
129SvEv ES cell backcrossing to C57BL/6J for more than 6 generations |
129SvR1 ES cell and backcrossing to C57BL/6J for one generations |
129 SvR1 ES cells and backcrossing to C57BL/6J for more than 1 generations |
129SvR1 ES cell and backcrossing to C57BL/6 for more than 10 generations, |
| Age of mouse | Biochemistry: 3 months Morphology:3 months Electrophysiology:3 months Behaviors:p21 days to 16 weeks |
Biochemistry:3-4 months. Morphology:P1 neuron culture and 1-3 months Electrophysiology:4-6 weeks. Behaviors:3-8 months |
Biochemistry: not stated Electrophysiology:6-7 weeks. Behaviors:5-6 weeks |
Biochemistry: not stated Morphology:5 weeks Electrophysiology:5-7 weeks Behaviors:5-6 weeks; |
Biochemical:p25 and p70 days Morphology: adult Electrophysiology:N/A Behaviors: N/A |
|
Transcripts not disrupted |
Shank3c,d,e, f | Shank3c,d,e,f | Shank3c,d,e,f | Shank3e, f | Shank3d, e, f |
|
Genotype of mice analyzed |
Heterozygous /Homozygous |
Homozygous | Homozygous | Homozygous | Homozygous |
|
Altered synaptic proteins |
Reduction of GluA1 | Reduction of GKAP, Homer1 b/c, GluA1, GluN2A |
N/A | Reduction of SAPAP3/GKAP3, Homer1, PSD-93, GluA2, GluN2A, GluN2B |
Increased GluN2B and SHANK2 |
|
Brain and synaptic morphology |
CA1 Hippocampus Activity dependent spine remodeling was affected |
CA1 Hippocampus Longer dendritic spines. Decreased spine density. No change in length and thickness of PSD |
N/A | Striatum Increase in striatal volume, dendritic length and surface area. Decreased spine density, length, and thickness of PSD |
CA1 hippocampus No defect identified |
|
Synaptic physiology |
CA1 Hippocampus Reduced AMPAR- mediated basal transmission. Decreased mEPSC amplitude. Increased mEPSC frequency, Decrease in paired- pulse ratio. Reduced LTP. No change in NMDAR- or mGluR- mediated LTD. |
CA1 Hippocampus No change in basal synaptic transmission. No change in amplitude or frequency of mEPSCs or mIPSCs. No change in paired- pulse ratio, I/O, fiber volley. Reduced LTP. |
Striatum Slight reduction in corticostriatal synaptic transmission |
CA1 Hippocampus No change in field recordings of population spikes, paired-pulse ratio, mEPSC frequency and amplitude Striatum No change in paired- pulse ratio. Reduced field population spikes, Reduced mEPSC frequency and amplitude. |
N/A |
|
Social behaviors |
Reduced social sniffing by males in male-female interactions, mild social impairment in reciprocal interactions in juveniles, normal three chamber test for adult mice |
Reduced interest in novel mice in non- social vs novel social pairing in 3 chamber test, females performed better than males. Decreased bidirectional social interactions in dyadic test. |
Normal initiation of social interaction. Perturbed recognition of social novelty during 3 chamber test |
Perturbed recognition of social novelty during 3 chamber test, Decreased reciprocal interactions in dyadic test. Decreased frequency of nose-to-nose interaction. Decreased anogenital sniffing |
N/A |
|
Ultrasonic vocalizations (USV calls) |
Reduced calls observed in some cohorts of adult mice during social interaction but no difference in newborn pups |
Males made more calls while females made fewer calls. Altered frequency, complexity, and duration of calls |
Not mentioned | N/A | N/A |
|
Repetitive behaviors |
Increased self- grooming, inflexible behavior in reversal water maze observed in some cohorts |
Increased head pokes in hole-board test, increased self- grooming. Stereotypic object manipulation in novel object test |
No increase in self- injurious grooming, |
Self-injurious grooming, causing skin lesions |
self-injurious grooming, causing skin lesion |
|
Learning & Memory |
Impaired novel object recognition. Normal Morris water maze, normal fear conditioning |
Impaired in acquisition and reversal in Morris water maze. Impaired short- and long-term memory |
N/A | No difference observed in Morris water maze. |
N/A |
|
Schizophrenia- related behaviors |
Normal sensory gating and startle reflex |
No difference in PPI. Not hyperactive in the open field |
N/A | N/A | N/A |
AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; LTP, long-term potentiation; LTD, long-term depression; NMDA, N-methyl-D-asparate; mEPSC, miniature excitatory postsynaptic current; mIPSC, miniature inhibitory postsynaptic current; mGluR5, metabotropic glutamate receptor 5; N/A, not analyzed; I/O, input/output; PPI, prepulse inhibition; USV, ultrasonic vocalization.
Synaptic Proteins in Shank3-Deficient Mouse Brain
PSD proteins were altered in different brain regions of all Shank3 mutant mice but to varying degrees in the hippocampus of Δex4-9B+/−, Δex4-9J−/−, and Δex11−/−; the striatum of Δex11−/− and Δex13-16−/−; and neocortex of Δex11−/− mice. Homer1b/c and GKAP1/SAPAP1 were reduced in the PSD fraction but not in the cytosolic fraction of Δex4-9J−/− hippocampus (Wang et al., 2011). Homer1, GKAP/SAPAP, and PSD-93 were reduced in PSD fractions isolated from the striatum of Δex13-16−/− mice (Peca et al., 2011) but GKAP/SAPAP was not reduced in striatum of Δex11−/− mice (Schmeisser et al., 2012). Interestingly, Shank2 was increased in the synaptosomal fraction of Δex11−/− striatum, while Shank3 was found to be increased in Shank2 Δex7−/− mutant mice (Schmeisser et al., 2012). This compensatory mechanism may contribute to the reciprocal changes in Shank2 Δex7−/− and Shank3 Δex11−/− mice. It will be interesting to examine whether the same phenomena occurs in other Shank3 mutant mice. Many of these proteins were either not altered in the neocortex or not examined in neocortex in these mutant mice. The corresponding changes of Homer1 and Shank3 in several mouse lines and several brain regions are consistent with the known multimeric network formed between these two proteins (Hayashi et al., 2009; Tu et al., 1999).
Mutations of Shank3 altered the levels of synaptic glutamate receptors. The AMPA receptor subunit GluA1 was reduced in hippocampal neurons examined in culture and hippocampal tissues from Δex4-9J−/− (Wang et al., 2011) and Δex4-9B+/− mice (Bozdagi et al., 2010), and GluA2 was reduced in the striatum of Δex13-16−/− mice (Peca et al., 2011). In the case of NMDA receptors, GluN2A subunit was reduced in the hippocampus of Δex4-9J−/− mice (Wang et al., 2011). Both GluN2A and GluN2B subunits were reduced in the striatum of Δex13-16−/− mice (Peca et al., 2011) but they were unchanged in the stratum of Δex11−/− mice (Schmeisser et al., 2012). In contrast, GluN2B was increased in synaptosomal fractions from Δex11−/− hippocampus (Schmeisser et al., 2012). In nearly all mouse lines and brain areas examined, changes in the level of these synaptic proteins and receptors was relatively modest, and many other known Shank3 interacting proteins listed in Table 2 were not altered or not examined in mutant mice. The specific patterns of altered synaptic proteins varied among different mutant mice lines with similar mutations. Such variation may be due to isoform-specific effects of different mutations. However, a direct comparison, ideally by running the same experiments head-to-head for each line of mutant mice with matched genotypes and age, will be important for a quantitative comparison of the effects of Shank3 mutations on synaptic protein composition at synapses of different brain regions.
Synaptic Structure in Shank3 Mutant Mice
The ultrastructure of glutamatergic synapses was examined by electron microscopy (EM) in all mutant mice except the Δex4-9B+/− line. Decreased PSD thickness and length were observed at corticostriatal synapses in Δex13-16−/− mice (Peca et al., 2011), but not in the hippocampal CA1 synapses in ex4-9J−/− mice (Wang et al., 2011) or Δex11−/− mice (Schmeisser et al., 2012). Dendritic branching and spine area was increased in medium spiny neurons (MSNs) of the striatum of Δex13-16−/− mice (Peca et al., 2011), but not examined in striatum of mice carrying other Shank3 mutations (Peca et al., 2011; Schmeisser et al., 2012; Wang et al., 2011). Spine length was increased in CA1 hippocampus of Δex4-9J−/− mice (Wang et al., 2011), and spine density was decreased in the striatum and CA1 hippocampus of Δex13-16−/− and Δex4-9J−/− mice, respectively. The reduction of spine density visualized by Golgi impregnation was developmental stage-specific in Δex4-9J−/− mice, with significant spine loss observed at 4 weeks but not at 10 weeks of age (Wang et al., 2011). Activity-induced spine growth by theta burst stimulation in cultured brain slices was attenuated at CA1 synapses of Δex4-9B+/− mice (Bozdagi et al., 2010).
The totality of ultrastructural and morphological analysis in Shank3 mutant mice indicates complex regulation of glutamatergic synapse size, shape, and structure. In general, mutation of Shank3 leads to loss of spines, a reduction in spine volume, and decreased PSD thickness in the adult. These effects, together with spine elongation, suggests a phenotype of reduced or delayed synapse maturation that is reminiscent of the phenotypes observed in mouse models of fragile X syndrome (Comery et al., 1997; Irwin et al., 2001), Rett syndrome (Armstrong, 2005; Belichenko et al., 2009; Chao et al., 2007), and Angelman syndrome (Dindot et al., 2008; Sato and Stryker, 2010; Yashiro et al., 2009). Notably, synapse structure phenotypes vary with specific Shank3 mutations, are different in different brain regions, and display developmental heterogeneity, perhaps due to differential spatial and temporal expression of other Shank family members or to compositional variation across different populations of glutamatergic synapses.
Synapse Function and Plasticity in Shank3 Mutant Mice
Shank proteins regulate the abundance and signaling of ionotropic glutamate receptors at excitatory synapses. Accordingly, synaptic transmission and plasticity were examined in different brain regions in all Shank3 except Δex11mutant mice. Measurements of miniature excitatory postsynaptic current (mEPSC) frequency and amplitude, paired pulse ratio, input/output (I/O) curves, fiber volley, and population spikes indicated that synaptic transmission is reduced at hippocampal CA1 synapses of Δex4-9B+/− (Bozdagi et al., 2010) and Δex4-9B−/− (Yang et al., 2012), but not in Δex4-9J−/− (Wang et al., 2011) or Δex13-16−/− mice (Peca et al., 2011), and was not examined in Δex11−/− mice (Schmeisser et al., 2012). The explanation for the difference between Δex4-9B (Bozdagi et al., 2010; Yang et al., 2012) and Δex4-9J−/− (Wang et al., 2011) is not immediately clear. One possibility is that these mutations induce different cryptic splicing as described above in Δex4-9J−/− mice (Wang et al., 2011). Another possibility is that heterozygous mutations may produce a dominant gain-of-function phenotype. In addition, mouse genetic background, animal age, and specific protocols used for the studies may contribute to the variability.
In striatum, the frequency of mEPSCs and amplitude of population spikes were significantly decreased in Δex13-16−/− mice, but only mildly affected in Δex4-7−/− mice (Peca et al., 2011). Presynaptic responses measured by paired pulse ratio and input/output curves were not altered at corticostriatal synapses in Δex13-16−/− or Δex4-7−/− mice (Peca et al., 2011). The different degree of synaptic transmission defects in mice with specific Shank3 mutations supports the notion of an isoform-specific contribution to synaptic function.
Hippocampal LTP was reduced at CA1 synapses of Δex4-9J−/− and Δex4-9B but not examined in Δex11−/− and Δex13-16−/− animals (Bozdagi et al., 2010; Peca et al., 2011; Schmeisser et al., 2012; Wang et al., 2011; Yang et al., 2012). NMDA receptor-dependent long-term depression (LTD) induced by low frequency stimulus and mGluR-dependent LTD induced by PP-LFS were not affected in CA1 hippocampus of ex4-9B mice (Bozdagi et al., 2010; Yang et al., 2012), suggesting an alteration in the set-point for bidirectional Hebbian synaptic plasticity (Cho and Bear, 2010). The same analysis was not conducted in other mutant lines (Peca et al., 2011; Wang et al., 2011).
Collectively, these data support circuit defects mediated by glutamate receptors in Shank3 mutant mice that appear to be both synapse- and mutation-specific. It is not yet clear whether there are common core synaptic defects in the various mutant mice, but the phenotypic heterogeneity itself appears consistent with the clinical heterogeneity of patients harboring SHANK3 mutations. Since different mutations affect different isoforms of Shank3, some of the observed phenotypes may arise from isoform-specific effects on synaptic transmission. Firm conclusions in this regard are complicated by the fact that the different Shank3 isoforms are probably expressed in various Shank3 mutant mice, which were analyzed at different ages using slightly different protocols. Moreover, acute knockdown of Shank3 in cultured neurons decreases mGluR-dependent plasticity (Verpelli et al., 2011), suggesting differences in effects of Shank3 on mGluR1/5 signaling over development and pointing to the need for cautious interpretation regarding the pathogenic versus compensatory roles of synaptic and circuit phenotypes observed in Shank3 mutant mice.
Behavioral Phenotypes in Shank3 Mutant Mice
Based on the strong genetic evidence for SHANK3 defects as a cause of human ASD, Shank3 mutant mice offer an opportunity to model autism-like behaviors in rodents. Extensive behavioral analyses were performed in Shank3 Δex4-9B (Bozdagi et al., 2010; Yang et al., 2012), Δex4-9J−/− (Wang et al., 2011), Δex4-7−/−, and Δex13-16−/− (Peca et al., 2011) mutant mice at different ages, on different genetic backgrounds, and using different protocols. The most notable and consistent observation was reduced social interaction and affiliation behaviors using different testing methods (Bozdagi et al., 2010; Peca et al., 2011; Wang et al., 2011; Yang et al., 2012). Variable performances were noted in different cohorts of Δe4-9B−/− mice (Yang et al., 2012). Repetitive behaviors measured by increased self-grooming in the home cage and behavioral inflexibility in the reverse Morris water maze were observed in Δex4-9J−/− (Wang et al., 2011) and Δex4-9B−/− mice (Bozdagi et al., 2010; Yang et al., 2012) but were not apparent in Δex4-7−/− mice (Peca et al., 2011). A more marked increase in self-grooming and self-injurious behaviors was observed in Δex11−/− and Δex13-16−/− mice (Peca et al., 2011; Schmeisser et al., 2012). Different severity of similar behaviors with different mutations may reflect Shank3 isoform-specific contributions to specific behaviors. The number, frequency, and duration of ultrasonic vocalizations were altered in a sex-specific manner in Δex4-9J−/− mice (Wang et al., 2011). Reduced ultrasonic vocalizations adult were also reported in Δex4-9B (Bozdagi et al., 2010; Yang et al., 2012). The interpretation and predictive value of aberrant ultrasonic vocalizations in mice relative to communication behaviors in human ASD remains the subject of investigation (Holy and Guo, 2005; Scattoni et al., 2009).
Despite the consistent intellectual disability reported in patients with multiple types of SHANK3 mutations, Shank3 mutant mice differed significantly in performance of learning and memory tasks. Impaired performance in the Morris water maze task was seen in Δex4-9J−/− mice (Wang et al., 2011) but not in Δex13-16−/− (Peca et al., 2011) and Δex4-9B−/− animals (Yang et al., 2012). Short-term and long-term memory in a social transmission test were impaired in Δex4-9J−/− mice (Wang et al., 2011). Prepulse inhibition (PPI) is not affected in Δex4-9J−/− (Wang et al., 2011) and Δex4-9B−/− (Yang et al., 2012) mice but has not been examined in other mice.
Deciphering the relationship between phenotypic diversity and the molecular diversity of Shank3 mutations remains a significant challenge. It is tempting to speculate that the phenotypic diversity in Shank3 mutant mice reflects the clinical heterogeneity in patients with SHANK3 defects. In a strict sense, none of these Shank3 mouse mutations are equivalent to SHANK3 mutations found in human ASD patients. The Δex4-9 (Bozdagi et al., 2010) and Δex4-7 mutations in mouse (Peca et al., 2011) are closest to patients with intragenic exon 1-9 deletion and splice mutation of intron 5 in SHANK3 (Bonaglia et al., 2011; Hamdan et al., 2011) (Table 1). Mutations in the SH3 and PDZ domains are missense mutations in humans (Boccuto et al., 2012; Waga et al., 2011), but mouse mutation of Δex11 and Δex13-16 are exon deletion and frame shift mutations (Peca et al., 2011; Schmeisser et al., 2012).
Since each mutation has a different impact on Shank3 isoform expression, a simple hypothesis is that the diversity of phenotypes in Shank3 mutant mice reflects the molecular diversity of Shank3. However, analysis of heterozygotes versus homozygotes, different measurements in different brain regions, as well as different genetic backgrounds could all contribute to phenotypic heterogeneity. Regarding genetic background, different strains used included Bruce4 C57BL/6 (Δ4-9B) (Bozdagi et al., 2010; Yang et al., 2012), mixed 129SvEv and C57BL/6J backcrossed to C57BL/6J F7 generations (Δ4-9J) (Wang et al., 2011), mixed 129SvR1 and C57BL/6 background (Δex4-7, Δex11, and Δex13-16) (Peca et al., 2011; Schmeisser et al., 2012) (Table 4). A naturally occurring Disc1 (Disrupted in schizophrenia) mutation in the 129 strain of ES cells (Clapcote et al., 2007; Clapcote and Roder, 2006) was segregated from the Δex4-9J−/− deletion (Wang et al., 2011) but was not reported in other Shank3 mutant mice using mouse 129 ES cells (Peca et al., 2011; Schmeisser et al., 2012). Synaptic dysfunction and abnormal behaviors have been documented in mice harboring Disc1 mutations (Clapcote et al., 2007; Kim et al., 2009), and this may present a confounding factor for phenotypic analysis.
Translation of Shank Mutant Mice to Human ASD
It is a big leap from mouse behavioral phenotypes to human clinical presentations of neurobehavioral disorders like ASD (Bucan and Abel, 2002; Moy et al., 2006; Silverman et al., 2010). In human patients, ASD is a behavioral diagnosis with considerable clinical heterogeneity. There is currently no reliable biomarker, pathology, anatomical finding, or functional neuroimaging change that can be considered pathognomonic or predictive for ASD (Anagnostou and Taylor, 2011; Bauman and Kemper, 2005; Courchesne et al., 2007; Lord et al., 2000a). Remarkably little is known about the neurological basis of ASD, and many brain regions and circuits have been implicated in ASD (Amaral et al., 2008; Anagnostou and Taylor, 2011; Bauman and Kemper, 2005; Courchesne et al., 2007). Several competing hypotheses have been proposed to account for core deficits and ancillary symptomatic domains in ASD, but none have been widely accepted (Belmonte et al., 2004; Courchesne et al., 2007; Geschwind and Levitt, 2007; Rubenstein, 2010; Zoghbi, 2003). Because of the molecular and clinical heterogeneity documented in ASD, the challenge of interpreting any human data from heterogeneous patient populations is obvious. In mouse behavioral studies, testing paradigms for learning and memory have been widely accepted (Crawley, 2008; Crawley and Paylor, 1997; Morris, 1981). However, to date none of the various social and communication behaviors have been validated as robustly translatable from rodents to humans (Silverman et al., 2010). This may be due to the high degree of specialization and diverse strategies for ethologically relevant social behaviors in mammals, particularly primates (Bucan and Abel, 2002; Flint and Mott, 2008; Kas et al., 2007). Although the triad of impaired social interaction, communication, and stereotypical behaviors is recognized as core to ASD, the clinical presentation of these impairments is highly varied in humans. In fact, there are few clinical tools available to evaluate behavioral features quantitatively in humans that could guide more basic neurobiological studies in model systems (Lord et al., 2001; Lord et al., 2000b; Lord et al., 1994).
A burning issue in the field is the extent to which common pathophysiology underlies ASD. Analyses of the Shanks mutant mice indicate that subtle differences in mutations within a given ASD risk gene can produce overlapping but non-identical cellular, synaptic, and behavioral phenotypes. One approach for the future will be to tailor specific Shank mutations in the mouse to correspond precisely to human mutations where patients have undergone extensive clinical evaluation. Another important element for translating observations from Shank3 mutant mice will be to couple in vivo physiology and imaging in the mouse to functional neuroimaging in human patients to help identify conserved circuit phenotypes. Perhaps most significantly, the successful development of pharmacological therapies for core clinical features of ASD would open the door to more comprehensive data-driven validation of mouse models to better enable forward translation.
Future Directions
Numerous questions have emerged from the analysis of SHANK defects in human ASD patients and Shank mutant mice. In human patients, natural history studies of genotype and phenotype in patients with various SHANK mutations are critical. A detailed description and comparison of clinical features in patients with mutations in different SHANK genes will provide guidance for modeling human disease in animal models. Because of the similar protein domain structure among SHANK family proteins, it will be interesting to determine whether ASD patients with analogous mutations in SHANK genes have significant overlapping clinical features or whether different SHANK family members influence distinct phenotypes. At the molecular level, it will be important to know the full complement of SHANK1, SHANK2 and SHANK3 isoforms and how various ASD-linked mutations, particularly point mutations or intragenic deletions, alter SHANK2 and SHANK3 isoform expression in humans. To date, most of the expression and subcellular localization data for Shank3 have used a single RNA probe and single antibody which may fail to detect difference among Shank3 isoforms.
There is a critical need to directly compare the different Shank2 and Shank3 mutant mice head to head for cellular, synaptic, circuit, and behavioral phenotypes. Such direct comparisons will allow for more definitive identification of common synaptic defects, circuit endophenotypes, and behaviors. Can mutations in Shank2 and Shank3 open the door to a molecular pathway that provides novel therapeutic targets? Study of Shank2 Δex6-7 mice has offered a promising start (Won et al., 2012). For example, it will be important to examine whether NMDA receptor agonists and mGluRs positive allosteric modulators reverse phenotypes in Shank2 Δex7−/− mice (Schmeisser et al., 2012) or in other Shank mutant mice. Perhaps more importantly, the diverse and often non-congruent phenotypes in various Shank mutant mice highlight the fact that most of the current mouse models do not carry the human mutations. Specific mutations are likely to produce specific phenotypes in patients and hence must be modeled accordingly in mice for the mutant mice to have full translational potential. Much remains to be learned, but it is tempting to consider SHANK3 “restoration” in a loose sense as a therapeutic strategy for Phelan-McDermid syndrome, and perhaps more broadly in ASD. Yet, anthropomorphizing rodent behavior in the hope of analogizing with symptomatic improvement in neuropsychiatric disease is fraught with cautionary tales. Interventions that improve cognitive performance, behaviors, or neuropathology in rodents have frequently not led to accelerated therapeutic development, most notably in Alzheimer’s disease where reports abound of cured mouse models despite repeated clinical failures (Cisse et al., 2011; Gandy and DeKosky, 2013; Malenka and Malinow, 2011; Vaillend et al., 2002). Ultimately, the value of Shank mutant mice will depend critically on the ability to use human patients to validate their predictive utility. Recently, whole genome sequencing technology has successfully identified a list of candidate genes in ASD (Bi et al., 2012; Chahrour et al., 2012; Iossifov et al., 2012; Neale et al., 2012; O’Roak et al., 2012a; Sanders et al., 2012), and this list will likely expand in the future. Because of the rarity of sequence variants across the population, it has been challenge to establish a causal role for specific variants in human disease. Functional studies are thus a critical component to determine the pathogenicity of specific genetic variants. The lesson learned from modeling SHANK mutations in mice will almost certainly be valuable to modeling other ASD candidate genes in the future.
Acknowledgements
We thank Juliet Hernandez, Benjamin Philpot, Dan Smith, Julia Sommer, and William Wetsel for critical review of the manuscript. We thank Xiaoming Wang and Alexandra Bey for help preparing Tables and comments. Work in the lab of YHJ is supported by Autism Speaks, NIH grants 5K12-HD0043494-08 and R01MH098114-01. Work in the lab of MDE is supported by Pfizer, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Anagnostou E, Taylor MJ. Review of neuroimaging in autism spectrum disorders: what have we learned and where we go from here. Mol Autism. 2011;2:4. doi: 10.1186/2040-2392-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderlid BM, Schoumans J, Anneren G, Tapia-Paez I, Dumanski J, Blennow E, Nordenskjold M. FISH-mapping of a 100-kb terminal 22q13 deletion. Hum Genet. 2002;110:439–443. doi: 10.1007/s00439-002-0713-7. [DOI] [PubMed] [Google Scholar]
- Armstrong DD. Neuropathology of Rett syndrome. J Child Neurol. 2005;20:747–753. doi: 10.1177/08830738050200090901. [DOI] [PubMed] [Google Scholar]
- Asperger H. Archive fur psychiatrie und Nervenkrankheiten. 1944. Die “Autistichen Psychopathen” im Kindersalter; pp. 76–136. [Google Scholar]
- Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU. An architectural framework that may lie at the core of the postsynaptic density. Science. 2006;311:531–535. doi: 10.1126/science.1118995. [DOI] [PubMed] [Google Scholar]
- Bauman ML, Kemper TL. Neuroanatomic observations of the brain in autism: a review and future directions. Int J Dev Neurosci. 2005;23:183–187. doi: 10.1016/j.ijdevneu.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Belichenko PV, Wright EE, Belichenko NP, Masliah E, Li HH, Mobley WC, Francke U. Widespread changes in dendritic and axonal morphology in Mecp2-mutant mouse models of Rett syndrome: evidence for disruption of neuronal networks. J Comp Neurol. 2009;514:240–258. doi: 10.1002/cne.22009. [DOI] [PubMed] [Google Scholar]
- Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ. Autism and abnormal development of brain connectivity. J Neurosci. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beri S, Tonna N, Menozzi G, Bonaglia MC, Sala C, Giorda R. DNA methylation regulates tissue-specific expression of Shank3. J Neurochem. 2007;101:1380–1391. doi: 10.1111/j.1471-4159.2007.04539.x. [DOI] [PubMed] [Google Scholar]
- Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42:489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- Berkel S, Tang W, Trevino M, Vogt M, Obenhaus HA, Gass P, Scherer SW, Sprengel R, Schratt G, Rappold GA. Inherited and de novo SHANK2 variants associated with autism spectrum disorder impair neuronal morphogenesis and physiology. Hum Mol Genet. 2012;21:344–357. doi: 10.1093/hmg/ddr470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- Bi C, Wu J, Jiang T, Liu Q, Cai W, Yu P, Cai T, Zhao M, Jiang YH, Sun ZS. Mutations of ANK3 identified by exome sequencing are associated with Autism susceptibility. Hum Mutat. 2012 doi: 10.1002/humu.22174. [DOI] [PubMed] [Google Scholar]
- Boccuto L, Lauri M, Sarasua SM, Skinner CD, Buccella D, Dwivedi A, Orteschi D, Collins JS, Zollino M, Visconti P, et al. Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur J Hum Genet. 2012 doi: 10.1038/ejhg.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccuto L, Lauri M, Sarasua SM, Skinner CD, Buccella D, Dwivedi A, Orteschi D, Collins JS, Zollino M, Visconti P, et al. Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur J Hum Genet. 2013;21:310–316. doi: 10.1038/ejhg.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockers TM, Mameza MG, Kreutz MR, Bockmann J, Weise C, Buck F, Richter D, Gundelfinger ED, Kreienkamp HJ. Synaptic scaffolding proteins in rat brain. Ankyrin repeats of the multidomain Shank protein family interact with the cytoskeletal protein alpha-fodrin. J Biol Chem. 2001;276:40104–40112. doi: 10.1074/jbc.M102454200. [DOI] [PubMed] [Google Scholar]
- Bockers TM, Segger-Junius M, Iglauer P, Bockmann J, Gundelfinger ED, Kreutz MR, Richter D, Kindler S, Kreienkamp HJ. Differential expression and dendritic transcript localization of Shank family members: identification of a dendritic targeting element in the 3′ untranslated region of Shank1 mRNA. Mol Cell Neurosci. 2004;26:182–190. doi: 10.1016/j.mcn.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Boeckers TM, Liedtke T, Spilker C, Dresbach T, Bockmann J, Kreutz MR, Gundelfinger ED. C-terminal synaptic targeting elements for postsynaptic density proteins ProSAP1/Shank2 and ProSAP2/Shank3. J Neurochem. 2005;92:519–524. doi: 10.1111/j.1471-4159.2004.02910.x. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Beri S, De Agostini C, Novara F, Fichera M, Grillo L, Galesi O, Vetro A, Ciccone R, et al. Molecular Mechanisms Generating and Stabilizing Terminal 22q13 Deletions in 44 Subjects with Phelan/McDermid Syndrome. PLoS Genet. 2011;7:e1002173. doi: 10.1371/journal.pgen.1002173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Mani E, Aceti G, Anderlid BM, Baroncini A, Pramparo T, Zuffardi O. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet. 2005 doi: 10.1136/jmg.2005.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Mani E, Aceti G, Anderlid BM, Baroncini A, Pramparo T, Zuffardi O. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13. 3 deletion syndrome. J Med Genet. 2006;43:822–828. doi: 10.1136/jmg.2005.038604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan M, Abel T. The mouse: genetics meets behaviour. Nat Rev Genet. 2002;3:114–123. doi: 10.1038/nrg728. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD. Multiple rare variants in the etiology of autism spectrum disorders. Dialogues Clin Neurosci. 2009;11:35–43. doi: 10.31887/DCNS.2009.11.1/jdbuxbaum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour MH, Yu TW, Lim ET, Ataman B, Coulter ME, Hill RS, Stevens CR, Schubert CR, Greenberg ME, Gabriel SB, Walsh CA. Whole-exome sequencing and homozygosity analysis implicate depolarization-regulated neuronal genes in autism. PLoS Genet. 2012;8:e1002635. doi: 10.1371/journal.pgen.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Ching TT, Maunakea AK, Jun P, Hong C, Zardo G, Pinkel D, Albertson DG, Fridlyand J, Mao JH, Shchors K, et al. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat Genet. 2005;37:645–651. doi: 10.1038/ng1563. [DOI] [PubMed] [Google Scholar]
- Cho KK, Bear MF. Promoting neurological recovery of function via metaplasticity. Future Neurol. 2010;5:21–26. doi: 10.2217/fnl.09.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse M, Halabisky B, Harris J, Devidze N, Dubal DB, Sun B, Orr A, Lotz G, Kim DH, Hamto P, et al. Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature. 2011;469:47–52. doi: 10.1038/nature09635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapcote SJ, Lipina TV, Millar JK, Mackie S, Christie S, Ogawa F, Lerch JP, Trimble K, Uchiyama M, Sakuraba Y, et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron. 2007;54:387–402. doi: 10.1016/j.neuron.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Clapcote SJ, Roder JC. Deletion polymorphism of Disc1 is common to all 129 mouse substrains: implications for gene-targeting studies of brain function. Genetics. 2006;173:2407–2410. doi: 10.1534/genetics.106.060749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci U S A. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Jr., Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–923. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Pierce K, Schumann CM, Redcay E, Buckwalter JA, Kennedy DP, Morgan J. Mapping early brain development in autism. Neuron. 2007;56:399–413. doi: 10.1016/j.neuron.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Behavioral phenotyping strategies for mutant mice. Neuron. 2008;57:809–818. doi: 10.1016/j.neuron.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Crawley JN, Paylor R. A proposed test battery and constellations of specific behavioral paradigms to investigate the behavioral phenotypes of transgenic and knockout mice. Horm Behav. 1997;31:197–211. doi: 10.1006/hbeh.1997.1382. [DOI] [PubMed] [Google Scholar]
- Denayer A, Van Esch H, de Ravel T, Frijns JP, Van Buggenhout G, Vogels A, Devriendt K, Geutjens J, Thiry P, Swillen A. Neuropsychopathology in 7 Patients with the 22q13 Deletion Syndrome: Presence of Bipolar Disorder and Progressive Loss of Skills. Mol Syndromol. 2012;3:14–20. doi: 10.1159/000339119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SU, del Gaudio D, German JR, Peters SU, Ou Z, Bader PI, Berg JS, Blazo M, Brown CW, Graham BH, et al. 22q13.3 deletion syndrome: clinical and molecular analysis using array CGH. Am J Med Genet A. 2010;152A:573–581. doi: 10.1002/ajmg.a.33253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17:111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, Montcouquiol M, Sans N. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Synapse structure: glutamate receptors connected by the shanks. Curr Biol. 1999;9:R848–850. doi: 10.1016/s0960-9822(00)80043-3. [DOI] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Flint J, Mott R. Applying mouse complex-trait resources to behavioural genetics. Nature. 2008;456:724–727. doi: 10.1038/nature07630. [DOI] [PubMed] [Google Scholar]
- Folstein SE, Rosen-Sheidley B. Genetics of autism: complex aetiology for a heterogeneous disorder. Nat Rev Genet. 2001;2:943–955. doi: 10.1038/35103559. [DOI] [PubMed] [Google Scholar]
- Gandy S, DeKosky ST. Toward the treatment and prevention of Alzheimer’s disease: rational strategies and recent progress. Annu Rev Med. 2013;64:367–383. doi: 10.1146/annurev-med-092611-084441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Champagne N, Lafreniere RG, Xiong L, Spiegelman D, Brustein E, Lapointe M, Peng H, Cote M, Noreau A, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci U S A. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Spiegelman D, Piton A, Lafreniere RG, Laurent S, St-Onge J, Lapointe L, Hamdan FF, Cossette P, Mottron L, et al. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:421–424. doi: 10.1002/ajmg.b.30822. [DOI] [PubMed] [Google Scholar]
- Gejman PV, Sanders AR, Kendler KS. Genetics of Schizophrenia: New Findings and Challenges. Annu Rev Genomics Hum Genet. 2011 doi: 10.1146/annurev-genom-082410-101459. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Gong X, Jiang YW, Zhang X, An Y, Zhang J, Wu Y, Wang J, Sun Y, Liu Y, Gao X, et al. High proportion of 22q13 deletions and SHANK3 mutations in Chinese patients with intellectual disability. PLoS One. 2012;7:e34739. doi: 10.1371/journal.pone.0034739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabrucker AM, Knight MJ, Proepper C, Bockmann J, Joubert M, Rowan M, Nienhaus GU, Garner CC, Bowie JU, Kreutz MR, et al. Concerted action of zinc and ProSAP/Shank in synaptogenesis and synapse maturation. EMBO J. 2011a;30:569–581. doi: 10.1038/emboj.2010.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabrucker AM, Schmeisser MJ, Schoen M, Boeckers TM. Postsynaptic ProSAP/Shank scaffolds in the cross-hair of synaptopathies. Trends Cell Biol. 2011b;21:594–603. doi: 10.1016/j.tcb.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Gundelfinger ED, Boeckers TM, Baron MK, Bowie JU. A role for zinc in postsynaptic density asSAMbly and plasticity? Trends Biochem Sci. 2006;31:366–373. doi: 10.1016/j.tibs.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, Higashi K, Park AR, Spiegelman D, Dobrzeniecka S, Piton A, et al. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet. 2011;88:306–316. doi: 10.1016/j.ajhg.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MK, Tang C, Verpelli C, Narayanan R, Stearns MH, Xu RM, Li H, Sala C, Hayashi Y. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell. 2009;137:159–171. doi: 10.1016/j.cell.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holy TE, Guo Z. Ultrasonic songs of male mice. PLoS Biol. 2005;3:e386. doi: 10.1371/journal.pbio.0030386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, et al. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Sung CC, Brito IL, Sheng M. Degradation of postsynaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLoS One. 2010;5:e9842. doi: 10.1371/journal.pone.0009842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Iskenderian-Epps WS, Imperiali B. Modulation of Shank3 PDZ domain ligand-binding affinity by dimerization. Chembiochem. 2010;11:1979–1984. doi: 10.1002/cbic.201000246. [DOI] [PubMed] [Google Scholar]
- Jeffries AR, Curran S, Elmslie F, Sharma A, Wenger S, Hummel M, Powell J. Molecular and phenotypic characterization of ring chromosome 22. Am J Med Genet A. 2005;137:139–147. doi: 10.1002/ajmg.a.30780. [DOI] [PubMed] [Google Scholar]
- Kanner L. Autistic disturbances of affective contact. Nervous. 1943;Child:2–217. [PubMed] [Google Scholar]
- Kas MJ, Fernandes C, Schalkwyk LC, Collier DA. Genetics of behavioural domains across the neuropsychiatric spectrum; of mice and men. Mol Psychiatry. 2007;12:324–330. doi: 10.1038/sj.mp.4001979. [DOI] [PubMed] [Google Scholar]
- Kim JY, Duan X, Liu CY, Jang MH, Guo JU, Pow-anpongkul N, Kang E, Song H, Ming GL. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron. 2009;63:761–773. doi: 10.1016/j.neuron.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreienkamp HJ. Scaffolding proteins at the postsynaptic density: shank as the architectural framework. Handb Exp. 2008;Pharmacol:365–380. doi: 10.1007/978-3-540-72843-6_15. [DOI] [PubMed] [Google Scholar]
- Krueger DD, Bear MF. Toward fulfilling the promise of molecular medicine in fragile X syndrome. Annu Rev Med. 2011;62:411–429. doi: 10.1146/annurev-med-061109-134644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G, Konyukh M, Chaste P, Ey E, Rastam M, et al. Genetic and Functional Analyses of SHANK2 Mutations Suggest a Multiple Hit Model of Autism Spectrum Disorders. PLoS Genet. 2012;8:e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M, Kim E. Characterization of the Shank family of synaptic proteins. Multiple. 1999;J:Biol–Chem. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- Lord C, Cook EH, Leventhal BL, Amaral DG. Autism spectrum disorders. Neuron. 2000a;28:355–363. doi: 10.1016/s0896-6273(00)00115-x. [DOI] [PubMed] [Google Scholar]
- Lord C, Leventhal BL, Cook EH., Jr. Quantifying the phenotype in autism spectrum disorders. Am J Med Genet. 2001;105:36–38. [PubMed] [Google Scholar]
- Lord C, Risi S, Lambrecht L, Cook EH, Jr., Leventhal BL, DiLavore PC, Pickles A, Rutter M. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 2000b;30:205–223. [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Malinow R. Alzheimer’s disease: Recollection of lost memories. Nature. 2011;469:44–45. doi: 10.1038/469044a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams RR, Gidey E, Fouassier L, Weed SA, Doctor RB. Characterization of an ankyrin repeat-containing Shank2 isoform (Shank2E) in liver epithelial cells. Biochem J. 2004;380:181–191. doi: 10.1042/BJ20031577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Sudhof TC. Neurexins: three genes and 1001 products. Trends Genet. 1998;14:20–26. doi: 10.1016/S0168-9525(97)01324-3. [DOI] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, Scherer SW. Contribution of SHANK3 Mutations to Autism Spectrum Disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-De-Luca D, Mulle JG, Kaminsky EB, Sanders SJ, Myers SM, Adam MP, Pakula AT, Eisenhauer NJ, Uhas K, Weik L, et al. Deletion 17q12 is a recurrent copy number variant that confers high risk of autism and schizophrenia. Am J Hum Genet. 2010;87:618–630. doi: 10.1016/j.ajhg.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RG. Spatial localization does not require the presence of local cues. Learning and Motivation. 1981;12:239–260. [Google Scholar]
- Moy SS, Nadler JJ, Magnuson TR, Crawley JN. Mouse models of autism spectrum disorders: the challenge for behavioral genetics. Am J Med Genet C Semin Med Genet. 2006;142:40–51. doi: 10.1002/ajmg.c.30081. [DOI] [PubMed] [Google Scholar]
- Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2012a;44:471. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012b doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Kubota T, Nakamura Y, Murakami R, Nishikubo T, Tanaka I, Takahashi Y, Hayashi S, Imoto I, Inazawa J, et al. 22q13 Microduplication in two patients with common clinical manifestations: a recognizable syndrome? Am J Med Genet A. 2007;143A:2804–2809. doi: 10.1002/ajmg.a.31771. [DOI] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011 doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan K. 22q13.3 Deletion Syndrome. In: Pagon Roberta A, Bird Thomas C, Dolan Cynthia R, Stephens Karen., editors. Genetest, E.-i.-c. 2007. Seattle, Internet. [Google Scholar]
- Phelan MC. Deletion 22q13.3 syndrome. Orphanet J Rare Dis. 2008;3:14. doi: 10.1186/1750-1172-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe A, Boddaert N, Vaivre-Douret L, Robel L, Danon-Boileau L, Malan V, de Blois MC, Heron D, Colleaux L, Golse B, et al. Neurobehavioral profile and brain imaging study of the 22q13. 3 deletion syndrome in childhood. Pediatrics. 2008;122:e376–382. doi: 10.1542/peds.2007-2584. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redecker P, Bockmann J, Bockers TM. Expression of postsynaptic density proteins of the ProSAP/Shank family in the thymus. Histochem Cell Biol. 2006 doi: 10.1007/s00418-006-0199-9. [DOI] [PubMed] [Google Scholar]
- Roussignol G, Ango F, Romorini S, Tu JC, Sala C, Worley PF, Bockaert J, Fagni L. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J Neurosci. 2005;25:3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL. Three hypotheses for developmental defects that may underlie some forms of autism spectrum disorder. Curr Opin Neurol. 2010;23:118–123. doi: 10.1097/WCO.0b013e328336eb13. [DOI] [PubMed] [Google Scholar]
- Sala C, Piech V, Wilson NR, Passafaro M, Liu G, Sheng M. Regulation of dendritic spine morphology and synaptic function by Shank and Homer. Neuron. 2001;31:115–130. doi: 10.1016/s0896-6273(01)00339-7. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple Recurrent De Novo CNVs, Including Duplications of the 7q11.23 Williams Syndrome Region, Are Strongly Associated with Autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, Dilullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012 doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasua SM, Dwivedi A, Boccuto L, Rollins JD, Chen CF, Rogers RC, Phelan K, Dupont BR, Collins JS. Association between deletion size and important phenotypes expands the genomic region of interest in Phelan-McDermid syndrome (22q13 deletion syndrome) J Med Genet. 2011 doi: 10.1136/jmedgenet-2011-100225. [DOI] [PubMed] [Google Scholar]
- Sato D, Lionel AC, Leblond CS, Prasad A, Pinto D, Walker S, O’Connor I, Russell C, Drmic IE, Hamdan FF, et al. SHANK1 Deletions in Males with Autism Spectrum Disorder. Am J Hum Genet. 2012;90:879–887. doi: 10.1016/j.ajhg.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Stryker MP. Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. Proc Natl Acad Sci U S A. 2010;107:5611–5616. doi: 10.1073/pnas.1001281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scattoni ML, Crawley J, Ricceri L. Ultrasonic vocalizations: a tool for behavioural phenotyping of mouse models of neurodevelopmental disorders. Neurosci Biobehav Rev. 2009;33:508–515. doi: 10.1016/j.neubiorev.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CP, Sabo A, Sakai Y, Crosby J, Muzny D, Hawes A, Lewis L, Akbar H, Varghese R, Boerwinkle E, et al. Oligogenic heterozygosity in individuals with high-functioning autism spectrum disorder. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A, Janssen AL, Udvardi PT, Shiban E, Spilker C, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113(Pt 11):1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- Silverman JL, Turner SM, Barkan CL, Tolu SS, Saxena R, Hung AY, Sheng M, Crawley JN. Sociability and motor functions in Shank1 mutant mice. Brain Res. 2011;1380:120–137. doi: 10.1016/j.brainres.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- State MW. Another piece of the autism puzzle. Nat Genet. 2010a;42:478–479. doi: 10.1038/ng0610-478. [DOI] [PubMed] [Google Scholar]
- State MW. The genetics of child psychiatric disorders: focus on autism and Tourette syndrome. Neuron. 2010b;68:254–269. doi: 10.1016/j.neuron.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao-Cheng JH, Dosemeci A, Gallant PE, Smith C, Reese T. Activity induced changes in the distribution of Shanks at hippocampal synapses. Neuroscience. 2010;168:11–17. doi: 10.1016/j.neuroscience.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- Uchino S, Wada H, Honda S, Nakamura Y, Ondo Y, Uchiyama T, Tsutsumi M, Suzuki E, Hirasawa T, Kohsaka S. Direct interaction of post-synaptic density-95/Dlg/ZO-1 domain-containing synaptic molecule Shank3 with GluR1 alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor. J Neurochem. 2006;97:1203–1214. doi: 10.1111/j.1471-4159.2006.03831.x. [DOI] [PubMed] [Google Scholar]
- Vaillend C, Rampon C, Davis S, Laroche S. Gene control of synaptic plasticity and memory formation: implications for diseases and therapeutic strategies. Curr Mol Med. 2002;2:613–628. doi: 10.2174/1566524023361952. [DOI] [PubMed] [Google Scholar]
- Verhoeven WM, Egger JI, Cohen-Snuijf R, Kant SG, de Leeuw N. Phelan-McDermid syndrome: Clinical report of a 70-year-old woman. Am J Med Genet A. 2013;161:158–161. doi: 10.1002/ajmg.a.35597. [DOI] [PubMed] [Google Scholar]
- Verhoeven WM, Egger JI, Willemsen MH, de Leijer GJ, Kleefstra T. Phelan-McDermid syndrome in two adult brothers: atypical bipolar disorder as its psychopathological phenotype? Neuropsychiatr Dis Treat. 2012;8:175–179. doi: 10.2147/NDT.S30506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpelli C, Dvoretskova E, Vicidomini C, Rossi F, Chiappalone M, Schoen M, Di Stefano B, Mantegazza R, Broccoli V, Boeckers TM, et al. Importance of shank3 in regulating metabotropic glutamate receptor 5 (mGluR5) expression and signaling at synapses. J Biol Chem. 2011 doi: 10.1074/jbc.M111.258384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmar FR, State M, Klin A. Autism and autism spectrum disorders: diagnostic issues for the coming decade. J Child Psychol Psychiatry. 2009;50:108–115. doi: 10.1111/j.1469-7610.2008.02010.x. [DOI] [PubMed] [Google Scholar]
- Waga C, Okamoto N, Ondo Y, Fukumura-Kato R, Goto Y, Kohsaka S, Uchino S. Novel variants of the SHANK3 gene in Japanese autistic patients with severe delayed speech development. Psychiatr Genet. 2011;21:208–211. doi: 10.1097/YPG.0b013e328341e069. [DOI] [PubMed] [Google Scholar]
- Wang X, McCoy P, Rodriguiz RM, Pan Y, Je HS, Roberts A, Kim C, Berrios J, Colvin JS, Bousquet-Moore D, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson HL, Crolla JA, Walker D, Artifoni L, Dallapiccola B, Takano T, Vasudevan P, Huang S, Maloney V, Yobb T, et al. Interstitial 22q13 deletions: genes other than SHANK3 have major effects on cognitive and language development. Eur J Hum Genet. 2008 doi: 10.1038/ejhg.2008.107. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Wong AC, Shaw SR, Tse WY, Stapleton GA, Phelan MC, Hu S, Marshall J, McDermid HE. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohr M, Roullet FI, Hung AY, Sheng M, Crawley JN. Communication impairments in mice lacking Shank1: reduced levels of ultrasonic vocalizations and scent marking behavior. PLoS One. 2011;6:e20631. doi: 10.1371/journal.pone.0020631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, Ha S, Chung C, Jung ES, Cho YS, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- Wong AC, Ning Y, Flint J, Clark K, Dumanski JP, Ledbetter DH, McDermid HE. Molecular characterization of a 130-kb terminal microdeletion at 22q in a child with mild mental retardation. Am J Hum Genet. 1997;60:113–120. [PMC free article] [PubMed] [Google Scholar]
- Yang M, Bozdagi O, Scattoni ML, Wohr M, Roullet FI, Katz AM, Abrams DN, Kalikhman D, Simon H, Woldeyohannes L, et al. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. J Neurosci. 2012;32:6525–6541. doi: 10.1523/JNEUROSCI.6107-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, Prakash R, Weinberg RJ, Ehlers MD, Philpot BD. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12:777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]