Abstract
The recently developed field of ligand homology modeling, LHM, that extends the ideas of protein homology modeling to the prediction of ligand binding sites and for use in virtual ligand screening has emerged as a powerful new approach. Unlike traditional docking methodologies, LHM can be applied to low-to-moderate resolution predicted as well as experimental structures with little if any diminution in performance; thereby enabling ~75% of an average proteome to have potentially significant virtual screening predictions. In large scale benchmarking, LHM is able to predict off-target ligand binding. Thus, despite the widespread belief to the contrary, low-to-moderate resolution predicted structures have considerable utility for biochemical function prediction.
Introduction
Over the past decade, the field of protein structure prediction has matured to the point where a significant fraction of the proteins in a given proteome can be modeled at low-to-moderate resolution [1]. On the other hand, the biochemical function of many proteins in a proteome, most especially those associated with ligand binding and other intermolecular interactions, are only partially known [2]. For example, the metabolic enzymes of well-studied organisms such as yeast are not fully characterized [3,4]. Thus, a key question facing the field is can predicted protein structures be successfully employed for the prediction of protein function? Of course, function is multifaceted, but clearly the inference of biochemical function would be the most direct application of structural information. In this review, we focus on the utility of predicted protein structures in the identification of ligand binding sites, and having identified these sites, their usefulness in virtual ligand screening to assist in drug discovery. But, before embarking on a discussion of the utility of lower resolution structures, a brief summary of the status of the field when high-resolution structures are used is appropriate as it provides the standard by which newly developed approaches must be assessed.
Binding site detection in high-resolution structures
Having a three-dimensional structure in hand, one would like to identify its small molecule binding sites. Some approaches locate binding sites by a geometric match to three-dimensional descriptors or templates of biologically relevant sites [5,6]. More powerful is the evolutionary trace methodology that combines protein structure with conserved residue patterns mapped onto the protein’s surface [7–9]. There are also geometric methods that locate binding residues by searching for cavities/pockets in a protein’s structure [10,11]. Among the best pocket detection algorithms is LIGSITECSC [12] that calculates surface-accessibility on the protein’s Connolly surface [13] and then re-ranks the pockets by the degree of conservation of select surface residues. Other methods calculate titration curves [14] or identify electrostatically destabilized residues[15]. These methods strictly focus on the protein’s sequence and structural features and ignore the identity of the ligand, but they are a necessary first step.
Virtual ligand screening using high-resolution structures
Having identified a binding site in a structure, the next step is to identify its binding ligands. Most traditional approaches are docking-based and prioritize compounds by predicting their binding mode [16] and then binding affinity [17]. Here, high-resolution structures of the target protein receptor, preferably in its ligand-bound conformational state, are generally required [18]. There are many successful self-docking studies where the ligand is excised from its crystal structure and then redocked [19]. However, many proteins exhibit significant motion upon ligand binding [20,21], and even small motions diminish docking accuracy. For example, for trypsin, HIV-1 protease and thrombin, ~90% of initial docking accuracy is lost when the mean protein structural rearrangement exceeds 1.5 Å [22]. These results raise the following questions: Are ligand binding sites really so structurally unique in nature and if not, why are high-resolution structures needed for ligand docking?
Does the need for high-resolution structures in binding site prediction and virtual screening reflect physical principles or is it just a technical limitation?
There is the widespread belief that predicted structures whose backbone RMSD ranges from 2–6 A are useless for either ligand binding site prediction or for virtual ligand screening [22]. For example, the performance of the LigsiteCSC [12] pocket detection algorithm deteriorates dramatically as one goes from crystal structures to predicted models in large scale benchmark tests [20]. However, local structural distortions are routine in nature [23]. For example, the binding sites of distantly related native proteins that bind very similar, if not identical ligands, with similar residues have an average pairwise backbone RMSD of 2.15 ± 0.77 Å [24]. As a specific example, for the subset of the kinome having holo crystal structures, the structural variation of the “conserved” ATP-binding site is ~2.4 Å [25]. Thus, there is significant structural plasticity of ligand binding sites [23,26]; it is unlikely that there is a unique ligand-protein conformation, with other nearby conformations having an entirely unfavorable binding free energy. The observed ensemble of native ligand binding conformations also suggests that low-resolution models might be useful for binding site identification/virtual screening provided that they capture the majority of the structural features and essential interactions.
Why then do extant docking methods [16,27–31] require high-resolution structures? One underlying cause is the fact that they are driven by steric and van der Waals interactions [32]. A slight conformational inaccuracy could cause dramatic interaction change. If a ligand fits into the binding site, then ligand ranking is dominated by the molecular weight of the ligand, independent of whether the cognate ligand or a randomized version is used [32]. Thus, there is the need for a more accurate atomic force to be developed. However, if the resulting force field is too complex, it would have limited practical utility as it must be able to screen millions of compounds across the thousands of proteins in the human or other proteomes [33].
Ligand Homology Modeling: Binding site detection and virtual ligand screening
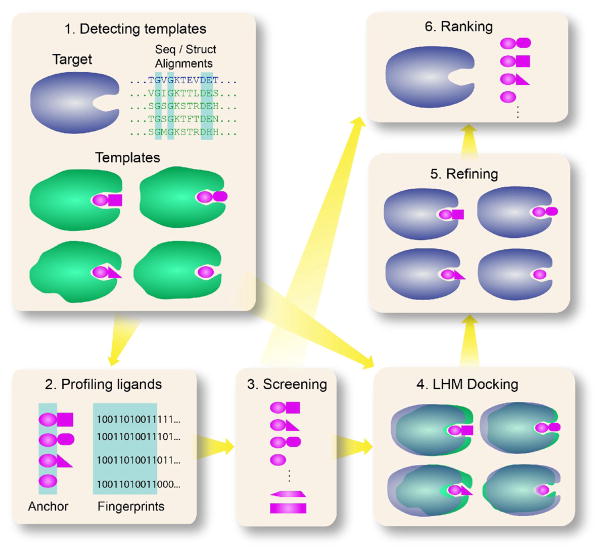
To employ protein models requires approaches that can accommodate binding site structural variations without a significant diminution in accuracy. As a first approximation, one might imagine that global structural similarity between proteins would be sufficient to infer protein function [34], most especially, common binding sites. In a recent study [35], for structurally related proteins whose pairwise sequence identity is in the twilight zone, we concluded that even at quite high levels of structural similarity, less than 25% of the targets share a common binding pocket. Thus, structural similarity alone is insufficient to transfer binding site location. A class of methods that exhibits the desired insensitivity to receptor structure deformation and which allows one to infer binding site location and type of ligands bound is Ligand Homology Modeling (LHM) [36–43]. LHM exploits the fact that the ideas of homology modeling, as applied to protein structure prediction [44], are applicable to functional inference, ligand binding pose prediction and virtual ligand screening. As shown schematically in Figure 1, LHM consists of six steps:
Figure 1.
Flowchart of Ligand Homology Modeling (LHM). Target and template proteins are colored in blue and green, respectively, and ligands are colored in purple.
Functional relationships between evolutionarily distant proteins are detected by sequence profile-driven threading to identify common ligand binding pockets, functionally important residues and structural conservation (anchors) of their ligand binding modes [37].
These conserved features are used to construct a ligand fingerprint profile from the identified template ligands [45].
Initial virtual screening of ligands is then done via fingerprint scanning.
The small molecule ligands are placed in the protein’s predicted binding site using the conserved ligand anchor regions identified in (1) [37]. Interestingly, the pose of the anchor in the ligand binding site tends to be strongly conserved, as are the residues contacting the ligand. Furthermore, the B-factors of the residues touching the ligand’s anchor are lower than those outside the anchor region.
The ligand’s pose is readjusted to optimize its interactions with the protein’s structure [40]. We further found that the positions of the side chain functional groups in contact with the ligand anchor functional groups tend to be strongly conserved and act together as a structural unit [46]. Indeed, they can refine the backbone geometry. This is in contrast to traditional ligand docking where the protein’s structure is held fixed and the ligand conformation is adjusted to accommodate the protein’s structure [32,47].
Using the refined conformations, the ligand library is then re-ranked via a machine learning procedure [37,42].
One of the advantages of LHM is that binding site detection is quite insensitive to structural quality. For example, consider the results when FINDSITE [20] was applied to a representative benchmark set, none of whose templates are closer than 35% identical. We consider the prediction of a binding site to be successful when the centers of mass of the predicted and observed binding sites are < 4 Å. Using crystal structures, for the best of top five predicted ligand-binding sites, the success rate for FINDSITE is 70.9% versus 51.3% for LIGSITECSC. For TASSER [48] predicted models, FINDSITE has a 67.3% success rate, whereas LIGSITECSC’s success rate is 32.5%. Similar results have been reported for binding site detection by other LHM variants [41,43,49]. LHM has also been applied to predict metal binding sites [1,50]. For example, FINDSITE-metal identifies the metal binding site in TASSER models in 59.4% of the cases. Moreover, when the metal is iron, copper, zinc, calcium, and magnesium ions, the identity of the binding site metal can be predicted with 70% – 90% accuracy.
What happens when holo templates are unavailable for the target of interest?
While contemporary structure prediction approaches provide sufficiently accurate models for about 76% of the proteins in the human proteome < 1000 residues in length [1], because of the relative scarcity of solved holo template structures in the PDB [51,52], one can only infer ligand binding information for ~26% of the human proteome [53]. Thus, methods that do not require holo template structures must be developed. To address this, FINDSITEX [53], an extension of FINDSITE [20], was developed that uses predicted structures for template proteins having experimental ligand binding information but which lack solved structures. Thus, pseudo holo templates are generated. To provide predicted protein structures, a fast and accurate version of TASSERVMT [54], TASSERVMT-lite, for template-based structural modeling was developed and tested, with comparable performance as the best CASP9 servers [55]. Then, a hybrid approach that combines structure alignments with an evolutionary similarity score for identifying functional relationships between target and template proteins with binding data was formulated.
FINDSITEX was applied to all identified human G-protein coupled receptors (GPCRs). First, TASSERVMT-lite improved models of all previously modeled human GPCR structures [56]. We then used these structures to screen against the ZINC8 [57] non-redundant (Tanimoto coefficient [58], TC<0.7) ligand set of 88,949 compounds combined with ligands from the GLIDA database[59]. Testing FINDSITEX (excluding GPCRs from the binding data library whose sequence identity > 30% to the target protein) on a 168 protein human GPCR set with known binders, the average enrichment factor in the top 1% of the compound library (EF0.01) is 22.7, with encouraging results for off-target interaction predictions. All 998 predicted human GPCR structures, virtual screening results and predicted off-target interactions are available at [60].
Combined LHM approaches to proteome scale virtual ligand screening
To combine the advantages of information provided by distant holo templates when they are available with experimental data and using pseudo holo templates when they are not, FINDSITEcomb was developed [61]. A significant component of FINDSITEcomb, is an improved version of FINDSITE, FINDSITEfilt that filters out false positive ligands in threading identified templates by a better binding site detection procedure that includes binding site amino acid similarity. For virtual ligand screening, FINDSITEcomb combines FINDSITEfilt with FINDSITEX that uses the ChEMBL[2] and DrugBank[62] ligand binding databases. The rank of each screened ligand is the best of its three ranks to ligands using fingerprints derived from the PDB, ChEMBL, and DrugBank libraries. In what follows, we summarize the results of FINDSITEcomb in benchmarking mode, where all template proteins with > 30% sequence identity to a target are excluded. We note that in large scale testing FINDSITEcomb produces significant virtual screening predictions for about 75% of an average proteome [33].
Comparison of LHM to traditional docking approaches
The DUD set is designed to help test docking algorithms by providing challenging decoys [63]. For each active, there are 36 decoys with similar physical properties (e.g. molecular weight, calculated LogP) but dissimilar chemical topology. Table 1 compares the relative performance of FINDSITEcomb with traditional docking methods, including AUTODOCK Vina [47] and DOCK 6 [31], in cross docking (a realistic scenario), where all 97,974 non-redundant DUD ligands are screened against all targets, as well as in non cross docking, where screening is just done against the experimentally determined active and inactive molecules. Results are presented for crystal structures and TASSERVMT-lite modeled structures. For both cases, each FINDSITE component performs better than AUTODOCK Vina or DOCK 6. While the performance of traditional methods deteriorates when models are used, FINDSITE based approaches do not. Finally, in [64], several docking programs were compared for virtual screening accuracy in non cross docking on experimental structures on DUD. FINDSITEcomb, whose mean average area under the ROC curve, the Area Under the Accumulation Curve, AUAC=0.77, performs as well as the best performing GLIDE (v4.5) [28] (mean AUAC=0.72). FINDSITEcomb performs better than all other compared methods: DOCK 6 (mean AUAC=0.55), FlexX [30] (mean AUAC=0.61), ICM [27] (mean AUAC=0.63), PhDOCK (mean AUAC=0.59) [29,65,66] and Surflex [64] (mean AUAC=0.66). Table 2 shows the AUAC values using both experimental and modeled structures for FINDSITEcomb with AUAC =0.77 and 0.75 respectively, as well as its constituent components for both experiment and modeled structures. As in Table 1, the dominant contribution to the success of FINDSITEcomb is due to FINDSITEfilt whose AUAC=0.74 for experimental and modeled structures is the same.
Table 1.
Comparison of virtual screening approaches on the DUD benchmark using experimental and modeled structures
| Cross Docking | Non Cross Docking | |||
|---|---|---|---|---|
| Method | Average EF0.01a Experimental Structures | Average EF0.01a Modeled Structures | Average EF0.01a Experimental Structures | Average EF0.01a Modeled Structures |
| FINDSITEX | 16.89 | 20.05 | 5.92 | 8.24 |
| FINDSITEfilt | 22.32 | 21.26 | 11.0 | 11.3 |
| FINDSITEcomb | 27.69 | 23.10 | 14.1 | 13.3 |
| AUTODOCK | 8.92 | 2.17 | 5.45 | 2.48 |
| Vina | ||||
| DOCK 6 | 3.14 | 3.05 | 3.82 | 1.29 |
EF0.01 is the enrichment factor relative to random for the top 1% of ranked molecules.
Table 2.
AUAC values of different FINDSITE methods for DUD and 3,576 DrugBank targets
| DUD non cross docking | DrugBank | ||
|---|---|---|---|
| Experimental structures | Modeled structures | Modeled structures | |
| FINDSITEcomb | 0.77 | 0.75 | 0.87 |
| FINDSITEfilt | 0.74 | 0.74 | 0.86 |
| FINDSITEX | 0.67 | 0.70 | 0.69 |
| FINDSITE | - | 0.60 | |
In addition to being broadly applicable, FINDSITEcomb is considerably faster than traditional docking methods. On a single state of the art CPU, for a 325 residue protein screened against 100,000 compounds, FINDSITEcomb is ~ 30 times faster than AUTODOCK Vina [47] and ~160 times faster than DOCK 6 [31]. Thus, FINDSITEcomb can be applied to screen millions of compounds on a proteomic scale. Despite the fact that predicted models rather than high-resolution crystal structures are used, LHM methods are very strongly competitive with traditional docking approaches.
Large scale benchmarking tests on drug target proteins and the prediction of off-target interactions
FINDSITEcomb was tested in benchmarking mode on all 3,576 DrugBank [67] targets <1000 residues in length. Target and template structures are modeled with TASSERVMT-lite [53]. The screened compound library consists of all 6,507 drugs (the true binders of all targets) plus 67,871 ZINC8 non-redundant (culled to TC<0.7) compounds [57] as background. The results of FINDSITEcomb along with its component methods and the original FINDSITE [20] are compiled in Table 3. FINDSITEcomb is better than any of its component methods. Table 3 also shows that FINDSITEfilt is better than FINDSITE [20] by a significant ~45% for EF0.01 (46.0 vs. 31.7), as well as in its coverage of targets with EF0.01> 1 (58% vs. 43%). FINDSITEcomb has an average EF0.01 of 52.1 and is better than random (EF0.01> 1) for 65% of the targets. Finally, Table 2, column 4, shows the AUAC results for FINDSITE FINDSITEcomb where AUAC=0.87 and its constituent components. As in the DUD benchmark, the performance of FINDSITEcomb is dominated by FINDSITEfilt.
Table 3.
Performance of different FINDSITE based methods for 3,576 DrugBank targets
| Method | Average EF0.01 | # (%) of targets having EF0.01 > 1 |
|---|---|---|
| FINDSITE(PDB) | 31.7 | 1526 (43%) |
| FINDSITEX | 36.6 | 1714 (48%) |
| FINDSITEfilt(PDB) | 46.0 | 2080 (58%) |
| FINDSITEcomb | 52.1 | 2333 (65%) |
Another application of the LHM approach was in the structural and functional characterization of the entire human kinome [25]. Encouraging virtual screening results were presented for ligands predicted to bind to the conserved ATP-binding pocket [57]. In a more rigorous test, cross-reactivity virtual profiling of the human kinome was done. For almost 70% of the inhibitors, their alternate molecular targets can be effectively identified in the human kinome with a high (>0.5) sensitivity, yet relatively low false positive rate (<0.5) [68].
Conclusions
Just as the field of protein structure prediction has greatly benefited by the development of template based approaches [55,69], we argue that the ligand homology modeling [1,20,37,38,41–43] has matured to the point where LHM is a powerful method for the prediction of ligand binding sites and virtual ligand screening. It offers the advantages that predicted as well as high-resolution structures can be successfully used, with minor diminution in performance. While certainly not perfect, in virtual screening LHM results are often considerably better than random and could be used to guide experimental screening approaches.
As noted by Bourne and coworkers [70,71] and is evident from an analysis of DrugBank [62] targets, the binding of ligand to a protein target other than the one for which the drug was designed is quite common [67,72]. Moreover, in PDB structures, very similar binding sites are found in globally unrelated proteins [73]. The challenge will be to extend these observations to predicted low-to-moderate resolution protein structures and then to apply them on a proteomic scale. If so, LHM could be a powerful tool to help repurpose FDA approved drugs and could help with the elucidation of metabolic pathways [74]. These and other related applications will undoubtedly be pursued in the near future.
Highlights.
Ligand homology modeling, LHM, can use low-to-moderate resolution models for binding site predictions and virtual screening.
LHM performs better with protein models than traditional approaches do for crystal structures.
LHM is applicable to ~75% of an average proteome.
LHM is a promising approach to repurpose FDA approved drugs.
Acknowledgments
This work was supported in part by grant Nos. GM-48835, GM-37048 and GM-084222 of the Division of General Medical Sciences of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brylinski M, Skolnick J. FINDSITE-metal: Integrating evolutionary information and machine learning for structure-based metal-binding site prediction at the proteome level. Proteins-Structure Function and Bioinformatics. 2011;79:735–751. doi: 10.1002/prot.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic acids research. 2011;39:D691–697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobson PD, Smallbone K, Jameson D, Simeonidis E, Lanthaler K, Pir P, Lu CA, Swainston N, Dunn WB, Fisher P, et al. Further developments towards a genome-scale metabolic model of yeast. Bmc Systems Biology. 2010:4. doi: 10.1186/1752-0509-4-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiehn O, Barupal DK, Kind T. Extending Biochemical Databases by Metabolomic Surveys. Journal of Biological Chemistry. 2011;286:23637–23643. doi: 10.1074/jbc.R110.173617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pazos F, Sternberg MJ. Automated prediction of protein function and detection of functional sites from structure. Proc Natl Acad Sci U S A. 2004;101:14754–14759. doi: 10.1073/pnas.0404569101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleming K, Kelley LA, Islam SA, MacCallum RM, Muller A, Pazos F, Sternberg MJ. The proteome: structure, function and evolution. Philos Trans R Soc Lond B Biol Sci. 2006;361:441–451. doi: 10.1098/rstb.2005.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •7.Erdin S, Ward RM, Venner E, Lichtarge O. Evolutionary trace annotation of protein function in the structural proteome. J Mol Biol. 2010;396:1451–1473. doi: 10.1016/j.jmb.2009.12.037. Describes the proteome scale application of the powerful Evolutionary Trace approach that enables the GO functions of proteins to be assigned for proteins of unknown function solved by the Structural Genomics projects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lichtarge O, Wilkins A. Evolution: a guide to perturb protein function and networks. Curr Opin Struct Biol. 2010;20:351–359. doi: 10.1016/j.sbi.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkins A, Erdin S, Lua R, Lichtarge O. Evolutionary trace for prediction and redesign of protein functional sites. Methods in molecular biology. 2012;819:29–42. doi: 10.1007/978-1-61779-465-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glaser F, Morris RJ, Najmanovich RJ, Laskowski RA, Thornton JM. A method for localizing ligand binding pockets in protein structures. Proteins. 2006;62:479–488. doi: 10.1002/prot.20769. [DOI] [PubMed] [Google Scholar]
- 11.Hendlich M, Rippmann F, Barnickel G. LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J Mol Graph Model. 1997;15:359–363. 389. doi: 10.1016/s1093-3263(98)00002-3. [DOI] [PubMed] [Google Scholar]
- •12.Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. A widely used method for identifying binding sites in experimental structures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly M. Analytical molecular surface calculation. Journal of Applied Crystallography. 1983;16:548–558. [Google Scholar]
- 14.Ondrechen MJ, Clifton JG, Ringe D. THEMATICS: a simple computational predictor of enzyme function from structure. Proc Natl Acad Sci U S A. 2001;98:12473–12478. doi: 10.1073/pnas.211436698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elcock AH. Prediction of functionally important residues based solely on the computed energetics of protein structure. J Mol Biol. 2001;312:885–896. doi: 10.1006/jmbi.2001.5009. [DOI] [PubMed] [Google Scholar]
- 16.Meiler J, Baker D. ROSETTALIGAND: protein-small molecule docking with full side-chain flexibility. Proteins. 2006;65:538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 17.Kroemer RT. Structure-based drug design: docking and scoring. Curr Protein Pept Sci. 2007;8:312–328. doi: 10.2174/138920307781369382. [DOI] [PubMed] [Google Scholar]
- 18.McGovern SL, Shoichet BK. Information decay in molecular docking screens against holo, apo, and modeled conformations of enzymes. J Med Chem. 2003;46:2895–2907. doi: 10.1021/jm0300330. [DOI] [PubMed] [Google Scholar]
- 19.Kroemer RT, Vulpetti A, McDonald JJ, Rohrer DC, Trosset JY, Giordanetto F, Cotesta S, McMartin C, Kihlen M, Stouten PF. Assessment of docking poses: interactions-based accuracy classification (IBAC) versus crystal structure deviations. J Chem Inf Comput Sci. 2004;44:871–881. doi: 10.1021/ci049970m. [DOI] [PubMed] [Google Scholar]
- ••20.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci U S A. 2008;105:129–134. doi: 10.1073/pnas.0707684105. One of the first papers that described Ligand Homology Modeling and that showed the widespread utility of predicted structures for GO functional inference and binding site identification. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karthikeyan S, Zhou Q, Osterman AL, Zhang H. Ligand binding-induced conformational changes in riboflavin kinase: structural basis for the ordered mechanism. Biochemistry. 2003;42:12532–12538. doi: 10.1021/bi035450t. [DOI] [PubMed] [Google Scholar]
- 22.Erickson JA, Jalaie M, Robertson DH, Lewis RA, Vieth M. Lessons in molecular recognition: the effects of ligand and protein flexibility on molecular docking accuracy. J Med Chem. 2004;47:45–55. doi: 10.1021/jm030209y. [DOI] [PubMed] [Google Scholar]
- 23.Amemiya T, Koike R, Kidera A, Ota M. PSCDB: a database for protein structural change upon ligand binding. Nucleic Acids Research. 2012;40:D554–D558. doi: 10.1093/nar/gkr966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skolnick J, Brylinski M. FINDSITE: a combined evolution/structure-based approach to protein function prediction. Briefings in Bioinformatics. 2009;10:378–391. doi: 10.1093/bib/bbp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •25.Brylinski M, Skolnick J. Comprehensive Structural and Functional Characterization of the Human Kinome by Protein Structure Modeling and Ligand Virtual Screening. Journal of Chemical Information and Modeling. 2010;50:1839–1854. doi: 10.1021/ci100235n. Presents structural models of the entire human kinome and shows that such models are useful in virtual ligand screening by LHM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brylinski M, Skolnick J. What is the relationship between the global structures of apo and holo proteins? Proteins-Structure Function and Bioinformatics. 2008;70:363–377. doi: 10.1002/prot.21510. [DOI] [PubMed] [Google Scholar]
- 27.Abagyan R, Totrov M, Kuznetsov D. ICM - a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comput Chem. 1994;15:488–506. [Google Scholar]
- 28.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 29.Jain AN. Surflex-Dock 2.1: Robust performance from ligand energetic modeling, ring flexibility, and knowledge-based search. J Comput-Aided Mol Des. 2007;21:281–306. doi: 10.1007/s10822-007-9114-2. [DOI] [PubMed] [Google Scholar]
- 30.Kramer B, Rarey M, Lengauer T. Evaluation of the FLEXX incremental construction algorithm for protein-ligand docking. Proteins. 1999;37:228–241. doi: 10.1002/(sici)1097-0134(19991101)37:2<228::aid-prot8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 31.Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V, Rizzo RC, Case DA, James TL, Kuntz ID. DOCK 6: combining techniques to model RNA-small molecule complexes. RNA. 2009;15:1219–1230. doi: 10.1261/rna.1563609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim R, Skolnick J. Assessment of programs for ligand binding affinity prediction. Journal of Computational Chemistry. 2008;29:1316–1331. doi: 10.1002/jcc.20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.SUNPRO: A Database of Structure & FUNction Predictions of Proteins from Representative Organisms on World Wide Web. URL: http://cssb.biology.gatech.edu/sunpro/index.html.
- 34.Fischer M, Zhang QC, Dey F, Chen BY, Honig B, Petrey D. MarkUs: a server to navigate sequence-structure-function space. Nucleic Acids Res. 2011;39:W357–361. doi: 10.1093/nar/gkr468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brylinski M, Skolnick J. Comparison of structure-based and threading-based approaches to protein functional annotation. Proteins. 2010;78:118–134. doi: 10.1002/prot.22566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brylinski M, Skolnick J. Q-Dock: Low-resolution flexible ligand docking with pocket-specific threading restraints. J Comput Chem. 2008 doi: 10.1002/jcc.20917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •37.Brylinski M, Skolnick J. FINDSITE(LHM): a threading-based approach to ligand homology modeling. PLoS Comput Biol. 2009;5:e1000405. doi: 10.1371/journal.pcbi.1000405. Demonstrates the existence of structural and chemically conserved anchor regions in ligands that can be used to increase the accuracy and speed of ligand docking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brylinski M, Skolnick J. FINDSITE: a threading-based approach to ligand homology modeling. PLoS computational biology. 2009;5:e1000405. doi: 10.1371/journal.pcbi.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skolnick J, Brylinski M. FINDSITE: a combined evolution/structure-based approach to protein function prediction. Brief Bioinform. 2009;10:378–391. doi: 10.1093/bib/bbp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brylinski M, Skolnick J. Q-Dock(LHM): Low-resolution refinement for ligand comparative modeling. J Comput Chem. 2010;31:1093–1105. doi: 10.1002/jcc.21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •41.Wass MN, Kelley LA, Sternberg MJE. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Research. 2010;38:W469–W473. doi: 10.1093/nar/gkq406. A powerful LHM approach for ligand binding site prediction that performed very well in CASP8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HS, Zhang Y. BSP-SLIM: A blind low-resolution ligand-protein docking approach using predicted protein structures. Proteins-Structure Function and Bioinformatics. 2012;80:93–110. doi: 10.1002/prot.23165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •43.Roy A, Yang JY, Zhang Y. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Research. 2012;40:W471–W477. doi: 10.1093/nar/gks372. An LHM approach to binding site identification that improves upon FINDSITE by better describing the ligand binding site and that performed well in CASP9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aliso Viejo CA, editor. Daylight Theory Manual. 4.9. Daylight Chemical Information Systems, Inc; 2007. [Google Scholar]
- 46.Brylinski M, Lee SY, Zhou HY, Skolnick J. The utility of geometrical and chemical restraint information extracted from predicted ligand-binding sites in protein structure refinement. Journal of Structural Biology. 2011;173:558–569. doi: 10.1016/j.jsb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •47.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. A widely used traditional approach to molecular docking and virtual ligand screening. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci U S A. 2004;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roy A, Zhang Y. Recognizing Protein-Ligand Binding Sites by Global Structural Alignment and Local Geometry Refinement. Structure. 2012;20:987–997. doi: 10.1016/j.str.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mallick M, Vidyarthi AS, Shankaracharya Tools for Predicting Metal Binding Sites in Protein: A Review. Current Bioinformatics. 2011;6:444–449. [Google Scholar]
- 51.Aloy P, Querol E, Aviles FX, Sternberg MJE. Automated structure-based prediction of functional sites in proteins: Applications to assessing the validity of inheriting protein function from homology in genome annotation and to protein docking. Journal of Molecular Biology. 2001;311:395–408. doi: 10.1006/jmbi.2001.4870. [DOI] [PubMed] [Google Scholar]
- 52.Aloy P, Pichaud M, Russell RB. Protein complexes: structure prediction challenges for the 21st century. Curr Opin Struct Biol. 2005;15:15–22. doi: 10.1016/j.sbi.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 53.Zhou HY, Skolnick J. FINDSITEx: A Structure-Based, Small Molecule Virtual Screening Approach with Application to All Identified Human GPCRs. Molecular Pharmaceutics. 2012;9:1775–1784. doi: 10.1021/mp3000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou H, Skolnick J. Template-based protein structure modeling using TASSER(VMT) Proteins. 2011 doi: 10.1002/prot.23183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kryshtafovych A, Fidelis K, Moult J. CASP9 results compared to those of previous CASP experiments. Proteins. 2011;79 (Suppl 10):196–207. doi: 10.1002/prot.23182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Devries ME, Skolnick J. Structure Modeling of All Identified G Protein-Coupled Receptors in the Human Genome. PLoS Comput Biol. 2006;2:e13. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Irwin JJ, Shoichet BK. ZINC--a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanimoto T. An elementary mathematical theory of classification and prediction. IBM Interanl Report 1958. 1958:1958. [Google Scholar]
- 59.Okuno Y, Tamon A, Yabuuchi H, Niijima S, Minowa Y, Tonomura K, Kunimoto R, Feng C. GLIDA: GPCR--ligand database for chemical genomics drug discovery--database and tools update. Nucleic Acids Res. 2008;36:D907–912. doi: 10.1093/nar/gkm948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Predictions of all human GPCR structures, virtual screening and predicted off-target interactions on World Wide Web. URL: http://cssb.biology.gatech.edu/skolnick/webservice/gpcr/index.html.
- ••61.Zhou H, Skolnick J. FINDSITEcomb: A threading/structure-based, proteomic-scale virtual ligand screening approach. Journal of Chemical Information and Modeling. 2012 doi: 10.1021/ci300510n. in press. A combined, next generation, LHM approach that uses both holo templates and pseudo holo templates for virtual ligand screening that gives quite good performance on the DUD benchmark as well as for DrugBank drug targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •63.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J Med Chem. 2006;49:6789–6801. doi: 10.1021/jm0608356. A much needed and widely used benchmark set to assess the utility of ligand docking algorithms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cross JB, Thompson DC, Rai BK, Baber JC, Fan KY, Hu Y, Humblet C. Comparison of Several Molecular Docking Programs: Pose Prediction and Virtual Screening Accuracy. J Chem Inf Model. 2009;49:1455–1474. doi: 10.1021/ci900056c. [DOI] [PubMed] [Google Scholar]
- 65.Jain AN. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 66.Pham TA, Jain AN. Parameter Estimation for Scoring Protein-Ligand Interactions Using Negative Training Data. J Med Chem. 2006;49:5856–5868. doi: 10.1021/jm050040j. [DOI] [PubMed] [Google Scholar]
- 67.Wishart DS. DrugBank and its relevance to pharmacogenomics. Pharmacogenomics. 2008;9:1155–1162. doi: 10.2217/14622416.9.8.1155. [DOI] [PubMed] [Google Scholar]
- 68.Brylinski M, Skolnick J. Cross-Reactivity Virtual Profiling of the Human Kinome by X-React(KIN): A Chemical Systems Biology Approach. Molecular Pharmaceutics. 2010;7:2324–2333. doi: 10.1021/mp1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu D, Zhang J, Roy A, Zhang Y. Automated protein structure modeling in CASP9 by I-TASSER pipeline combined with QUARK-based ab initio folding and FG-MD-based structure refinement. Proteins. 2011;79 (Suppl 10):147–160. doi: 10.1002/prot.23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ren JY, Xie L, Li WW, Bourne PE. SMAP-WS: a parallel web service for structural proteome-wide ligand-binding site comparison. Nucleic Acids Research. 2010;38:W441–W444. doi: 10.1093/nar/gkq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •71.Xie L, Xie L, Bourne PE. Structure-based systems biology for analyzing off-target binding. Current Opinion in Structural Biology. 2011;21:189–199. doi: 10.1016/j.sbi.2011.01.004. Argues that the off-targeting of drugs is the norm rather than the exception and describes proteome scale approaches to drug repurposing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Z, Li Y, Lin B, Schroeder M, Huang B. Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics. 2011;27:2083–2088. doi: 10.1093/bioinformatics/btr331. [DOI] [PubMed] [Google Scholar]
- ••73.Durrant JD, Amaro RE, Xie L, Urbaniak MD, Ferguson MAJ, Haapalainen A, Chen ZJ, Di Guilmi AM, Wunder F, Bourne PE, et al. A Multidimensional Strategy to Detect Polypharmacological Targets in the Absence of Structural and Sequence Homology. Plos Computational Biology. 2010;6 doi: 10.1371/journal.pcbi.1000648. A convicing and important paper which demonstrates that the notion of the “one-gene-one drug-one disease” paradigm of traditional drug discovery significantly underestimates the likelihood that a single drug interacts with multiple targets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Janga SC, Diaz-Mejia JJ, Moreno-Hagelsieb G. Network-based function prediction and interactomics: The case for metabolic enzymes. Metabolic Engineering. 2011;13:1–10. doi: 10.1016/j.ymben.2010.07.001. [DOI] [PubMed] [Google Scholar]