Abstract
Purpose
Magnetic resonance spectroscopy (MRS) of hyperpolarized substrates allows for the observation of label exchange catalyzed by enzymes providing a powerful tool to investigate tissue metabolism and potentially kinetics in vivo. However, the accuracy of current methods to calculate kinetic parameters have been limited by T1 relaxation effects, extracellular signal contributions, and reduced precision at lower SNR.
Theory and Methods
To address these challenges, we investigated a new modeling technique using Metabolic Activity Decomposition-Stimulated Echo Acquisition Mode (MAD-STEAM). The MAD-STEAM technique separates exchanging from non-exchanging metabolites providing twice the information as conventional techniques.
Results
This allowed for accurate measurements of rates of conversion and of multiple T1 values simultaneously using a single acquisition.
Conclusion
The additional measurement of T1 values for the reaction metabolites provides further biological information about the cellular environment of the metabolites. The new technique was investigated through simulations and in vivo studies of transgenic mouse models of cancer demonstrating improved assessments of kinetic rate constants and new T1 relaxation value measurements for hyperpolarized 13C-pyruvate, 13C-lactate and 13C -alanine.
Keywords: cancer, hyperpolarized Carbon-13 MRS, metabolism, stimulated-echo, STEAM, kinetics
Introduction
Magnetic resonance spectroscopy (MRS) of hyperpolarized substrates shows great promise in the development of new clinically relevant diagnostic indicators of disease [1–3]. Hyperpolarized (HP) MRS is advantageous over conventional radionuclide imaging in that it detects not only the injected substrate, but also the products of its biochemical reactions providing additional biologically important information [3, 4]. Prior to the introduction of HP MRS, noninvasive measures of flux through specific enzyme-catalyzed reactions have been limited by low SNR. Measurement of these fluxes could profoundly aid in the understanding of physiology and clinical medicine [4]. The 10,000 fold sensitivity enhancement gained by hyperpolarization of 13C nuclei offers the possibility to non-invasively measure fluxes through individual enzyme-catalyzed reactions in vivo and in real-time [4, 5].
Recently, time-resolved methods have been used to observe kinetics providing improved sensitivity to altered metabolism [6–16]. In ex vivo experiments, hyperpolarized 13C substrates have been used to measure flux through single enzyme-catalyzed steps [4]. In vivo, however, the acquired signal is affected by flow, perfusion, diffusion, and membrane transport, in addition to metabolism [1, 6–8]. To address these confounding factors, the stimulated echo acquisition mode (STEAM) approach included a diffusion preparation scheme, which can suppress signals from the vasculature and improve contrast for tissue metabolism [16,17].
Even with isolation of the signal to stationary tissue, robustly quantifying metabolism with kinetic parameters using HP MRS in vivo is difficult due to the lack of complexity of the dynamic curves and the number of free parameters that need to be estimated [9]. This problem arises from spin-relaxation occurring concurrently with exchange, making it difficult to differentiate metabolic conversion from relaxation [9]. This allows for linear dependency between parameters, which can contribute to poor conditioning for least squares minimization (LSM), and ultimately leading to reduced accuracy in estimation of rates of conversion and spin-relaxation.
Furthermore, the precision and accuracy of the quantitative analysis deteriorate as the signal-to-noise ratio (SNR) decreases [18]. This is important even in the field of hyperpolarized MRS, in which spatial resolution and time constraints in vivo may lead to lower SNR. Recently, saturation or inversion magnetization transfer (MT) [9,10], has been applied to hyperpolarized Carbon-13 MR to improve accuracy in the measurement of rates of conversion and observe real-time conversion. MT adds more linearly independent equations, requiring at least one extra excitation. Here, we applied a new modeling technique using Metabolic Activity Decomposition for reconstruction and a STEAM sequence for acquisition (MAD-STEAM). MAD-STEAM can be used to directly observe real-time conversion and separate exchanging from non-exchanging metabolites within a single acquisition [19]. The additional information from the MAD-STEAM experiment has the potential to improve the accuracy and precision of kinetic measurements even with low SNR by adding more linearly independent equations. The goal of this study was to investigate this new approach in both simulations and in vivo animal studies with quantitative comparisons to prior methods.
Hyperpolarized [1-13C]-pyruvate, in particular, has shown great promise as a potential probe for the presence of disease and its response to treatment [20–23]. Pyruvate is an important product of the glycolytic pathway, playing a central role in cellular metabolism. In most normal tissues under aerobic conditions, pyruvate dehydrogenase catalyzes the decarboxylation of a significant fraction of pyruvate to produce acetyl-coenzyme A, which is then used in the tricarboxylic acid cycle (TCA). Alternatively, pyruvate can be reduced to lactate in the reaction catalyzed by lactate dehydrogenase (LDH). In the presence of disease, however, the relative fluxes through these enzymes can be significantly altered [3]. For instance, even under aerobic conditions, the glycolytic flux in tumors is often upregulated, a phenomenon known as the Warburg effect, which leads to higher concentrations of lactate and an acidic tumor microenvironment [24–26]. This increased LDH flux is thought to be responsible for the increased labeling of lactate observed in tumors, as compared to the normal surrounding tissue, following injection of hyperpolarized [1-13C]-pyruvate [1–3]. Recently, it has been shown that there is a reduction in HP lactate detection in tumors responding to therapy [20–23]. It is widely accepted that these changes observed in lactate labeling are due to flux and exchange through the LDH enzyme [3]. To accurately quantify these changes in metabolism, a new modeling method was developed utilizing the extra information available with MAD-STEAM.
In addition to the high conversion to [1-13C]-lactate observed in many tumors, increased [1-13C]-alanine has been observed during tumor formation in a Myc-driven liver cancer model [27]. While it is well known that most cancers depend on a high rate of aerobic glycolysis for survival and proliferation [11, 12], surprisingly some cancers display glutamine addiction, having high glutamine uptake into the cell despite the fact that glutamine can be produced from glucose [28]. The conversion of [1-13C]-pyruvate to [1-13C]-alanine via alanine transaminase (ALT) is affected by the availability of the glutamate cofactor, produced from conversion of glutamine via glutaminase inside the cell. Using the new information available with MAD-STEAM, we sought to investigate the rate of conversion of pyruvate-to-alanine for the first time during disease progression in a Myc-driven model, in which both the Warburg effect and glutamine addiction have been reported [24,27–29].
This project was designed to increase specificity and accuracy in the measurement of spin-relaxation and multiple conversion pathways by applying MAD-STEAM to kinetic modeling and then evaluate the new information gained in transgenic models of cancer. A key benefit of measuring kinetics with the additional information from MAD-STEAM is that spin-relaxation and multiple rates of conversion can be measured simultaneously within a single acquisition. The additional parameters provided, such as the T1 spin-relaxation for the reaction metabolites, may yield further information about the cellular environment experienced by the metabolites. This new technique is also desirable since it improves accuracy by reducing sensitivity to background noise and moving spins within the vasculature. In this study, we applied our new modeling technique to measure real-time metabolic conversion and T1 spin-relaxation as kinetic markers of malignancy in vivo in transgenic mouse cancer models.
Theory
The MAD-STEAM Experiment
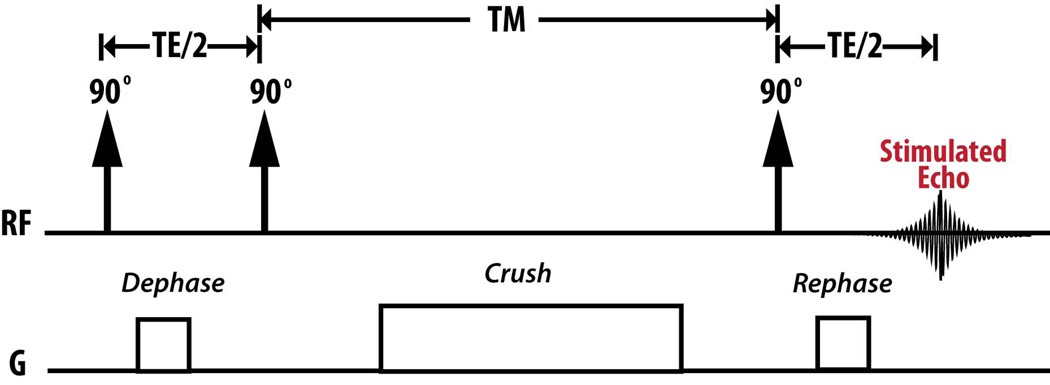
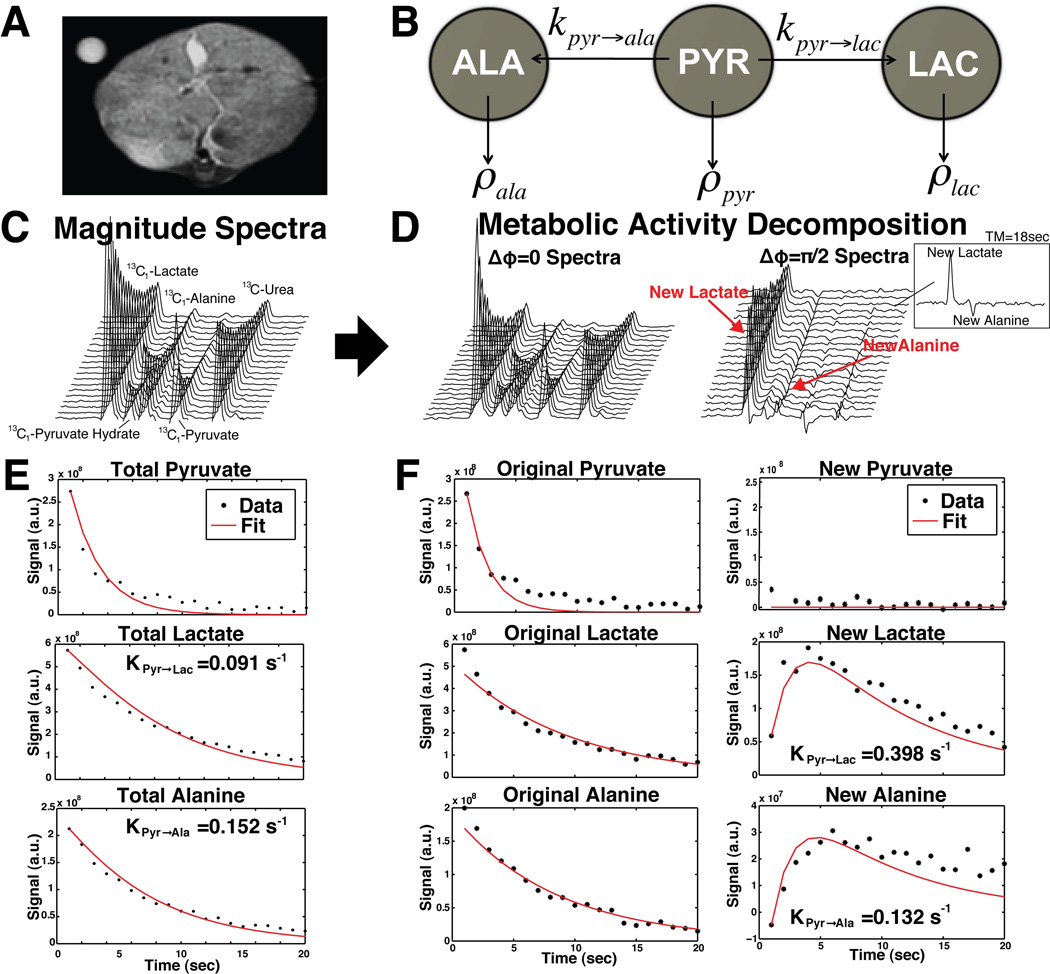
Recently Larson et al. showed that STEAM in the presence of metabolic conversion creates a phase shift that depends on the resonance frequency shift and echo time (TE), Δφ = 2πΔf (TE / 2)[19]. By choosing Δφ = ±π / 2, the metabolic conversion and exchange during a mixing time (TM) can be directly detected within a single acquisition [19]. The pulse sequence is shown in Figure 1. MAD-STEAM, which only uses a single acquisition, is advantageous for detecting hyperpolarized signals in vivo because magnetization cannot be renewed making it difficult to acquire multiple encoding steps. Ultrafast 2D NMR [30–32] could also be used to acquire similar information, however spatial inhomogeneity in vivo may cause distortion of the data [19].
Figure 1.
Metabolic Activity Decomposition with Stimulated Echo Acquisition Mode (MAD-STEAM) pulse sequence.
The MAD-STEAM experiment can be viewed as an extension of a 2D Exchange Spectroscopy (EXSY) NMR experiment [19,33]. The EXSY experiment also uses a stimulated-echo from three 90° pulses, in which the time between the first two 90° pulses, t1, is incremented to sample a entire 2D spectrum [19]. Instead of acquiring multiple acquisitions of incremented t1 durations, the MAD-STEAM experiment only samples t1 = TE / 2 such that the phase shift between the resonances of interest is ±π / 2 [19]. In other words, MAD-STEAM acquires the projection of a 2D EXSY spectrum storing cross peak information in phase. The MAD-STEAM approach is similar to 1D EXSY experiment except that it stores cross peak information in phase rather than interpreting spectra after an inversion [34]. A spin is “phase tagged” when there is an exchange reaction available and the acquired phase after exchange is Δφ = ±π / 2.
Here we used the “phase tagging” information encoded through the MAD-STEAM experiment to separate the total signal of a metabolite, Mx (t), into the signal contributions from spins that have not gone through exchange, Orig{Mx (t)}, and spins that have gone through exchange, New{Mx (t)}, to improve kinetic measurements for the first time. Recently Chen et al. and Larson et al. showed that a stimulated echo based MRS approaches could also be used to isolate signal in stationary tissue from spins within the vasculature [16,17,19], providing improved observation of active cellular metabolic conversion for hyperpolarized carbon-13 MR in vivo.
Kinetic Models
Previously, it has been shown that the lactate and pyruvate peak intensities can be fit to a simple two-site exchange model to give apparent rates of conversion [4, 7, 10, 11, 14],
| (1) |
where MPyr and MLac denote the pyruvate and lactate peak integrals, t is time, ρPyr and ρLac are pyruvate and lactate spin-relaxation rates, 1/ T1,x. KPyr→Lac and KLac→Pyr are the pyruvate and lactate apparent rates of conversion. Using MAD-STEAM the two-site exchange system can be described by the following equation:
| (2) |
where Orig{MPyr (t)} and Orig{MLac (t)} denote the pyruvate and lactate peak integrals with no phase shift Δφ = 0. New{MPyr (t)} and New{MLac (t)} denote the pyruvate and lactate peak integrals of z-magnetizations with phase shifts Δφ = −π / 2 and Δφ = +π / 2, respectively. The ±π / 2 phase shift will put the Orig{MPyr (t)} and Orig{MLac (t)} in the real channel and the New{MLac (t)} in the imaginary channel, such that they can easily be separated during reconstruction. For all models we assume that pyruvate and lactate are at steady state such that there is no significant influx into the slab. This is a valid assumption because the vascular component was suppressed by the STEAM sequence [16, 17], and the acquisition was started after the arrival of pyruvate into the tissue.
This technique can be expanded to a three-site exchange system, such as the conversion of pyruvate to either lactate or alanine. Using the total signal from the peak areas for each metabolite, the following equations can be used to describe the system:
| (3) |
Because pyruvate generated from lactate and pyruvate generated from alanine are in small amounts in the liver relative to the forward reactions, they were assumed to be negligible, and thus New{MPyr (t)} is not part of the model. This constrains the number of free parameters to the number of linearly independent equations per time point to a 1:1 ratio. Using MAD-STEAM, the three-site exchange system can be described by the following system of equations:
| (4) |
The Orig{MAla (t)} and New{MAla (t)} were calculated based on the complex alanine peak integral and separated using the phase shift, ΔφPyr→Ala = −π / 2.21 at 3T and TE=14ms, using the equations below:
| (5) |
and
| (6) |
Parameter Estimation
Common methods, such as Gauss-Newton algorithm (GNA), Levenberg-Marquardt algorithm (LMA), the Newton method and Generalized Gradient Descent, to estimate parameters from non-linear systems use the Jacobian Matrix, J, iteratively to linearize the least squares problem Jx = b, where x is the matrix of parameters to be solved and b is the data. When the condition number, , is large (e is error), even a small error in b (noise) may cause a large error in the solution, x. Thus, if the Jacobian becomes ill-conditioned, the least squares estimate amplifies the noise by a factor of κ, and results may be grossly inaccurate [35,36].
Linearization has been used to simplify calculations in exchange systems [15]. Linearization could also be used along with MAD-STEAM to simplify calculations. In these comparisons, linearization was not used to avoid error in sum of squares minimization. Because the error on the linearized data is different than the error on the actual data, the linear least squares solution to linearized models minimizes the sum of squares of residuals for the nonlinear form of the dependent variables rather than the actual dependent variable.
The number of parameters a system can estimate reproducibly and accurately is related to the number of unique eigenvalues of the matrix K from the general equation, . The following matrix was obtained with using only the total peak area of the data (Eq 1),
| (7) |
This matrix has two distinct real eigenvalues for the following range of parameters: KPyr→Lac = 0.05 − 0.5s−1, KPyr→Lac = 0.001− 0.05s−1, T1,Pyr = 10 − 50s, and T1,Lac =10 − 50s. Meanwhile, the following matrix obtained from using Metabolic Activity Decomposition (Eq. 2) has four distinct real eigenvalues:
| (8) |
This matrix should be much more reliable for the prediction of more parameters because it has more unique eigenvalues.
Methods
Data Fitting
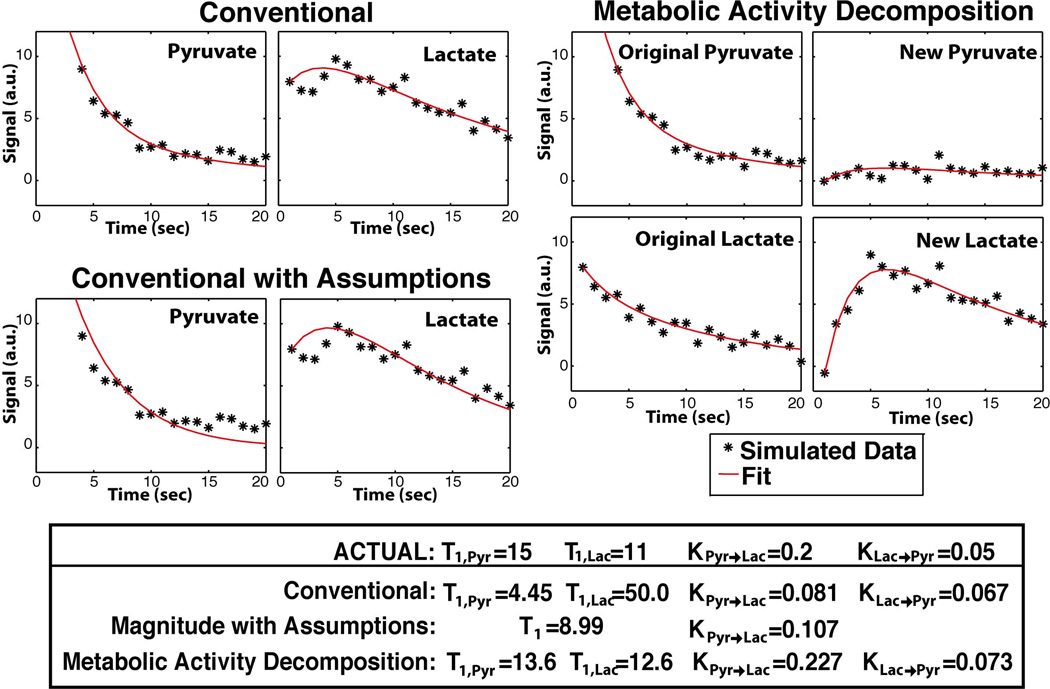
To solve the non-linear inverse problem, an iterative LMA was used in Matlab (The MathWorks Inc., Natick, MA), which interpolates between the method of gradient descent and the GNA [37–39]. This algorithm uses a trust region such that a reasonable approximation is always produced. Reasonable starting parameters, such as T1, x = 20s, KPyr→Lac = 0.05s−1, and KLac→Pyr = 0.01s−1, were used to speed up computation and improve convergence. A single simulation and fit are shown in Figure 2. All dynamic curves were normalized to the total carbon during the experiment not including 13C-Urea.
Figure 2.
Single simulation data and fitting results with Metabolic Activity Decomposition and simple two-site exchange with and without assumptions. Two-site Exchange with Assumptions assumes that KPyr→Lac = 0 and T1,Pyr = T1,Lac.
Simulated Data
Data were simulated for a 20s experiment over a range of parameters: KPyr→Lac = 0.01 − 0.1s−1, KLac→Pyr = 0.002 − 0.02s−1, KPyr→Ala = 0.005 − 0.1s−1, T1,Pyr = 10 − 40s, T1,Pyr = 10 − 40s, and T1,Ala =10 − 40s. Each combination was simulated over a range of SNRs (25–250 with five iterations each). The SNR was determined using the average signal of pyruvate at the first time point and dividing by the noise root mean square (rms) for both simulated data and in vivo data.
Hyperpolarization of [1-13C ]-Pyruvate and 13C -Urea
[1-13C]-Pyruvate mixed with the trityl radical OX063 (Tris[8-carboxyl-2,2,6,6tetra[2-(1hydroxyethyl)]-benzo(1,2-d:4,5-d)bis(1,3)dithiole-4-yl]methyl sodium salt, Oxford Instruments, Abingdon UK) and 13C-urea mixed with the trityl radical OX063 were simultaneously hyperpolarized [40] for phase correction [19] using conventional dynamic nuclear polarization (DNP) methods and a HyperSense DNP polarizer (Oxford Instruments, Abingdon, UK) operating at 3.3T and a temperature of 1.3K. This yields more than 40,000 fold increases in the pyruvate and urea signals [5]. The sample was dissolved to produce solutions with 80mM pyruvate and 80mM urea and a biologically appropriate pH (~7.4) with TRIS/NaOH/EDTA dissolution media.
Acquisition and Reconstruction
All data were acquired with a STEAM sequence slab selection, TE = 14ms, TMs starting at 50ms, 20 acquisitions, 1s temporal resolution, 256 spectral points, 2.5 kHz spectral bandwidth, a progressive flip angle scheme, and an adiabatic double spin echo [41]. A symmetrically sampled full echo was acquired to preserve phase information [19]. Copolarized 13C-urea was used as a phase reference to correct for phase shifts caused by homogeneous, bulk motion such as respiration, which would affect all metabolites [19].
For animal experiments, a 3T MRI system (GE Healthcare, Waukesha, WI, USA) was used with a dual-tuned mouse birdcage coil based on a design used previously [2, 27, 42]. Given the main field of 3T, the echo time TE=14ms was chosen. With TE=14ms, the phase of pyruvate to lactate conversion will be ΔφPyr→Lac = +π / 2 such that the new and original lactate are in quadrature. Since new and original alanine are not exactly in quadrature, the magnitude was broken into its components (Equation 5 and 6) based on ΔφPyr→Ala = −π / 2.21 in order to separate original and new alanine. This information was used for modeling three-site exchange in the liver cancer model, in which there is known high alanine production.
Animal Experiments
All animal studies were performed under a protocol approved by the UCSF Institutional Animal Care and Utilization Committee. Mice were anesthetized with 1–1.5% isoflurane and placed on a pad heated to 37°C during the MR experiment. Transgenic prostate cancer (TRAMP) mouse models at different stages of progression (n=5) and normal mice (n=5) were imaged for the two-site exchange study [2]. Data were acquired from a slab containing predominately the tumor or from a 20mm slab of the abdomen for normal mice. In the reproducibility study, data were acquired from a slab containing the liver of a normal mouse. For the three-site exchange study, a switchable transgenic liver cancer model was used (n=5). Following a baseline scan and hyperpolarized 13C study, doxycycline was removed from the diet to allow induction of expression of the human oncogenes Myc [43, 44] or Myc and Ras transgenes [45] and initiation of tumorigenesis. Data were acquired from a slab containing the entire liver but excluding the lungs and abdomen. During each study, 350 µL of the hyperpolarized [1-13C]-pyruvate solution was injected into the mouse over a 12s period, followed by a 0.15 mL normal saline flush. To reduce artifacts from the inflow of pyruvate, data acquisition began 25–30s following the start of injection of pyruvate. Axial, coronal, and sagittal T2-weighted fast spin-echo (FSE) images were acquired as anatomical references.
Enzyme Assays
LDH activity was measured in the transgenic liver cancer model (n=5) by observing the decrease in absorbance of reduced nicotinamide adenine dinucleotide (NADH) at 340 nm using a spectrophotometrical microplate reader (Saphire2, Tecan, Maennesorf, Switzerland). Multiple samples from tumor and adjacent tissue were frozen in liquid nitrogen and homogenized in modified RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5 % Na-deoxycholate, 1% Triton X 100, 0.1 % SDS, 2mM EDTA). Protein determination was performed using a BCA Protein Assay Kit (Pierce, Rockford, IL, USA). For determination of LDH activity, a standard curve was acquired using L-LDH from bovine heart (1000 units × mL−1, Sigma-Aldrich, St. Louis, MO, USA). Activity was measured in the presence of 0.15 mM NADH (Sigma-Aldrich) and 0.8 mM pyruvate (Sigma-Aldrich) in 50 mM Tris-HCl, pH 7.4 for 5 min.
Results
Simulations
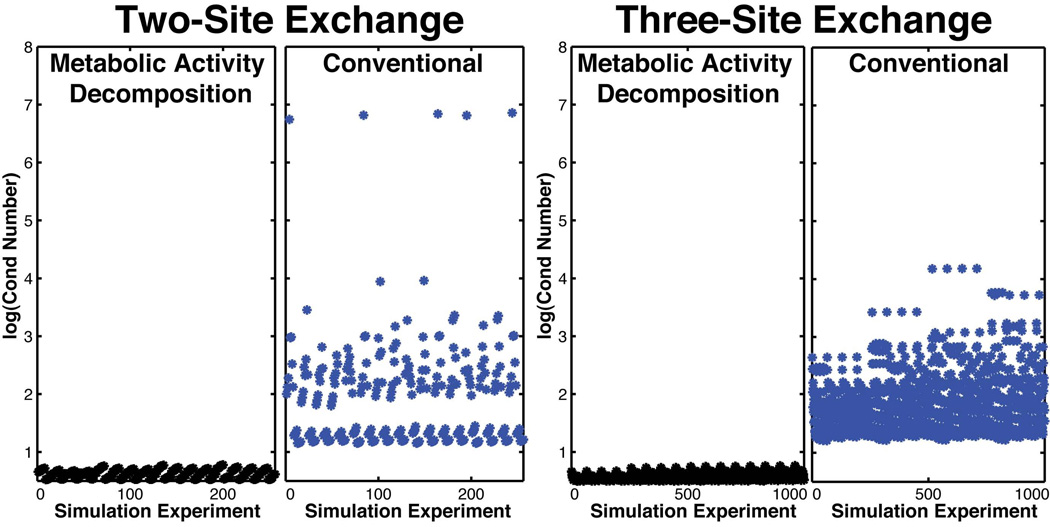
Conventional two-site exchange yields four unknowns but only two equations at each time point, causing unstable conditioning of the Jacobian matrix (Figure 3). When using conventional modeling, the condition number of the Jacobian matrix dramatically worsened when both the forward and backward rates are slow and the relaxation rates are similar, suggesting linear dependency between parameters. Intuitively, in these parameter ranges, conventional modeling is unable to distinguish forward from backward conversion and relaxation. Simulated data, shown in Figure 3 and Table 1, demonstrated that using only the total peak areas for a two-site exchange and three-site exchange model results in a miscalculation of the rate of conversion and is less accurate in calculating multiple effective T1s even when the model fits the data well (coefficients of correlation >0.8). For these nonlinear systems, goodness of fit does not necessarily correlate with accuracy.
Figure 3.
Comparison of stability and condition of (Left) two-site exchange with Eq. 1 using the total metabolite signal and two-site exchange with Eq. 2 using Metabolic Activity Decomposition and (Right) three-site exchange with Eq. 3 using the total metabolite signal versus three-site exchange with Eq. 4 using Metabolic Activity Decomposition. Comparison is over a wide range of parameters and all combinations in that range: T1,Pyr =10 − 40s, T1,Lac =10 − 40s, T1,Ala =10 − 40s, KPyr→Lac = 0.01 − 0.5s−1, KPyr→Ala = 0.005 − 0.1s−1, and KLac→Pyr = 0.01 − 0.1s−1. The condition number comes from Jacobian Matrix at the solution x.
Table 1.
Accuracy in Estimated Kinetics Parameters obtained using Metabolic Activity Decomposition and other Approaches to Kinetic Modeling.
| Two-Site Exchange | Three-Site Exchange | ||||
|---|---|---|---|---|---|
| SNR = 25 |
Metabolic Activity Decomposition |
Conventional |
Conventional with Assumptions a |
Metabolic Activity Decomposition |
Conventional |
| %Difference : KPyr→Lac | 8.7% | 338.6% | 43.7% | 2.9% | 30.8% |
| %Difference : KLac→Pyr | 21.9% | 570.7% | - | - | - |
| %Difference : KPyr→Ala | - | - | - | 8.1% | 65.6% |
| %Difference : T1,Pyr | 11.6% | 31.7% | - | 4.4% | 25.7% |
| %Difference : T1,Lac | 15.4% | 36.% | - | 3.1% | 16.6% |
| %Difference : T1,Ala | - | - | - | 4.6% | 22.7% |
| SNR = 50 | |||||
| %Difference : KPyr→Lac | 5.0% | 258.6% | 44.3% | 1.4% | 19.0% |
| %Difference : KLac→Pyr | 11.6% | 351.2% | - | - | - |
| %Difference : KPyr→Ala | - | - | - | 4.0% | 41.1% |
| %Difference : T1,Pyr | 7.3% | 26.2% | - | 2.4% | 15.6% |
| %Difference : T1,Lac | 8.6% | 27.4% | - | 1.5% | 11.4% |
| %Difference : T1,Ala | - | - | - | 2.3% | 15.0% |
| SNR = 100 | |||||
| %Difference : KPyr→Lac | 2.7 % | 113.2% | 44.7 % | 1.0% | 13.2% |
| %Difference : KLac→Pyr | 6.4 % | 205.6%d | - | - | - |
| %Difference : KPyr→Ala | - | - | - | 2.1% | 25.9% |
| %Difference : T1,Pyr | 4.5% | 19.4% | - | 1.1% | 9.5% |
| %Difference : T1,Lac | 5.3% | 26.8% | - | 0.7% | 7.2% |
| %Difference : T1,Ala | - | - | - | 1.2% | 10.0% |
Assumes T1,Pyr = T1,Lac and KLac→Pyr = 0.
Data is reported as average percent difference error between actual value and estimated value. Two-site exchange with Metabolic Activity Decomposition used Eq. 2 and conventional modeling used Eq. 1. Three-site exchange with Metabolic Activity Decomposition used Eq. 3 and conventional modeling used Eq. 4. The average percent difference, was found for n=5 simulation experiments per parameter combination, and SNR = 25–250 and without noise.
Comparison was over a range of physiologically expected parameters and all combinations in that range: KPyr→Lac = 0.01 − 0.1s−1, KLac→Pyr = 0.002 − 0.02s−1, KPyr→Ala = 0.005 − 0.1s−1, and T1,x = 10 − 40s.
Even small errors in the back reaction may appear large when the denominator is small: %Difference = (|Actual − Estimated| / Actual)*100%
To improve the fit, the number of free parameters can be constrained by assuming T1,Pyr = T1,Lac and KLac→Pyr = 0 [10,20]. However, in the case of low SNR, this approach can often underestimate the conversion rate KPyr→Lac compensating with inaccurate relaxation rates which do not describe the actual kinetics of the system, as shown in Table 1. At SNRs values typically seen in vivo [6] using Metabolic Activity Decomposition, the precision of quantitative analysis is high as shown in Table 1. Even at very low SNRs, the error measured was less than 20%.
Two-Site Exchange In Vivo
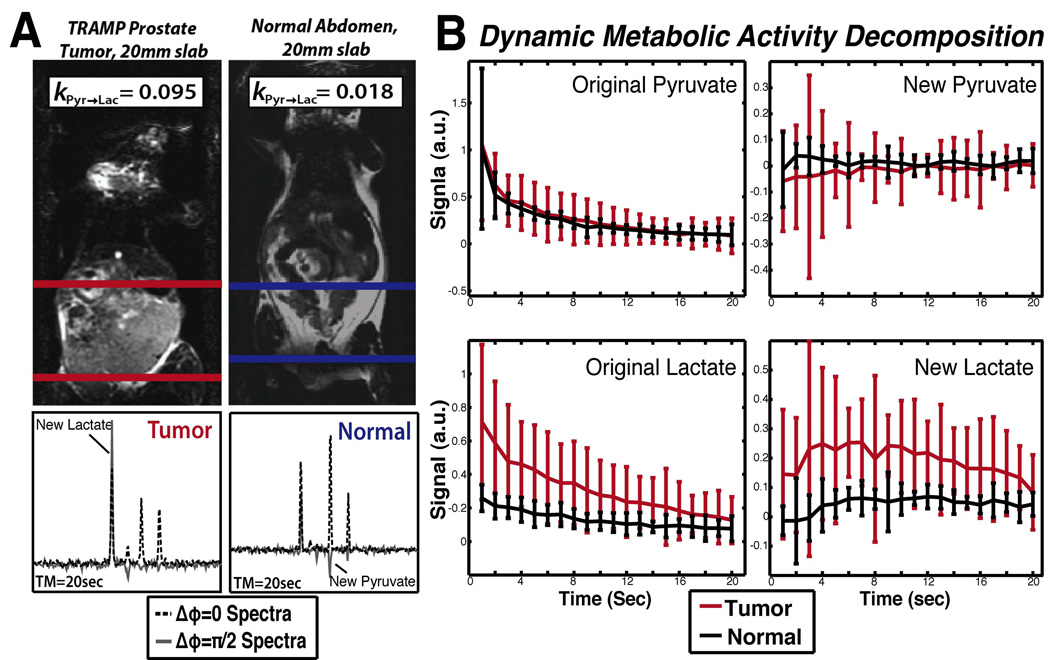
Using Metabolic Activity Decomposition, higher production of lactate in the tumor can be observed in the time series data, as shown in Figure 4. We observed high rates of conversion of pyruvate to lactate in a prostate tumor model (0.110±0.014s−1) versus the abdomen of a normal mouse (0.033±0.008s−1). In vivo, calculations with MAD-STEAM yielded apparent KPyr→Lac values with a larger effect size compared to conventional modeling (Table 2). Decreased apparent KLac→Pyr rates of conversion were also observed in tumors suggesting an increased unlabeled lactate pool-size and a high availability of NADH. Not only were the rates of conversion different in tumor tissue, but also, the effective T1,Lac s were different in tumors as shown in Table 2.
Figure 4.
(a) Sample spectra from tumor and normal locations and their corresponding slab locations. (b) Dynamic curves from Metabolic Activity Decomposition showing increased conversion of lactate to pyruvate and faster decay of the original lactate signal in TRAMP tumors versus normal tissue. Data was normalized to total carbon, excluding 13C-urea (n=5).
Table 2.
Comparison of rates on conversion in tumors versus in normal with Metabolic Activity Decomposition and conventional Two-site Exchange.
| KPyr→Lac | Tumor | Normal | P-value |
|---|---|---|---|
| Metabolic Activity Decomposition: | 0.110 ± 0.014 | 0.033 ± 0.008 | 0.005* |
| Conventional with Assumptionsb: | 0.068 ± 0.011 | 0.054 ± 0.008 | 0.385 |
| Conventional: | 0.150 ± 0.061 | 0.264 ± 0.069 | 0.164 |
| KLac→Pyr | |||
| Metabolic Activity Decomposition: | 0.001 ± 6.11×10−4 | 0.025 ± 0.007 | 0.028* |
| Conventional: | 0.153 ± 0.073 | 0.264 ±0.145 | 0.080 |
| T1,Pyr | |||
| Metabolic Activity Decomposition: | 0.120 ± 0.020 | 0.198 ± 0.035 | 0.123 |
| Conventional: | 0.197 ± 0.033 | 0.169 ± 0.020 | 0.271 |
| T1,Lac | |||
| Metabolic Activity Decomposition: | 0.105 ± 0.009 | 6.13×10−2 ± 0.007 | 0.026* |
| Conventional: | 0.038 ± 0.009 | 5.26×10−2 ±0.027 | 0.500 |
| T1,Both | |||
| Conventional with Assumptionsb: | 0.137 ± 0.011 | 0.170 ± 0.027 | 0.274 |
Data is reported as the mean ± mse with units s−1.
Assumes KLac→Pyr = 0 and T1,Pyr = T1,Lac
Two-sided unpaired students t-test (n=5,α=0.05).
Denotes significant differences (p-value < α).
Comparison of rates on conversion in tumor versus normal with Metabolic Activity Decomposition and conventional Two-site Exchange from slabs in the abdomen of TRAMP mice and normal mice. With this small sample size, significant differences were only detected when using Metabolic Activity Decomposition.
Multiple MAD-STEAM experiments were acquired in a single normal mouse liver using two-site exchange. This data showed reduced intrasubject variance in estimating KPyr→Lac using Metabolic Activity Decomposition (Mean = 0.079, σ = 0.012, Percent Error = 16.7%) versus using the magnitude of the spectra (Mean = 0.077, σ = 0.039, Percent Error = 50.2%) or the magnitude of the spectra with assumptions (Mean = 0.051, σ = 0.024, Percent Error = 46.9%). Large percent errors in parameter estimation in the three modeling techniques were attributed to the low SNR of lactate in normal tissue. Even with the lower SNR, Metabolic Activity Decomposition demonstrated improved reproducibility.
Three-Site Exchange In Vivo
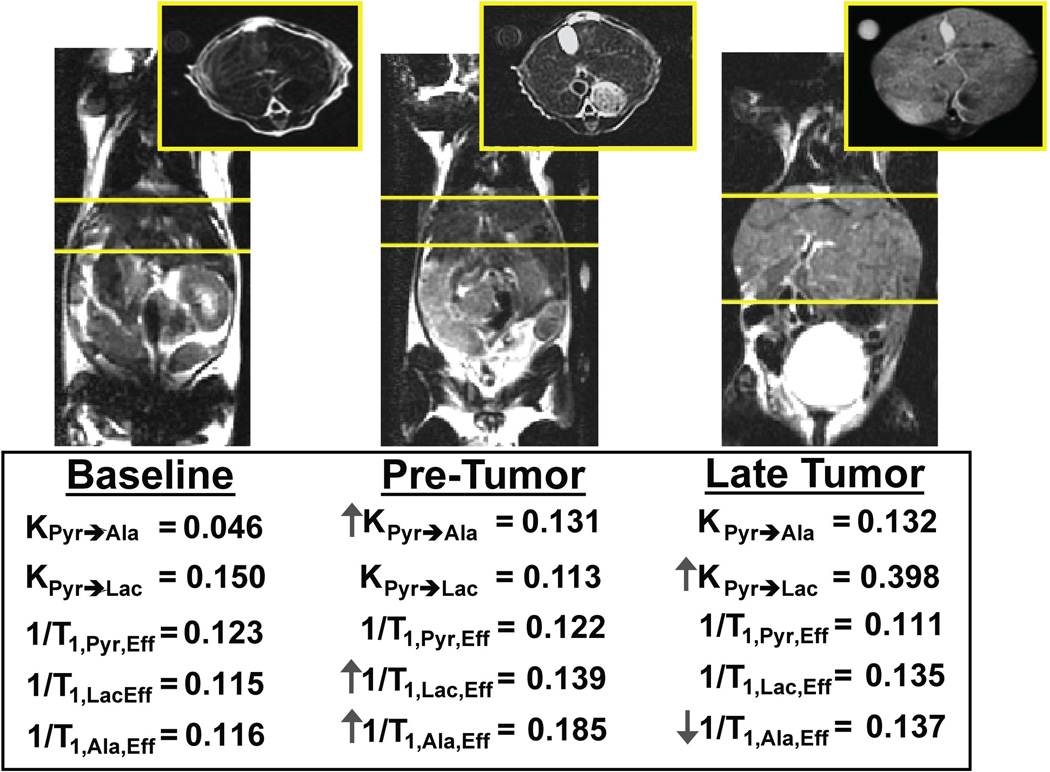
Representative data from a MAD-STEAM experiment and three-site exchange using Metabolic Activity Decomposition are shown in Table 2. Using Metabolic Activity Decomposition, real-time alanine conversion can be detected as seen in Figure 5D and 5F. Using Metabolic Activity Decomposition, changes in the rate of conversion of [1-13C]-pyruvate to [1-13C]-alanine were observed in addition to conversion to [1-13C]-lactate as shown in Figure 6. In this mouse model experiment, a three fold increase in KPyr→Ala was observed following oncogene expression in the pre-tumor state compared to baseline and a further three fold increase was detected when the tumor was at a late stage (baseline = 0.046, pre-tumor = 0.131, late tumor = 0.398). There was a 37.3% decrease in the effective T1,Ala from baseline to the pre-tumor state and a 26.8% increase in the effective T1,Ala from the pre-tumor to late state as shown in Figure 6. Additionally, there was a 17.3% decrease in the T1,Lac from baseline to the pre-tumor stage.
Figure 5.
(a) T2 -weighted axial anatomical image within the slab where data was acquired. (b) Schematic of the three-site exchange system. Stacked plots of the successively acquired (c) magnitude spectra, (d) real (Δφ = 0) and imaginary (Δφ = π/2) spectra from a MADSTEAM pulse sequence with TE=14ms. The areas under the curve from (e) the magnitude spectra with the corresponding three-site exchange fit Eq. 3 using only the total amount of each metabolite and (f) the areas under the curve from the real and imaginary spectra with the corresponding three-site exchange fit to Eq. 4 using original and new metabolite information provided by Metabolic Activity Decomposition.
Figure 6.
T2-weighted axial and coronal anatomical images showing progression in a switchable oncogene driven model of liver cancer and the corresponding rates of conversion calculated from three-site exchange using MAD-STEAM and Eq. 4 at each time point: baseline, two weeks (pre-tumor) and five weeks (late tumor) off doxycycline. Yellow lines contain the liver where data was acquired.
As supporting data, KPyr→Lac measurements were compared to enzymatic assays. KPyr→Lac measured with Metabolic Activity Decomposition (R2 = 0.809) demonstrated a higher correlation with LDH activity than modeling with the total signal of each metabolite (R2 = 0.343). KPyr→Lac measured with Metabolic Activity Decomposition also had a higher correlation to LDH activity assays than metabolite ratios such as the lactate-to-total carbon ratio (R2 = 0.530) excluding 13C -urea and the lactate-to-pyruvate ratio (R2 = 0.662) over the entire dynamic experiment and the lactate-to-pyruvate (R2 = 0.687) after perfusion of the pyruvate bolus (t=3s after start of acquisition, acquisition started at 30 seconds after start of pyruvate injection).
Discussion
Metabolic Activity Decomposition kinetic modeling of hyperpolarized MR data demonstrated improved precision in estimation of both rates of conversion and relaxation rates (Table 1). This improvement is attributed to additional information from the Metabolic Activity Decomposition method, which provides a well-conditioned system and reduces sensitivity to linear dependency from similar parameters (Figure 3). There is some bias in the data for all methods presented due to fitting to the data magnitude. The magnitude was used for fitting in vivo data because it is commonly used in the literature [3,8,10], and a main goal of this work was to compare to previous approaches. This bias could be removed by using complex data instead of using the magnitude.
Simulations over a wide range of relaxation and conversion rates demonstrated that this new modeling technique not only more accurately calculated these parameters, but it also was robust to noise with SNRs down to 10:1. Even though all models showed high goodness of fit, a good fit did not necessarily correlate with accurate estimates, suggesting that caution should be taken when evaluating the accuracy of these nonlinear models based on goodness of fit alone [46].
Even with the inherent 50% loss in SNR using a STEAM sequence compared to some other acquisition strategies [16], the accuracy gained from new “phase tagging” information with Metabolic Activity Decomposition far overcame any loss of accuracy from decreased SNR. For instance, at an SNR=100 the average error in estimation of KPyr→Lac for the two-site exchange was 113.0% with conventional modeling, but only 2.7% using Metabolic Activity Decomposition both with simulated data. Using only the conventional modeling, accuracy was improved to 44.7% error by assuming KLac→Pyr = 0 and T1,Pyr = T1,Lac. Even at a 50% lower SNR (SNR=50) using Metabolic Activity Decomposition, the average error was far less than all conventional approaches, 5.0%, as shown in Table 1. Similarly, the average error in estimation of KPyr→Lac for three-site exchange model was 13.21% at an SNR=100 using conventional modeling, but only 1.4% at an SNR=50 using Metabolic Activity Decomposition. Also, alternate preparation schemes have been shown improve SNR for the stimulated echo acquisition [16] and thus could substantially reduce such losses for MADSTEAM experiments as well.
In a comparison of tumor versus normal, using Metabolic Activity Decomposition to solve for rates of conversion and relaxation times had a larger effect size in both conversion and relaxation rates, allowing the new method to better distinguish changes in metabolism and cellular environment. In tumors, the measured KPyr→Lac values were significantly higher than normal, which agrees with prior findings that cancerous tissues have higher LDH-A expression [47]. Interestingly, effective T1,Lac was shorter in tumors compared to normal tissues suggesting an altered cellular environment for lactate in the tumors. Because the b-value increases during the experiment, the effective T1s measured in these experiments are a combination of both diffusion weighting as well as the longitudinal relaxation.
Improved reproducibility was also observed. The KPyr→Lac measured using Metabolic Activity Decomposition had an improved correlation with the measured enzyme activity compared to conventional modeling. This suggests that the new technique more accurately reflects enzymatic activity. Even with isolation of the detected signal to stationary tissue and “phase tagging,” other factors such as membrane transport were not accounted for in this model. Prior studies have isolated intracellular and extracellular metabolites and have shown that membrane transport can play a role in the measured rate of conversion [7]. The combination of this work with these methods would provide even further specificity for enzyme activity. Another future direction is the minimization of the voxel volume, which would allow for improved localization of tissue areas and could enable the study of smaller tumors and smaller regions of normal tissues.
When the modeling technique was used in a liver cancer model known to have high pyruvate conversion to alanine in the pre-tumor stage [13], the two-site exchange model is no longer valid. Here, the conversion rate of [1-13C]-pyruvate to [1-13C]-alanine was investigated for the first time during tumor formation. Because the phase shift of [1-13C]-pyruvate to [1-13C]-alanine with a 14ms echo time at 3T is approximately −π / 2, Metabolic Activity Decomposition could be used to model the three-site exchange system to obtain improved measurements of KPyr→Ala. In a transgenic oncogene-driven liver cancer model, changes in KPyr→Ala were observed between baseline, two weeks (pre-tumor), and five weeks (late tumor) off doxycycline in addition to changes in KPyr→Lac during progression, as shown in Figure 6.
Conclusions
This technique improved accuracy, simultaneously measured conversion rates and effective T1s, and suppressed vascular effects, thus providing new quantitative measures of relaxation and conversion rates. The ability to quantify rates of conversion with increased accuracy can improve current hyperpolarized kinetic models and could aid in the understanding of metabolic alterations in diseases including cancer. Meanwhile the T1 relaxation measurements obtained in this approach provide additional parameters for the investigating the effects of the intracellular environment on the metabolites of interest.
In vivo, the increased accuracy resulted in a larger effect size between tumors and normal tissue. KPyr→Lac increased in tumors, the back reaction KLac→Pyr decreased, and T1,Lac was shorter in tumors suggesting an altered cellular environment in the tumors. Moreover, the ability to detect changes between different stages of progression suggests that KPyr→Ala may also be valuable in addition to KPyr→Lac in investigating metabolism alterations with disease, particularly in case of liver cancer. Since it provided better measures of tumor kinetic parameters, this new acquisition and modeling technique demonstrated biomedical potential for monitoring cancer progression and response to therapy.
Acknowledgements
The authors acknowledge: Dr. Cornelius Von Morze, Galen Reed, Dr. Hikari Yoshihara, Illwoo Park, Peter Shin, Lucas Carvajal, Romelyn Delos Santos, Kristen Scott, Kayvan Keshari, Sukumar Subramaniam, Dr. Emma Essocks-Burns, and Mark Van Criekinge for assistance performing experiments, as well as Dr. Michael Lustig, Dr. Ralph Hurd and Dr. Murat Arcak for their expertise and Dr. James Tropp for the 1H/13C mouse coil, as well as, funding from NIH P41-EB013598, National Science Foundation Graduate Research Fellowship, NIGMS-IMSD R25-GM56847, NIH K99 EB012064, and UCSF Liver Center grant NIH P30 DK026743.
References
- 1.Golman K, Petersson JS. Metabolic imaging and other applications of hyperpolarized 13C1. Academic Radiology. 2006;13:932–942. doi: 10.1016/j.acra.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Albers MJ, Bok R, Chen AP, Cunningham CH, Zierhut ML, Zhang VY, Kohler SJ, Tropp J, Hurd RE, Yen YF, Nelson SJ, Vigneron DB, Kurhanewicz J. Hyperpolarized 13C lactate, pyruvate, and alanine: Noninvasive biomarkers for prostate cancer detection and grading. Cancer Research. 2008;68:8607–8615. doi: 10.1158/0008-5472.CAN-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurhanewicz J, Vigneron DB, Brindle K, Chekmenev EY, Comment A, Cunningham CH, DeBerardinis RH, Green GG, Leach MO, Rajan SS, Rizi RR, Ross BD, Warren WS, Malloy CR. Analysis of cancer metabolism by imaging hyperpolarized nuclei: Prospects for translation to clinical research. Neoplasia. 2011;13:81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merritt ME, Harrison C, Storey C, Jeffrey FM, Sherry AD, Malloy CR. Hyperpolarized 13C allows a direct measure of flux through a single enzyme-catalyzed step by NMR. Proceedings of the National Academy of Sciences. 2007;104:19773–19777. doi: 10.1073/pnas.0706235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proceedings of the National Academy of Sciences. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larson PEZ, Bok R, Kerr AB, Lustig M, Hu S, Chen AP, Nelson SJ, Pauly JM, Kurhanewicz J, Vigneron DB. Investigation of tumor hyperpolarized [1-13C]-pyruvate dynamics using time-resolved multiband RF excitation echo-planar MRSI. Magnetic Resonance in Medicine. 2010;63:582–591. doi: 10.1002/mrm.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris T, Eliyahu G, Frydman L, Degani H. Kinetics of hyperpolarized 13C1-pyruvate transport and metabolism in living human breast cancer cells. Proceedings of the National Academy of Sciences. 2009;106:18131–18136. doi: 10.1073/pnas.0909049106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zierhut ML, Yen YF, Chen AP, Bok R, Albers MJ, Zhang V, Tropp J, Park I, Vigneron DB, Kurhanewicz J, Hurd RE, Nelson SJ. Kinetic modeling of hyperpolarized 13C1-pyruvate metabolism in normal rats and tramp mice. Journal of Magnetic Resonance. 2010;202:85–92. doi: 10.1016/j.jmr.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng H, Lee Y, Hilty C. Quantitative rate determination by dynamic nuclear polarization enhanced NMR of a diels-alder reaction. Analytical Chemistry. 2010;82:8897–8902. doi: 10.1021/ac101670n. [DOI] [PubMed] [Google Scholar]
- 10.Kettunen MI, Hu De, Witney TH, McLaughlin R, Gallagher FA, Bohndiek SE, Day SE, Brindle KM. Magnetization transfer measurements of exchange between hyperpolarized [1-13C]-pyruvate and [1-13C]-lactate in a murine lymphoma. Magnetic Resonance in Medicine. 2010;63:872–880. doi: 10.1002/mrm.22276. [DOI] [PubMed] [Google Scholar]
- 11.Harrison C, Yang C, Jindal A, DeBerardinis RJ, Hooshyar MA, Merritt M, DeanSherry A, Malloy CR. Comparison of kinetic models for analysis of pyruvate-to-lactate exchange by hyperpolarized 13C NMR. NMR in Biomedicine. 2012:1099–1492. doi: 10.1002/nbm.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menichetti L, Frijia F, Flori A, Wiesinger F, Lionetti V, Giovannetti G, Aquaro GD, Recchia FA, Ardenkjaer-Larsen JH, Santarelli MF, Lombardi M. Assessment of real-time myocardial uptake and enzymatic conversion of hyperpolarized [1-13C]-pyruvate in pigs using slice selective magnetic resonance spectroscopy. Contrast Media and Molecular Imaging. 2012;7:85–94. doi: 10.1002/cmmi.480. [DOI] [PubMed] [Google Scholar]
- 13.Schulte RF, Sperl JI, Weidl E, Menzel MI, Janich MA, Khegai O, Durst M, ArdenkjaerLarsen JH, Glaser SJ, Haase A, Schwaiger M, Wiesinger F. Saturation-recovery metabolic-exchange rate imaging with hyperpolarized [1-13C] pyruvate using spectral-spatial excitation. Magnetic Resonance in Medicine. 2012:1522–2594. doi: 10.1002/mrm.24353. [DOI] [PubMed] [Google Scholar]
- 14.Witney TH, Kettunen MI, Brindle KM. Kinetic modeling of hyperpolarized 13C label exchange between pyruvate and lactate in tumor cells. Journal of Biological Chemistry. 2011;286:24572–24580. doi: 10.1074/jbc.M111.237727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulte RF, Sperl JI, Weidl E, Menzel MI, Janich MA, Khegai O, Durst M, Ardenkjaer-Larsen JH, Glaser SJ, Haase A, Schwaiger M, Wiesinger F. Saturation-recovery metabolic-exchange rate imaging with hyperpolarized [1-13C] pyruvate using spectral-spatial excitation. Magnetic Resonance Medicine. 2012 doi: 10.1002/mrm.24353. [DOI] [PubMed] [Google Scholar]
- 16.Larson P, Kerr A, Reed G, Hurd R, Kurhanewicz J, Pauly J, Vigneron D. Generating super stimulated-echoes in MRI and their application to hyperpolarized 13C diffusion metabolic imaging. Medical Imaging, IEEE Transactions on. 2012;31:265–275. doi: 10.1109/TMI.2011.2168235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen AP, Hurd RE, Cunningham CH. Spin tagging for hyperpolarized 13C metabolic studies. Journal of Magnetic Resonance. 2012;214:319–323. doi: 10.1016/j.jmr.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Santarelli MF, Positano V, Giovannetti G, Frijia F, Menichetti L, ArdenkjaerLarsen JH, Marchi DD, Lionetti V, Aquaro G, Lombardi M, Landini L. How the signal-to-noise ratio influences hyperpolarized 13C dynamic MRS data fitting and parameter estimation. NMR in Biomedicine. 2011:1099–1492. doi: 10.1002/nbm.1813. [DOI] [PubMed] [Google Scholar]
- 19.Larson PEZ, Kerr A, Leon C, Pauly J, Vigneron DB. Hyperpolarized 13C metabolic activity decomposition with stimulated-echoes. Journal of Magnetic Resonance. 2012 [Google Scholar]
- 20.Day S, Kettunen M, Gallagher F, Hu DE, Lerche M, Wolber J, Golman K, ArdenkjaerLarsen J, Brindle K. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nature Medicine. 2007;13:1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- 21.Ward CS, Venkatesh HS, Chaumeil MM, Brandes AH, VanCriekinge M, Dafni H, Sukumar S, Nelson SJ, Vigneron DB, Kurhanewicz J, James CD, Haas-Kogan DA, Ronen SM. Noninvasive detection of target modulation following phosphatidylinositol 3-kinase inhibition using hyperpolarized 13C magnetic resonance spectroscopy. Cancer Research. 2010;70:1296–1305. doi: 10.1158/0008-5472.CAN-09-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dafni H, Larson PE, Hu S, Yoshihara HA, Ward CS, Venkatesh HS, Wang C, Zhang X, Vigneron DB, Ronen SM. Hyperpolarized 13C spectroscopic imaging informs on hypoxia-inducible factor-1 and myc activity downstream of platelet-derived growth factor receptor. Cancer Research. 2010;70:7400–7410. doi: 10.1158/0008-5472.CAN-10-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park I, Bok R, Ozawa T, Phillips JJ, James CD, Vigneron DB, Ronen SM, Nelson SJ. Detection of early response to temozolomide treatment in brain tumors using hyperpolarized 13C NMR metabolic imaging. Journal of Magnetic Resonance Imaging. 2011;33:1284–1290. doi: 10.1002/jmri.22563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Hu S, Balakrishnan A, Bok RA, Anderton B, Larson PE, Nelson SJ, Kurhanewicz J, Vigneron DB, Goga A. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-myc-induced tumor formation and regression. Cell Metabolism. 2011;14:131–142. doi: 10.1016/j.cmet.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends in Biochemical Sciences. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, DallaFavera R, Dang CV. c-myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proceedings of the National Academy of Sciences. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishkovsky M, Frydman L. Progress in hyperpolarized ultrafast 2D NMR spectroscopy. ChemPhysChem. 2008;9:2340–2348. doi: 10.1002/cphc.200800461. [DOI] [PubMed] [Google Scholar]
- 31.Zeng H, Bowen S, Hilty C. Sequentially acquired two-dimensional NMR spectra from hyperpolarized sample. Journal of Magnetic Resonance. 2009;199:159–165. doi: 10.1016/j.jmr.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Frydman L, Blazina D. Ultrafast two-dimensional nuclear magnetic resonance spectroscopy of hyperpolarized solutions. Nature Physics. 2007;3:415–419. [Google Scholar]
- 33.Brotin T, Devic T, Lesage A, Emsley L, Collet A. Synthesis of Deuterium-Labeled Cryptophane-A and Investigation of Xe@Cryptophane Complexation Dynamics by 1D-EXSY NMR Experiments. Chemistry - A European Journal. 2001;7(7):1561–1573. doi: 10.1002/1521-3765(20010401)7:7<1561::aid-chem1561>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 34.Jeener J, Meier BH, Bachmann P, Ernst RR. Investigation of exchange processes by two-dimensional nmr spectroscopy. The Journal of Chemical Physics. 1979;71:4546–4553. [Google Scholar]
- 35.Rivals I, Personnaz L. Jacobian conditioning analysis for model validation. Neural Compution. 2004;16:401–418. doi: 10.1162/089976604322742083. [DOI] [PubMed] [Google Scholar]
- 36.Park T, Haug EJ. Ill-conditioned equations in kinematics and dynamics of machines. International Journal for Numerical Methods in Engineering. 1988;26:217–230. [Google Scholar]
- 37.Li TCY. An interior, trust region approach for nonlinear minimization subject to bounds. SIAM Journal on Optimization. 1996;6:418–445. [Google Scholar]
- 38.Coleman TF, Li Y. On the convergence of interior-reflective newton methods for nonlinear minimization subject to bounds. Mathematical Programming. 1994;67:189–224. [Google Scholar]
- 39.Mor´e JJ. The levenberg-marquardt algorithm: Implementation and theory. 1978;630:105–116. [Google Scholar]
- 40.Wilson D, Keshari K, Larson P, Chen A, Criekinge M, Bok R, Nelson S, Macdonald J, Vigneron D, Kurhanewicz J. Multi-compound polarization by DNP allows simultaneous assessment of multiple enzymatic activities in vivo. Journal of Magnetic Resonance. 2010;205:141–147. doi: 10.1016/j.jmr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cunningham CH, Chen AP, Albers MJ, Kurhanewicz J, Hurd RE, Yen YF, Pauly JM, Nelson SJ, Vigneron DB. Double spin-echo sequence for rapid spectroscopic imaging of hyperpolarized 13C. Journal of Magnetic Resonance. 2007;187:357–362. doi: 10.1016/j.jmr.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Derby K, Tropp J, Hawryszko C. Design and evaluation of a novel dual-tuned resonator for spectroscopic imaging. Magnetic Resonance in Medicine. 1990;86:256262. [Google Scholar]
- 43.Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors over-expressing myc. Nature Medicine. 2007;13:820–827. doi: 10.1038/nm1606. [DOI] [PubMed] [Google Scholar]
- 44.Shachaf C, Kopelman A, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann M, Borowsky A, Ruebner B, Cardiff R, Yang Q, Bishop JM, Contag CH, Felsher DW. Myc inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 45.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, Horner JW, CordonCardo C, Ronald A, DePinho GDY. Essential role for oncogenic ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 46.Spiess AN, Neumeyer N. An evaluation of R2 as an inadequate measure for nonlinear models in pharmacological and biochemical research: a Monte Carlo approach. BMC Pharmacology. 2012;10:6. doi: 10.1186/1471-2210-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dang CV, Semenza GL. Oncogenic alterations of metabolism. Trends in Biochemical Sciences. 1999;24:68–72. doi: 10.1016/s0968-0004(98)01344-9. [DOI] [PubMed] [Google Scholar]