Abstract
In the Life Sciences ‘omics’ data is increasingly generated by different high-throughput technologies. Often only the integration of these data allows uncovering biological insights that can be experimentally validated or mechanistically modelled, i.e. sophisticated computational approaches are required to extract the complex non-linear trends present in omics data. Classification techniques allow training a model based on variables (e.g. SNPs in genetic association studies) to separate different classes (e.g. healthy subjects versus patients). Random Forest (RF) is a versatile classification algorithm suited for the analysis of these large data sets. In the Life Sciences, RF is popular because RF classification models have a high-prediction accuracy and provide information on importance of variables for classification. For omics data, variables or conditional relations between variables are typically important for a subset of samples of the same class. For example: within a class of cancer patients certain SNP combinations may be important for a subset of patients that have a specific subtype of cancer, but not important for a different subset of patients. These conditional relationships can in principle be uncovered from the data with RF as these are implicitly taken into account by the algorithm during the creation of the classification model. This review details some of the to the best of our knowledge rarely or never used RF properties that allow maximizing the biological insights that can be extracted from complex omics data sets using RF.
Keywords: Random Forest, variable importance, local importance, conditional relationships, variable interaction, proximity
BACKGROUND
Development of high-throughput techniques and accompanying technology to manage and mine large-scale data has led to a revolution of Systems Biology in the last decade [1–3]. ‘Omics’ technologies such as genomics, transcriptomics, proteomics, metabolomics, epigenomics and metagenomics allow rapid and parallel collection of massive amounts of different types of data for the same model system. Software tools to manage [4], visualize [5] and integratively analyse omics-scale data are crucial to deal with its inherent complexity and ultimately uncover new biology. For example, knowledge on both gene expression and protein abundance may better explain a phenotype than gene expression or protein abundance separately. Particularly machine learning algorithms play a central role in the process of knowledge extraction [6, 7]. They are applied for supervised pattern recognition in data sets: typically they are used to train a classification model that allows separating samples of different classes (e.g. healthy or ill) based on variables (e.g. SNPs in a Genome-Wide Association Study or GWAS), and to estimate which variables were important for this task (see below).
The Random Forest (RF) algorithm [8] has become very popular for pattern recognition in omics-scale data, mainly because RF provides two aspects that are very important for data mining: high prediction accuracy and information on variable importance for classification. The prediction performance of RF compares well to other classification algorithms [7] such as support vector machines (SVMs, [9, 10]), artificial neural networks [11–13], Bayesian classifiers [14, 15], logistic regression [16], k-nearest-neighbours [17], discriminant analysis such as Fisher’s linear discriminant analysis [18] and regularized discriminant analysis [19], partial least squares (PLS, [20]) and decision trees such as classification and regression trees (CARTs, [21]). The theoretical and practical aspects of many of those algorithms and their application in biology have been discussed elsewhere (for example [6, 22, 23]). SVM and RF are arguably the most widely used classification techniques in the Life Sciences. Comparisons between the prediction accuracy of SVM and RF have been made several times [e.g. 24–29]. Although the performance of carefully tuned SVMs is generally slightly better than RF [24], RF offers unique advantages over SVM (see below). Further comparisons between SVM and RF will not be discussed here.
Life Science data sets typically have many more variables than samples. This problem is known as the ‘curse of dimensionality’ or the small n large p problem [30]. For instance, genomics, transcriptomics, proteomics and GWAS data sets suffer from this problem with in general thousands of measurements of genes, transcripts, proteins or SNPs determined for only dozens of samples [31–33]. RF effectively handles these data sets by training many decision trees using subsets of the data. Furthermore, RF has the potential to unravel variable interactions, which are ubiquitous in data sets generated in the Life Sciences. Interactions can for example be expected between SNPs in GWAS [34], between microbiota in metagenomics [35], between physicochemical properties of peptides in proteomic biomarker discovery studies [36] and between cellular levels of gene-products in gene-expression studies [25]. Additionally, the combinations of variables that together define molecules, e.g. mass spectrometry m/z ratios or Nuclear Magnetic Resonance chemical shifts, can distinguish phenotypes in metabolomics and metabonomics [37]. A final example includes combinations of several protein characteristics influencing the success rate in structural genomics [38]. In summary, its versatility makes RF a very suitable technique to investigate high-throughput data in this omics era.
Recent reviews aimed towards a more specialized audience have discussed the use of RF in (i) a broad scientific context [7], (ii) genomics research [39] and (iii) genetic association studies [40]. Here, we focus on the application of RF for supervised classification in the Life Sciences. In addition to reviewing the different uses of RF, we provide ideas to make this algorithm even more suitable for uncovering complex interactions from omics data. First, we introduce the general characteristics of RF for the reader who is not familiar with RF, followed by its use to tackle problems in data analysis. We also discuss rarely used properties of RF that allow determining interaction between variables. RF even has the potential to characterize these interactions for sample subclasses (e.g. groups of patients for which a SNP combination is predictive, while for a different group of patients the same SNP combination is not). Here, we discuss several research strategies that may allow exploiting RF to its full potential.
HOW DOES RF WORK?
Predictive RF models (from now on referred to as RFM) are non-parametric, hard to over-train, relatively robust to outliers and noise and fast to train. The RF algorithm can be used without tuning of algorithm parameters, although a better classification model can often easily be obtained by optimization of very few parameters (see below) [8]. RF trains an ensemble of individual decision trees based on samples, their class designation and variables. Every tree in the forest is built using a random subset of samples and variables (Figure 1), hence the name RF. The RF description by Breiman serves as a general reference for this section [8, 41].
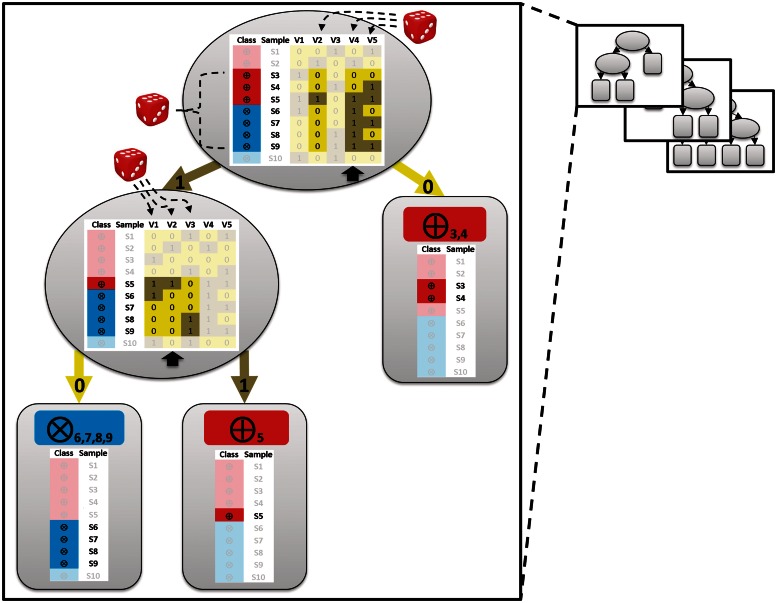
Figure 1:
Training of an individual tree of an RFM. The tree is built based on a data matrix (shown within the ellipses). This matrix consists of samples (S1–S10; e.g. individuals) belonging to two classes (encircled crosses or encircled plus signs; e.g. healthy and ill) and measurements for each sample for different variables (V1-V5; e.g. SNPs). Dice: random selection. Dashed lines: randomly selected samples and variables. For each tree, a bootstrap set is created by sampling samples from the data set at random and with replacement until it contains as many samples as there are in the data set. The random selection will contain about 63% of the samples in the original data set. In this example, the bootstrap set contains seven unique samples (samples S3–S9; non-selected samples S1, S2 and S10 are faded). For every node (indicated as ellipses) a few variables are randomly selected (here three; the other two non-selected variables are shown faded; by default RF selects the square root of the total number of variables) and evaluated for their ability to split the data. The variable resulting in the largest decrease in impurity is chosen to define the splitting rule. In case of the top node, this is V4 and for the second node on the left hand side this is V2 (indicated with the black arrows). This process is repeated until the nodes are pure (so called leaves; indicated with round-edged boxes): they contain samples of the same class (encircled cross or plus signs).
Suppose a forest of decision trees (e.g. CARTs) is constructed based on a given data set. For each tree, a different training set is created by randomly sampling samples (e.g. patient samples) from the data set with replacement resulting in a training set, or ‘bootstrap’ set, containing about two-third of the samples in the original data set. The remaining samples in the original data set are the ‘out-of-bag’ (OOB) samples. The tree is grown using the bootstrap data set by recursive partitioning (Figure 1). For every tree ‘node’, variables are randomly selected from the set of all variables and evaluated for their ability to split the data (Figure 1). The variable resulting in the largest decrease in impurity is chosen to separate the samples at each ‘parent node’, starting at the top node, into two subsets, ending up in two distinct ‘child nodes’. In RF, the impurity measure is the Gini impurity. A decrease in Gini impurity is related to an increase in the amount of order in the sample classes introduced by a split in the decision tree. After the bootstrap data has been split at the top node, the splitting process is repeated. The partitioning is finished when the final nodes, ‘terminal nodes’ or ‘leafs’, are either (i) ‘pure’, i.e. they contain only samples belonging to the same class or (ii) contain a specified number of samples. A classification tree is usually grown until the terminal nodes are pure, even if that results in terminal nodes containing a single sample. The tree is thus grown to its largest extent; it is not ‘pruned’. After a forest has been fully grown, the training process is completed. The RFM can subsequently be used to predict the class of a new sample. Every classification tree in the forest casts an unweighted vote for the sample after which the majority vote determines the class of the sample.
Although a single tree from the RFM is a weak classifier because it is trained on a subset of the data, the combination of all trees in a forest is a strong classifier [8]. Random selection of candidate variables for splitting ensures a low correlation between trees and prevents over-training of an RFM. Therefore, trees in an RFM need not be pruned, in contrast to classical decision trees that do not use random selection of variables [8]. The expected error rate of classification of new samples by a classifier, is usually estimated by cross-validation procedures, such as leave-one-out or K-fold cross-validation [42]. In K-fold cross-validation, the original data are randomly partitioned into K subsets (folds). Each of the K folds is once used as a test set while the other K − 1 folds are used as training data to construct a classifier. The average of the K error rates is the expected error rate of the classification of new samples when the classifier is built with all samples. In leave-one-out cross-validation a single sample is left out from the training set. General cross-validation procedures are unnecessary to predict the classification performance of a given RFM. A cross-validation is already built-in, as each tree in the forest has its own training (bootstrap) and test (OOB) data.
IMPORTANT VARIABLES FOR CLASS PREDICTION
In addition to an internal cross-validation RF also calculates estimates of variable importance for classification [8]. Importance estimates can be very useful to interpret the relevance of variables for the data set under study. The importance scores can for example be used to identify biomarkers [36] or as a filter to remove non-informative variables [25]. Two frequently used types of the RF variable importance measures exist. The mean decrease in classification is based on permutation. For each tree, the classification accuracy of the OOB samples is determined both with and without random permutation of the values of the variable. The prediction accuracy after permutation is subtracted from the prediction accuracy before permutation and averaged over all trees in the forest to give the permutation importance value. The second importance measure is the Gini importance of a variable and is calculated as the sum of the Gini impurity decrease of every node in the forest for which that variable was used for splitting. The use of different variable importance measures is discussed below in more detail.
The importance of variables for classification of a single sample is provided by RF as the local importance. It thus shows a direct link between variables and samples. As discussed in more detail below, the differences in local importance between samples can for example be used to detect variables that are important for a subset of samples of the same class (e.g. the important variables for a subtype of cancer in a data set with cancer patients and healthy subjects as classes). The local importance score is derived from all trees for which the sample was not used to train the tree (and is therefore OOB). The percentage of correct votes for the correct class in the permuted OOB data is subtracted from the percentage of votes for the correct class in the original OOB data to assign a local importance score for the variable of which the values were permuted. The score reflects the impact on correct classification of a given sample: negative, 0 (the variable is neutral) and positive. Local importances are rarely used and noisier than global importances, but a robust estimation of local importance values can be obtained by running the same classification several times [43] and for instance averaging the local importance scores.
PROXIMITY SCORES ALLOW DETERMINING SIMILARITY BETWEEN SAMPLES
RF not only generates variable-related information such as variable importance measures, but also calculates the proximity between samples. The proximity between similar samples is high. For proximity calculations, all samples in the original data set are classified by the forest. The proximity between two samples is calculated as the number of times the two samples end up in the same terminal node of a tree, divided by the number of trees in the forest. Provided sufficient variables are included in the RFM, outliers or mislabelled samples can be defined as samples whose proximity to all other samples from the same class is small. Identification of outliers or mislabelled samples serves as important feedback for the biologist who, if necessary, can correct for experimental mistakes. Similarly, subclasses can in principle be identified by finding samples that have similar proximities to all other samples of the same class. Subclasses in a data set with healthy and diseased subjects can for example be severe and mild subtypes of the disease. Proximity scores also allow the identification of prototypes, representative samples of a group of samples. The variable values of prototypes may explain how those variables relate to the classification of the group. Proximity scores may also be used to construct multidimensional scaling (MDS) plots. MDS plots aim to visualize the dissimilarity (calculated as 1 – proximity) between samples typically in a two-dimensional plot, so that the distances between data points are proportional to the dissimilarities. A good class separation may be obtained by plotting the first two scaling coordinates against each other, provided they capture sufficient information.
RF IMPLEMENTATIONS
The RF algorithm is available in many different open source software packages. Conveniently, the ‘randomForest’ package [44] is available as an R implementation [45] of the original RF code by Breiman and Cutler [41]. It is probably the most referred RF implementation because it is easy to use and the user benefits from other R data processing functionality. Recently, a framework for tree growing called Random Jungle (RJ) was developed [46]. It is currently the fastest implementation of RF, allows parallel computation of trees and is therefore very suited for the analysis of genome-wide data. The Willows package was also designed for tree-based analysis of genome-wide data by maximizing the use of computer memory [47]. The WEKA workbench [48] is a data mining environment that includes several machine learning algorithms including RF. The workbench allows for easy pre-processing of data and comparison between RF and other algorithms.
RF IN THE LIFE SCIENCES
Table 1 lists a non-exhaustive, yet in our opinion representative, number of studies that applied RF in different areas of the Life Sciences. A summary of the use of RF features in these areas is also provided in Table 1. The publications include many highly cited papers and papers that we included because they describe noteworthy use of RF properties. A detailed overview of the use of RF in these publications as well as meta data on them can be found in Supplementary Table S1.
Table 1:
Random Forest use in Life Sciences publications ordered by data type or origin
| Area | Publications | RF features |
Other possible uses of RF |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mtrya varied | Number of trees varied | Tree sizeb varied | Variable importance | Local importance | Proximity | Conditional importancec | Variable interactionsc | Alternative voting scheme | RF algorithm modified | RF in pipeline | ||
| Genomics | [26, 28, 49–63] | 4 | 5 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| Metabolomics | [64–66] | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Proteomics | [33, 36, 67–69] | 1 | 1 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Transcriptomics | [24, 25, 27, 70–76] | 2 | 4 | 1 | 8 | 1 | 3 | 0 | 0 | 0 | 1 | 4 |
| Other | [29, 38, 77–97] | 5 | 6 | 0 | 13 | 0 | 3 | 0 | 0 | 0 | 2 | 2 |
| Total | 58 papers | 13 | 16 | 1 | 41 | 1 | 7 | 0 | 0 | 0 | 3 | 11 |
The number of studies in different application areas that use RF properties or that report adaptations to RF is indicated. aNumber of variables to select for the best split at each node. bBy changing the number of splits or node size. cInferred from tree structure.
Three-quarters of the studies exploited the variable importance output of the RF algorithm (Table 1). For example, information on variable importance has been used to identify risk-associated SNPs in a genome-wide association study [56], to determine important genes and pathways for the classification of micro-array gene-expression data [27] and to identify factors that can be used to predict protein–protein interactions [29]. Very few studies report on the use of an iterative variable selection procedure [25] to select the most relevant variables and optimize the prediction accuracy of the RFM, although the classification accuracy improved when such a protocol was applied [24, 25, 68, 98] (Supplementary Table S1). In several data mining pipelines, important variables were selected from an RFM, which were subsequently used in other analysis techniques [50, 71].
Improving prediction accuracy has also been researched. In addition to a better separation of the samples of different classes, the variables of an accurate RFM are likely to be more relevant than those of a less accurate RFM. The number of variables to select for the best split at each node, mtry, was already marked as a tuning parameter by Breiman [6]. Varying the number of trees in the forest may also improve the OOB-error. One-fourth of the papers tuned and optimized the value of mtry and the number of trees. A single study not only regulated the size of the forest but also the size of the trees by varying the minimal node size [25]. The improvement of the prediction accuracy however was negligible. In contrast, Segal reported a better prediction accuracy may be achieved by regulation of the tree size via limiting the number of splits or the size of nodes for which splitting is allowed [99]. Boulesteix et al. [100] also recommended tuning tree depth and minimal node size in the context of genetic association studies. Alternative voting schemes, such as weighted voting, may improve classification accuracy [101] too, but have not been applied in the papers listed in Table 1.
Zhang and Wang pointed out that the interpretation of an RFM may be less practical than the interpretation of a single decision tree classifier due to the many trees in a forest. In a single tree, it is clear in which level of the tree and with what cut-off a variable is used to make a split. In a forest, a variable may or may not be present in a given tree, and if it is present, it may be so at different levels in the tree and have different cut-offs. They proposed to shrink a full forest to a smaller forest having a manageable number of trees and a level of prediction accuracy similar to the original RFM [102]. The smallest forest is one of the attempts to modify RF or use RF in combination with other methods in order to increase the prediction accuracy or model interpretability of RFMs (Table 1). Several other modifications were reviewed by Verikas et al. [7]. RF has not only been used in combination with other techniques, but several studies also combined multiple RFMs in a pipeline for better classification results (Table 1, [55, 72, 87]). RF has also been used in conjunction with dimension reduction techniques [33, 54]. For example, RF has been applied after PLS (PLS-RF, [33]). Sampson and colleagues argued the loadings (relative contribution of variables to the variability in the data) produced by PLS allow for meaningful interpretation of the association between variables and disease. De Lobel et al. [54] have used RF as a pre-screening method to remove noisy SNPs before multifactor-dimensionality reduction in genetic association studies. Additionally, RF has been incorporated in a transductive confidence machine [95], a framework that allows the prediction of classifiers to be complemented with a confidence value that can be set by the user prior to classification [103].
NEGLECTED RF PROPERTIES
RF has several properties that allow extracting relevant trends from data with complex variable relations, such as omics data sets. Nevertheless, these properties have according to our knowledge not yet been exploited to their full extent and only a few studies have explored their potential. Below we discuss the most important ones.
PROXIMITY
Proximity values are a measure of similarity between samples. A few studies used proximity values to detect outliers [27, 73, 74] resulting in an RFM before and after removal of outliers. The OOB prediction accuracy may improve after removing the outliers [74]. However, not in all cases a comparison was reported between the OOB errors of the second and the first model [73].
In addition to outlier detection, studies listed in Table 1 used proximity scores in MDS plots [27, 67, 96] and for class discovery from RF clustering results [91]. Analogous to their role in clustering, proximity scores also in supervised classification have the potential to allow discovering subclasses of data samples and even to identify corresponding prototypic variable values. However, we did not come across literature examples of utilization of the RF proximity measure for identification of subclasses or variable prototypes.
LOCAL IMPORTANCE
The global variable importance generated by RF captures classification impact of variables on all samples. The local variable importance is an estimate of the importance of a variable for the classification of a single sample. Local importance may therefore reveal specific variable importance patterns within groups of samples that may not be evident from global importance values. In other words, variables that are important for a subset of samples from the same class could show a clear local importance signal, while this signal would be lost in the global measure. Nevertheless, only one study in the Life Sciences reported the use of local importances in data analysis (Table 1). In this study, the local importance measure was exploited to predict microRNAs (miRNAs) that are significantly associated to the modification of expression of specific mRNAs [76]. Local importance instead of global importance was used in a regression RF analysis because the authors assumed that only a subset of miRNAs would significantly contribute to the regression fit. Recently, we developed PhenoLink, a method that links phenotypes to omics data sets [43]. Local importances were applied for variable selection using two criteria: (i) a removal criterion: having a negative or neutral local importance for the majority of class samples removing variables that do not positively contribute to the classification and (ii) a selection criterion: having a positive local importance for at least a few samples (typically 3) or for a percentage of samples (at least 10%) of a class. Classification of a metabolomics data set consisting of 9303 headspace (gas-phase) GC-MS metabolomics-based measurements (variables) for 45 different bacterial samples resulted in a classification (OOB) error of 71% (results not shown). After removal of 8587 ‘garbage’ variables the classification error was reduced to 18%. This dramatic reduction of classification error is due to the ‘garbage’ variables that make it more difficult for RF to recognize the informative variables. The positive selection criterion resulted in the same classification error but with an additional 210 variables removed and a total of 506 variables relevant for separating the bacterial samples based on headspace metabolites. PhenoLink was used effectively to remove redundant or even confusing variables and to detect variables that were important for a subset of samples in a number of studies ranging from gene-trait matching, metabolomics-transcriptomics matching and identification of biomarkers based on a variety of data sources [43]. Altogether, utilization of local importances is promising for many omics data sets and has the potential to uncover variables important for subsets of samples.
CONDITIONAL RELATIONSHIPS AND VARIABLE INTERACTIONS
For data sets generated in the Life Sciences, e.g. for metabolomics and proteomics measurements, gene expression data and GWAS studies, variables (e.g. SNPs in genetic association studies) are typically important for a subset of samples of the same class (e.g. patients) and conditional relations between variables might be important for a subset of samples. For example, certain SNPs or SNP combinations may be important for the first subgroup of patients and not important for the second subgroup.
Variable interactions have been reported to increase the global variable importance value [56]. The importance value itself however only provides the combined importance of the variable and all its interactions with other variables, but does not specify the actual variable interactions. Interactions between two variables can be inferred from a classification tree if a variable systematically makes a split on the other variable more likely or less likely than expected compared to variables without interactions. A recent paper reviewed the ability to identify SNP interactions by variations of logic regression, RF and Bayesian logistic regression [52]. For RF, an interaction importance measure was defined. However, the actual SNP interactions were not identified by the interaction importance, but rather by a relatively high variable importance measure. As Chen and colleagues discussed, the problem with their interaction importance measure was that two interacting SNPs need to be jointly selected in a tree branch relatively often. Furthermore, in the branches further down the tree the interaction of SNP A and B may have to be prominent in the presence of other variables in order to show a signal in the interaction importance [52].
Interactions between variables will often go hand in hand with conditional dependencies between the variables, i.e. variable B contributes to classification given that variable A is present above B in the tree. Conditional relations between variables are implicitly taken into account by the conditional inference forest algorithm (cforest, implemented in the party package [104–106] in R). cforest is a variant of RF that has been designed for unbiased variable selection (discussed below) [107]. Like RF, cforest generates a variable importance measure. Variable importance measures are currently subject of debate and rankings produced using permutation importance may be preferred over Gini importance rankings when variables: (i) are correlated [105, 108–110], (ii) vary in their scale of measurement (e.g. continuous and categorical variables) [104, 110] and (iii) vary in their number of categories [104, 110]. These variable characteristics are common in Life Science data sets, e.g. for patient parameters (for instance a categorical variable such as the dichotomous variable ‘has dog’: yes, no; another discrete variable such as ‘number of children’: 0, 1, 2, 3, 4; and a continuous variable ‘IgG blood level’: 0–20 g/l) and gene expression (continuous) versus SNP data (categorical). In combination with subsampling instead of bootstrap sampling, the splitting criterion of cforest has been reported to be less biased than the RF criterion [105]. The algorithm to determine the conditional importance measure generated by cforest explicitly takes into account the conditional relationships. However, like in RF conditional relationships are still implicit in the importance value output of cforest.
Analysis of individual RFM tree structures might be a good strategy to investigate interactions between variables. If variable A precedes variable B significantly more often than expected for variables without interactions, B is likely conditionally dependent on A. Recently, in a GWAS study the genetic variants underlying age-related macular degeneration (AMD) were investigated [111]. The authors analysed tree structures and proposed an importance measure based on associations between a variable (SNP) and the response variable (trait), conditional on other variables (other SNPs). For a given SNP, the forest was searched for nodes where that SNP was used as a splitting variable. A conditional Chi-square statistic was calculated for each of those nodes using SNPs that preceded the SNP in the same tree. The maximal conditional Chi-square (MCC) importance was defined as the highest Chi-square value of all nodes where the SNP was used as a splitting variable. The MCC value thus quantifies the relationship between a phenotype and a SNP given its preceding SNPs in the RFM.
The interactions between alleles of patients or healthy people in these SNPs were shown in a tree-like graph. The effects of the conditional relationships between variables for all samples of a given class are directly visible in these graphs. Partial dependence plots [112] may reveal the same information as they show how the classification of a data set is altered as a function of a subset of variables (usually one or two) after accounting for the average effects of all other variables in the model. CARTscans [113] allow visualization of conditional dependencies on categorical variables. However, multidimensional partial dependence plots or CARTscans have to be manually inspected to derive concrete interactions between variables.
The MCC importance can probably also be applied to other high-throughput data with numerous noisy and only a few important variables, as long as the node size is sufficient [111]. To date, however, no publicly available MCC implementation exists. Importantly, none of the above-described studies allow deriving a minimum set of variables and their interactions required to classify a given data set. Such minimum set is essential in reducing the complexity of a biomarker and increasing its interpretability. In addition, it could very well be that variable interactions are relevant only for a subset of samples of the same class. Generating this potentially crucial information for a given data set would require supplementing for instance the MCC algorithm of Wang and co-workers with, e.g. a clustering of samples based on, e.g. local variable importance or RF proximity scores and subsequently selecting the variables and/or variable interactions that explain the classification of a given subset of samples of the same class. A publicly available and validated MCC implementation might therefore be promising for the discovery of variable interactions in proteomics, metabolomics, genomics and transcriptomics data using RF, especially if the implementation would also include the determination of variable interactions for subsets of samples and visualization tools that support interpretation of such complex relationships.
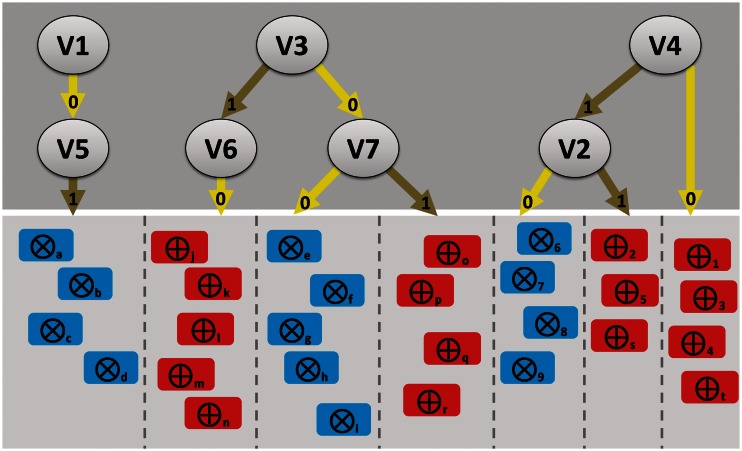
For inspiration, we provide a concept visualization of interacting variables, relevant for subsets of samples, different from the visualizations discussed earlier. The visualization might be a typical result from extensive omics data mining from the trees in an RFM (Figure 2). Linking the samples of the same subclass using evidence-based graphs, much like those from STRING [114], could furthermore allow the viewer to see and understand the (other) biological connection(s) between samples that are found to be linked by (interacting) variables identified in this data-driven approach.
Figure 2:
Concept visualization of how relations between variables and samples could be represented following the dissection of the trees in a random forest. In this hypothetical case, a supervised classification was performed on samples from two classes (encircled crosses or encircled plus signs; e.g. healthy individuals or patients). Dissection of the random forest trees might result in the further (unsupervised) distinction of subsets of samples. Top panel: variables (V1-Vn; e.g. SNPs in a GWAS study), their values (1 or 0) and interactions. Bottom panel: subsets (separated by the dashed lines) of samples from the pure classes that are predicted by a given interaction between variables. An interpretation example: provided that SNP4 (V4) is present, SNP2 (V2) allows the distinction between two subsets (consisting of healthy individuals 6, 7 8, 9 and patients 2, 5 and s). If SNP4 is absent, then the patient samples 1, 3, 4 and t can be classified. In case SNP1 (V1) is absent and SNP5 (V5) is present, a subset of healthy individuals consisting of samples a, b, c and d can be classified. Note that in this example, there can apparently no subset be distinguished if SNP1 (V1) is present or SNP5 (V5) is absent.
CONCLUSION
The RF algorithm has been widely used in the Life Sciences. It is suited for both regression and classification tasks, for example the prediction of disease state of patients (samples) using expression characteristics of genes (variables). However, RF has predominantly been used in a straight-forward way as a classifier without preceding variable selection and parameter tuning, or as a variable filter prior to using other prediction algorithms. RF is an elegant and powerful algorithm allowing the extraction of additional relevant knowledge from omics data, such as conditional relations between variables and interactions between variables for subsets of samples. Exploiting local importances, proximity values and analysis of individual trees could prove to be a compass to unlocking this information from complex omics data.
Key points.
RF is widely used in the Life Sciences because RF classification models are versatile, have a high prediction accuracy and provide additional information such as variable importances.
RF is often used as a black box, without parameter optimization, variable selection or exploitation of proximity values and local importances.
RF is a unique and valuable tool to analyse variable interactions and conditional relationships for data sets in which (combinations of) variables are important for subsets of samples, typically for omics data generated in the Life Sciences.
SUPPLEMENTARY DATA
Supplementary data are available online at http://bib.oxfordjournals.org/.
Biographies
Wouter Touw is a master student of Molecular Life Sciences at the Radboud University of Nijmegen, the Netherlands. He specializes in bioinformatics and structural biology.
Jumamurat Bayjanov is a postdoctoral researcher at the Radboud University Medical Centre, the Netherlands. He is involved in analyzing next-generation sequence data and developing machine-learning tools.
Lex Overmars is a PhD student at the Centre for Molecular and Biomolecular Informatics, Radboud University Medical Centre. His research focuses on the analysis of prokaryotic regulatory elements.
Lennart Backus is developing phylogenomics techniques for sequence-based prediction of microbial interactions at the Radboud University Medical Centre in a PhD project funded by TI Food and Nutrition.
Jos Boekhorst is a bioinformatician at NIZO food research. He uses computational tools to unravel links between microbes, food, health and disease.
Michiel Wels is group leader bioinformatics at NIZO food research and is involved in applying bioinformatics approaches to different food-related research questions.
Sacha van Hijum is a senior scientist bioinformatics at NIZO food research and group leader of the bacterial genomics group, Centre for Molecular and Biomolecular Informatics at the Radboud University Medical Centre. Bioinformatics research at the bacterial genomics group focuses on establishing the relation between microbial consortia and health.
References
- 1.Ideker T, Galitski T, Hood L. A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–72. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 2.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–4. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 3.Chuang H-Y, Hofree M, Ideker T. A decade of systems biology. Annu Rev Cell Dev Biol. 2010;26:721–44. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh S, Matsuoka Y, Asai Y, et al. Software for systems biology: from tools to integrated platforms. Nat Rev Genet. 2011;12:821–32. doi: 10.1038/nrg3096. [DOI] [PubMed] [Google Scholar]
- 5.Gehlenborg N, O’Donoghue SI, Baliga NS, et al. Visualization of omics data for systems biology. Nat Methods. 2010;7:S56–68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 6.Larranaga P. Machine learning in bioinformatics. Brief Bioinform. 2006;7:86–112. doi: 10.1093/bib/bbk007. [DOI] [PubMed] [Google Scholar]
- 7.Verikas A, Gelzinis A, Bacauskiene M. Mining data with random forests: a survey and results of new tests. Pattern Recognit. 2011;44:330–49. [Google Scholar]
- 8.Breiman L. Random Forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 9.Boser BE, Guyon IM, Vapnik VN. In: Proceedings of the fifth annual workshop on Computational learning theory - COLT ’92, 1992. A training algorithm for optimal margin classifiers; pp. 144–52. [Google Scholar]
- 10.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–97. [Google Scholar]
- 11.McCulloch WS, Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull Math Biophys. 1943;5:115–33. [PubMed] [Google Scholar]
- 12.Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev. 1958;65:386–408. doi: 10.1037/h0042519. [DOI] [PubMed] [Google Scholar]
- 13.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323:533–36. [Google Scholar]
- 14.Friedman N, Geiger D, Goldszmidt M. Bayesian network classifiers. Mach Learn. 1997;29:131–63. [Google Scholar]
- 15.Minsky M. Steps toward artificial intelligence. Proc IRE. 1961;49:8–30. [Google Scholar]
- 16.Kleinbaum DG, Kupper LL, Chambless LE. Logistic regression analysis of epidemiologic data: theory and practice. Commun Stat Theory. 1982;11:485–547. [Google Scholar]
- 17.Fixt E, Hodges JL. Discriminatory analysis-nonparametric discrimination: consistency properties. Int Stat Rev. 1989;57:238–47. [Google Scholar]
- 18.Fischer RA. The use of multiple measurements in taxonomic problems. Ann Hum Genet. 1936;7:179–88. [Google Scholar]
- 19.Friedman JH. Regularized discriminant analysis. J Am Stat Assoc. 1989;84:165–75. [Google Scholar]
- 20.Wold H. Perspectives in Probability and Statistics, Papers in Honour of M. S. Bartlett. 1975. Soft modeling by latent variables: the nonlinear iterative partial least squares approach. [Google Scholar]
- 21.Breiman L, Friedman JH, Olshen RA, et al. Classification and regression trees. The Wadsworth Statistics Probability Series. 1984;19:368. [Google Scholar]
- 22.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd edn. New York: Springer-Verlag; 2009. [Google Scholar]
- 23.Tarca AL, Carey VJ, Chen X-wen, et al. Machine learning and its applications to biology. PLoS Comput Biol. 2007;3:e116. doi: 10.1371/journal.pcbi.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Statnikov A, Wang L, Aliferis CF. A comprehensive comparison of random forests and support vector machines for microarray-based cancer classification. BMC Bioinformatics. 2008;9:319. doi: 10.1186/1471-2105-9-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Díaz-Uriarte R, Alvarez de Andrés S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang P, Wu H, Wang W, et al. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007;35:W339–44. doi: 10.1093/nar/gkm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang H, Lin A, Holford M, et al. Pathway analysis using random forests classification and regression. Bioinformatics. 2006;22:2028–36. doi: 10.1093/bioinformatics/btl344. [DOI] [PubMed] [Google Scholar]
- 28.Bao L, Cui Y. Prediction of the phenotypic effects of non-synonymous single nucleotide polymorphisms using structural and evolutionary information. Bioinformatics. 2005;21:2185–90. doi: 10.1093/bioinformatics/bti365. [DOI] [PubMed] [Google Scholar]
- 29.Qi Y, Bar-Joseph Z, Klein-seetharaman J. Evaluation of different biological data and computational classification methods for use in protein interaction. Bioinformatics. 2006;500:490–500. doi: 10.1002/prot.20865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellman RE RandCorporation. Dynamic Programming. Princeton: Princeton University Press; 1957. p. 342. [Google Scholar]
- 31.Somorjai RL, Dolenko B, Baumgartner R. Class prediction and discovery using gene microarray and proteomics mass spectroscopy data: curses, caveats, cautions. Bioinformatics. 2003;19:1484–91. doi: 10.1093/bioinformatics/btg182. [DOI] [PubMed] [Google Scholar]
- 32.Bureau A, Dupuis J, Falls K, et al. Identifying SNPs predictive of phenotype using random forests. Genet Epidemiol. 2005;28:171–82. doi: 10.1002/gepi.20041. [DOI] [PubMed] [Google Scholar]
- 33.Sampson DL, Parker TJ, Upton Z, et al. A comparison of methods for classifying clinical samples based on proteomics data: a case study for statistical and machine learning approaches. PLoS One. 2011;6:e24973. doi: 10.1371/journal.pone.0024973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore JH, Asselbergs FW, Williams SM. Bioinformatics challenges for genome-wide association studies. Bioinformatics. 2010;26:445–55. doi: 10.1093/bioinformatics/btp713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fusaro VA, Mani DR, Mesirov JP, et al. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat Biotechnol. 2009;27:190–8. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholson JK, Connelly J, Lindon JC, et al. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1:153–61. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 38.Goh C-S, Lan N, Douglas SM, et al. Mining the structural genomics pipeline: identification of protein properties that affect high-throughput experimental analysis. J Mol Biol. 2004;336:115–30. doi: 10.1016/j.jmb.2003.11.053. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Ishwaran H. Random forests for genomic data analysis. Genomics. 2012;99:323–9. doi: 10.1016/j.ygeno.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein BA, Polley EC, Briggs FBS. Random forests for genetic association studies. Stat Appl Genet Mol Biol. 2011;10:1–34. doi: 10.2202/1544-6115.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breiman L, Cutler A. Random Forests. http://www.stat.berkeley.edu/~breiman/RandomForests/ [Google Scholar]
- 42.Stone M. Cross-validatory choice and assessment of statistical predictions. J Roy Stat Soc B Met. 1974;36:111–47. [Google Scholar]
- 43.Bayjanov JR, Molenaar D, Tzeneva V, Siezen RJ, van Hijum SAFT. PhenoLink – a web-tool for linking phenotype to ∼omics data for bacteria: application to gene-trait matching for Lactobacillus plantarum strains. BMC Genomics. 2012;13:170. doi: 10.1186/1471-2164-13-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 45.R Development Core Team. R. A Language and Environment for Statistical Computing. 2012. [Google Scholar]
- 46.Schwarz DF, König IR, Ziegler A. On safari to Random Jungle: a fast implementation of Random Forests for high-dimensional data. Bioinformatics. 2010;26:1752–8. doi: 10.1093/bioinformatics/btq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H, Wang M, Chen X. Willows: a memory efficient tree and forest construction package. BMC Bioinformatics. 2009;10:130. doi: 10.1186/1471-2105-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frank E, Hall M, Trigg L, et al. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20:2479–81. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 49.Alvarez S, Diaz-Uriarte R, Osorio A, et al. A predictor based on the somatic genomic changes of the BRCA1/BRCA2 breast cancer tumors identifies the non-BRCA1/BRCA2 tumors with BRCA1 promoter hypermethylation. Clin Cancer Res. 2005;11:1146–53. [PubMed] [Google Scholar]
- 50.Briggs FBS, Bartlett SE, Goldstein BA, et al. Evidence for CRHR1 in multiple sclerosis using supervised machine learning and meta-analysis in 12,566 individuals. Hum Mol Genet. 2010;19:4286–95. doi: 10.1093/hmg/ddq328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caporaso JG, Lauber CL, Costello EK, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen CCM, Schwender H, Keith J, et al. Methods for identifying snp interactions: a review on variations of logic regression, random forest and bayesian logistic regression. IEEE/ACM Trans Comput Biol Bioinf. 2011;8:1580–91. doi: 10.1109/TCBB.2011.46. [DOI] [PubMed] [Google Scholar]
- 53.Christensen BC, Houseman EA, Godleski JJ, et al. Epigenetic profiles distinguish pleural mesothelioma from normal pleura and predict lung asbestos burden and clinical outcome. Cancer Res. 2009;69:227–34. doi: 10.1158/0008-5472.CAN-08-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Lobel L, Geurts P, Baele G, et al. A screening methodology based on Random Forests to improve the detection of gene-gene interactions. Eur J Hum Genet, 2010;18:1127–32. doi: 10.1038/ejhg.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dutilh BE, Jurgelenaite R, Szklarczyk R, et al. FACIL: fast and accurate genetic code inference and logo. Bioinformatics. 2011;27:1929–33. doi: 10.1093/bioinformatics/btr316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lunetta KL, Hayward LB, Segal J, et al. Screening large-scale association study data: exploiting interactions using random forests. BMC Genet. 2004;5:32. doi: 10.1186/1471-2156-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma D, Xiao J, Li Y, et al. Feature importance analysis in guide strand identification of microRNAs. Comput Biol Chem. 2011;35:131–6. doi: 10.1016/j.compbiolchem.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 58.Meijerink M, van Hemert S, Taverne N, et al. Identification of genetic loci in Lactobacillus plantarum that modulate the immune response of dendritic cells using comparative genome hybridization. PloS One. 2010;5:e10632. doi: 10.1371/journal.pone.0010632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rödelsperger C, Guo G, Kolanczyk M, et al. Integrative analysis of genomic, functional and protein interaction data predicts long-range enhancer-target gene interactions. Nucleic Acids Res. 2011;39:2492–502. doi: 10.1093/nar/gkq1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roshan U, Chikkagoudar S, Wei Z, et al. Ranking causal variants and associated regions in genome-wide association studies by the support vector machine and random forest. Nucleic Acids Res. 2011;39:e62. doi: 10.1093/nar/gkr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsou JA, Galler JS, Siegmund KD, et al. Identification of a panel of sensitive and specific DNA methylation markers for lung adenocarcinoma. Mol Cancer. 2007;6:70. doi: 10.1186/1476-4598-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Hemert S, Meijerink M, Molenaar D, et al. Identification of Lactobacillus plantarum genes modulating the cytokine response of human peripheral blood mononuclear cells. BMC Microbiol. 2010;10:293. doi: 10.1186/1471-2180-10-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vingerhoets J, Tambuyzer L, Azijn H, et al. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled Phase III clinical studies. AIDS. 2010;24:503–14. doi: 10.1097/QAD.0b013e32833677ac. [DOI] [PubMed] [Google Scholar]
- 64.Enot DP, Beckmann M, Draper J. On the interpretation of high throughput MS based metabolomics fingerprints with random forest. Metabolomics. 2006:226–35.. [Google Scholar]
- 65.Gupta S, Aires-de-Sousa J. Comparing the chemical spaces of metabolites and available chemicals: models of metabolite-likeness. Mol Divers. 2007;11:23–36. doi: 10.1007/s11030-006-9054-0. [DOI] [PubMed] [Google Scholar]
- 66.Pino Del Carpio D, Basnet RK, De Vos RCH, et al. Comparative methods for association studies: a case study on metabolite variation in a Brassica rapa core collection. PloS One. 2011;6:e19624. doi: 10.1371/journal.pone.0019624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Finehout EJ, Franck Z, Choe LH, et al. Cerebrospinal fluid proteomic biomarkers for Alzheimer’s disease. Ann Neurol. 2007;61:120–9. doi: 10.1002/ana.21038. [DOI] [PubMed] [Google Scholar]
- 68.Hettick JM, Kashon ML, Slaven JE, et al. Discrimination of intact mycobacteria at the strain level: a combined MALDI-TOF MS and biostatistical analysis. Proteomics. 2006;6:6416–25. doi: 10.1002/pmic.200600335. [DOI] [PubMed] [Google Scholar]
- 69.Munro NP, Cairns DA, Clarke P, et al. Urinary biomarker profiling in transitional cell carcinoma. Int J Cancer. 2006;119:2642–50. doi: 10.1002/ijc.22238. [DOI] [PubMed] [Google Scholar]
- 70.Gunther EC, Stone DJ, Gerwien RW, et al. Prediction of clinical drug efficacy by classification of drug-induced genomic expression profiles in vitro. Proc Natl Acad Sci USA. 2003;100:9608–13. doi: 10.1073/pnas.1632587100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo L, Ma Y, Ward R, et al. Constructing molecular classifiers for the accurate prognosis of lung adenocarcinoma. Clin Cancer Res. 2006;12:3344–54. doi: 10.1158/1078-0432.CCR-05-2336. [DOI] [PubMed] [Google Scholar]
- 72.Nannapaneni P, Hertwig F, Depke M, et al. Defining the structure of the general stress regulon of Bacillus subtilis using targeted microarray analysis and Random Forest classification. Microbiology. 2012;158:696–707. doi: 10.1099/mic.0.055434-0. [DOI] [PubMed] [Google Scholar]
- 73.Riddick G, Song H, Ahn S, et al. Predicting in vitro drug sensitivity using Random Forests. Bioinformatics. 2011;27:220–4. doi: 10.1093/bioinformatics/btq628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsuji S, Midorikawa Y, Takahashi T, et al. Potential responders to FOLFOX therapy for colorectal cancer by Random Forests analysis. Br J Cancer. 2011:1–7. doi: 10.1038/bjc.2011.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X, Simon R. Microarray-based cancer prediction using single genes. BMC Bioinformatics. 2011;12:391. doi: 10.1186/1471-2105-12-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wuchty S, Arjona D, Li A, et al. Prediction of associations between microRNAs and gene expression in glioma biology. PloS One. 2011;6:e14681. doi: 10.1371/journal.pone.0014681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bordner AJ. Predicting protein-protein binding sites in membrane proteins. BMC Bioinformatics. 2009;10:312. doi: 10.1186/1471-2105-10-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen X-wen, Jeong JC. Sequence-based prediction of protein interaction sites with an integrative method. Bioinformatics. 2009;25:585–91. doi: 10.1093/bioinformatics/btp039. [DOI] [PubMed] [Google Scholar]
- 79.Dybowski JN, Heider D, Hoffmann D. Prediction of co-receptor usage of HIV-1 from genotype. PLoS Comput Biol. 2010;6:e1000743. doi: 10.1371/journal.pcbi.1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han P, Zhang X, Norton RS, et al. Large-scale prediction of long disordered regions in proteins using random forests. BMC Bioinformatics. 2009;10:8. doi: 10.1186/1471-2105-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heider D, Verheyen J, Hoffmann D. Predicting Bevirimat resistance of HIV-1 from genotype. BMC Bioinformatics. 2010;11:37. doi: 10.1186/1471-2105-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hillenmeyer ME, Ericson E, Davis RW, et al. Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol. 2010;11:R30. doi: 10.1186/gb-2010-11-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Y, Fang Y, Fang J. Predicting residue-residue contacts using random forest models. Bioinformatics. 2011:1–7. doi: 10.1093/bioinformatics/btr579. [DOI] [PubMed] [Google Scholar]
- 84.Li Y, Wen Z, Xiao J, et al. Predicting disease-associated substitution of a single amino acid by analyzing residue interactions. BMC Bioinformatics. 2011;12:14. doi: 10.1186/1471-2105-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin N, Wu B, Jansen R, et al. Information assessment on predicting protein-protein interactions. BMC Bioinformatics. 2004;5:154. doi: 10.1186/1471-2105-5-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marino SR, Lin S, Maiers M, et al. Identification by random forest method of HLA class I amino acid substitutions associated with lower survival at day 100 in unrelated donor hematopoietic cell transplantation. Bone Marrow Transplant. 2012;47:217–26. doi: 10.1038/bmt.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Medema MH, Zhou M, van Hijum SAFT, et al. A predicted physicochemically distinct sub-proteome associated with the intracellular organelle of the anammox bacterium Kuenenia stuttgartiensis. BMC Genomics. 2010;11:299. doi: 10.1186/1471-2164-11-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nayal M, Honig B. On the nature of cavities on protein surfaces: application to the identification of drug-binding sites. Proteins. 2006;63:892–906. doi: 10.1002/prot.20897. [DOI] [PubMed] [Google Scholar]
- 89.Nimrod G, Szilágyi A, Leslie C, et al. Identification of DNA-binding proteins using structural, electrostatic and evolutionary features. J Mol Biol. 2009;387:1040–53. doi: 10.1016/j.jmb.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Radivojac P, Vacic V, Haynes C, et al. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 2010;78:365–80. doi: 10.1002/prot.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shi T, Seligson D, Belldegrun AS, et al. Tumor classification by tissue microarray profiling: random forest clustering applied to renal cell carcinoma. Mod Pathol. 2005;18:547–57. doi: 10.1038/modpathol.3800322. [DOI] [PubMed] [Google Scholar]
- 92.Slabbinck B, De Baets B, Dawyndt P, et al. Towards large-scale FAME-based bacterial species identification using machine learning techniques. Syst Appl Microbiol. 2009;32:163–76. doi: 10.1016/j.syapm.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 93.Springer C, Adalsteinsson H, Young MM, et al. PostDOCK: a structural, empirical approach to scoring protein ligand complexes. J Med Chem. 2005;48:6821–31. doi: 10.1021/jm0493360. [DOI] [PubMed] [Google Scholar]
- 94.Tognazzo S, Emanuela B, Rita FA, et al. Probabilistic classifiers and automated cancer registration: an exploratory application. J Biomed Inform. 2009;42:1–10. doi: 10.1016/j.jbi.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 95.Wang H, Lin C, Yang F, et al. Hedged predictions for traditional Chinese chronic gastritis diagnosis with confidence machine. Comput Biol Med. 2009;39:425–32. doi: 10.1016/j.compbiomed.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 96.Wiseman SM, Melck A, Masoudi H, et al. Molecular phenotyping of thyroid tumors identifies a marker panel for differentiated thyroid cancer diagnosis. Ann Surg Oncol. 2008;15:2811–26. doi: 10.1245/s10434-008-0034-8. [DOI] [PubMed] [Google Scholar]
- 97.Zhang G, Li H, Fang B. Discriminating acidic and alkaline enzymes using a random forest model with secondary structure amino acid composition. Process Biochem. 2009;44:654–60. [Google Scholar]
- 98.Kim Y, Wojciechowski R, Sung H, et al. Evaluation of random forests performance for genome-wide association studies in the presence of interaction effects. BMC Proc. 2009;3:S64. doi: 10.1186/1753-6561-3-s7-s64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Segal M. Technical Report, Center for Bioinformatics & Molecular Biostatistics, University of California, San Francisco, 2004. Machine learning benchmarks and random forest regression; pp. 1–14. [Google Scholar]
- 100.Boulesteix A-L, Bender A, Lorenzo Bermejo J, et al. Random forest Gini importance favours SNPs with large minor allele frequency: impact, sources and recommendations. Brief Bioinform. 2012;13:292–304. doi: 10.1093/bib/bbr053. [DOI] [PubMed] [Google Scholar]
- 101.Robnik-Sikonja M. Improving Random Forests. In: Boulicaut JF, et al., editors. Machine Learning: ECML 2004 Proceedings. Vol. 3201. Berlin: Springer; 2004. pp. 359–70. [Google Scholar]
- 102.Zhang H, Wang M. Search for the smallest random forest. Stat Interface. 2009;2:381. doi: 10.4310/sii.2009.v2.n3.a11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gammerman A, Vovk V, Vapnik V. In: Proceedings of the Fourteenth Conference on Uncertainty in Artificial Intelligence, 1998. Learning by transduction; pp. 148–55. [Google Scholar]
- 104.Strobl C, Boulesteix A-L, Zeileis A, et al. Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinformatics. 2007;8:25. doi: 10.1186/1471-2105-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Strobl C, Boulesteix A-L, Kneib T, et al. Conditional variable importance for random forests. BMC Bioinformatics. 2008;9:307. doi: 10.1186/1471-2105-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hothorn T, Bühlmann P, Dudoit S, et al. Survival ensembles. Biostatistics. 2006;7:355–73. doi: 10.1093/biostatistics/kxj011. [DOI] [PubMed] [Google Scholar]
- 107.Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: a conditional inference framework. J Comput Graph Stat. 2006;15:651–74. [Google Scholar]
- 108.Nicodemus KK, Malley JD. Predictor correlation impacts machine learning algorithms: implications for genomic studies. Bioinformatics. 2009;25:1884–90. doi: 10.1093/bioinformatics/btp331. [DOI] [PubMed] [Google Scholar]
- 109.Nicodemus KK, Malley JD, Strobl C, et al. The behaviour of random forest permutation-based variable importance measures under predictor correlation. BMC Bioinformatics. 2010;11:110. doi: 10.1186/1471-2105-11-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nicodemus KK. Letter to the Editor: On the stability and ranking of predictors from random forest variable importance measures. Brief Bioinform. 2011;12:369–73. doi: 10.1093/bib/bbr016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang M, Chen X, Zhang H. Maximal conditional chi-square importance in random forests. Bioinformatics. 2010;26:831–7. doi: 10.1093/bioinformatics/btq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001;29:1189–232. [Google Scholar]
- 113.Nason M, Emerson S, LeBlanc M. CARTscans: a tool for visualizing complex models. J Comput Graph Stat. 2004;13:807–25. [Google Scholar]
- 114.Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–8. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.