Abstract
Lipid bodies, also known as lipid droplets, are present in most eukaryotic cells. In leukocytes, lipid bodies are functionally active organelles with central roles in inflammation and are considered structural markers of inflammatory cells in a range of diseases. The identification of lipid bodies has methodological limitations because lipid bodies dissipate upon drying or dissolve upon fixation and staining with alcohol-based reagents. Here we discuss several techniques to detect and visualize lipid bodies within leukocytes by light microscopy. These techniques include staining with osmium or use of different fluorescent probes such as Nile red, BODIPY, Oil red, P96 and immunofluorescence labeling for adipose differentiation-related protein (ADRP).
Keywords: Lipid bodies, lipid droplets, leukocytes, bright field and fluorescence microscopy, osmium staining, nile red, oil red O, BODIPY, 1-pyrenedodecanoic acid, adipose differentiation-related protein (ADRP)
1. Introduction
Lipid bodies, also named lipid droplets or adiposomes, are now recognized as key organelles involved in lipid storage and metabolism, cell signaling and inflammation (1, 2). Lipid bodies are lipid-rich organelles distributed in the cytoplasm as roughly spherical organelles lacking a delimiting classical bilayer membrane (3–6), but surrounded by an outer monolayer of phospholipids, which at least in some cells may have a unique fatty acid composition (4, 7). The internal core of lipid bodies is rich in neutral lipids; and it is likely that more complex membranous domains, often obscured by overlying neutral lipids, are present within lipid bodies (8, 9). Indeed, studies of lipid bodies are providing functional, ultrastructural and protein compositional evidences that lipid bodies are not inert depots of neutral lipid. Rather, it has become evident that lipid bodies are highly regulated, dynamic and functional organelles. Over the past years substantial progresses have been made to demonstrate that lipid body biogenesis is a highly regulated process, which culminates in the compartmentalization of a specific set of proteins and lipids (reviewed in (1, 2)).
Lipid body accumulation within cells is observed in both clinical and experimental metabolic, infectious, neoplasic and other inflammatory conditions. Because lipid bodies can be destroyed by drying or fixation and staining with alcohol-based reagents, there are consequently some methodological limitations to their study (2, 10). Indeed, routinely used hematological staining as May–Grunwald–Giemsa staining lead to dissolution of lipid bodies commonly precluding their identification. However, using appropriate fixation procedures followed by methods of identification of lipids and/or of lipid body-specific proteins, lipid bodies can be readily identified within cells. In this chapter, we detail different techniques to visualize lipid bodies in different cell suspensions such as leukocytes isolated from the blood, cell lineages and peritoneal, pleural or bronchoalveolar cells.
2. Materials
2.1. Osmium Staining
Sodium cacodylate (cacodylic acid – sodium salt) is dissolved (4.28 g) in 180 mL of distilled water. Adjust pH to 7.4 with HCl and then make up to 200 mL with distilled water for 0.1 M final concentration.
Osmium tetroxide (see Note 1): to prepare a stock solution (1.5%), dissolve 1.5 g of osmium tetroxide in 100 mL of 0.1 M sodium cacodylate buffer. Aliquot in small glass tubes (~2 mL per tube) and store at 4–8°C. Protect from light.
Paraformaldehyde or formaldehyde solution (formalin) (see Note 2). For paraformaldehyde preparation, dilute paraformaldehyde to 2% in Hanks-buffered salt solution without calcium chloride and magnesium chloride (HBSS–/–) or phosphate-buffered saline (PBS). Dilutions should be made in fume hood and fresh dilutions of paraformaldehyde should be used in each experiment. Protect from light. For formalin preparation, dilute formalin (saturated solution of formaldehyde 37%) to 3.7% in PBS or HBSS–/–. Adjust to pH 7.4.
Thiocarbohydrazide solution 1% should be freshly prepared by dissolving 50 mg of thiocarbohydrazide in 5 mL of distilled water. The solution should be heated at a hot plate or microwave (30 s), followed by cooling to room temperature just prior to use.
Aqueous mounting medium.
Liquid blocker pen.
Glass microscope slides and coverslips.
2.2. Nile Red
Nile red: 9-diethylamino-5H-benzo [α] phenoxazine-5-one is a phenoxazone dye 1-Acho que é phenoxazine dye, 2-incluir espaço entre as palavras poorly soluble in water but it does dissolve in a wide variety of organic solvents (11). Stock solution: dissolve Nile red in acetone (1 mg/mL). Aliquot in small test tubes and store at –20°C. Working solution (prepare fresh): dilute at 1:10,000 in HBSS–/– or PBS from the stock solution. Keep protected from light (see Note 3).
Paraformaldehyde or formaldehyde solution (see Note 2). Refer to Section 2.1 (Item 3) for fixative preparation.
Anti-fading mounting medium for fluorescence microscopy.
Glass microscope slides and coverslips.
2.3. Oil Red O
Oil Red O: 1-([4-(Xylylazo)xylyl]azo)-2-naphthol, MW 408.49 is prepared at 0.5%: add 5 mL of propylene glycol (100%) to 0.5 g of oil red O with stirring and gradually complete the volume with propylene glycol to 100 mL. Heat the solution until 95°C, but do not allow going over 100°C. Filter through paper filter. The solution can be stored at room temperature.
Hematoxylin solution.
Aqueous mounting medium.
Paper filter.
2.4. BODIPY
BODIPY® 493/503: 4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene, (molecular weight: 262; Molecular Probes, cat no. D-3922) is stored at –20°C, protected from light (see Note 3). Stock solution: dissolve BODIPY in dimethyl sulfoxide (DMSO) at 1 mM. Aliquot in small test tubes (~10 μL per tube) and store at –20°C. Working solution (prepare fresh): dilute 1000× in HBSS–/–. All solutions must be protected from light (see Note 3).
Paraformaldehyde or formaldehyde solution (see Note 2). Refer to Section 2.1 (Item 3) for fixative preparation.
Anti-fading mounting medium for fluorescence microscopy.
Glass microscope slides and coverslips.
2.5. 1-Pyrenedodecanoic Acid
1-pyrenedodecanoic acid (molecular weight 400.56.2; Molecular Probes, cat. no. P-96). It is poorly soluble in water but it does dissolve in a variety of organic solvents including DMSO. Stock solution: dissolve P96 in DMSO (10 mM). Aliquot in small test tubes (~10 μL per tube) and store at –20°C, protected from light. Working solution (prepare fresh): dilute 1000× in HBSS–/– or Hanks-buffered salt solution with calcium chloride and magnesium chloride (HBSS–/–) (see Note 4). Keep protected from light (see Note 3).
Formaldehyde solution (formalin) prepared as Section 2.1 (Item 3).
Glass microscope slides and coverslips.
Anti-fading mounting medium for fluorescence microscopy.
2.6. Adipose Differentiation-Related Protein (ADRP, Adipophilin)
Monoclonal or polyclonal antibody to ADRP.
Fluorescent-labeled secondary antibodies.
Formaldehyde solution (formalin) prepared as Section 2.1 (Item 3).
Triton® X-100 (t-Octylphenoxypoly-ethoxyethanol).
Glass microscope slides and coverslips.
Anti-fading mounting medium for fluorescence.
3. Methods
3.1. Sample Preparation onto Slides
After obtaining a cell suspension (~0.5–1.0 × 106 cells/mL of medium), preparation of samples (cell suspensions) onto slides for lipid body staining can be done in two ways: using a cyto-centrifuge or by spreading a mixture of cells with melted agarose matrix onto a slide. For comparison of cell morphology observed with these two techniques, refer to Note 5. For cytospin preparations: label slides and cytocentrifuge a volume of 100 μL (~0.5–1.0 × 105 cells) of a cell suspension sample, at 18–23 g for 5 min. For agarose preparations: prepare first an agarose matrix. Weigh 0.125 g of agarose (low-melting point agarose; mp 65.5°C, gelling point 24°C) into a 125-mL Erlenmeyer flask and dilute to 2.5% by adding 5 mL of distilled water. Cover with aluminum foil. Mix well, but avoid swirling to prevent agarose binding to flask wall. Solubilize agarose in 70°C water bath for 15 min with gentle agitation. Aliquot in small test tubes (~1.5 mL) and store at 4°C. For slide preparation, resolubilize a tube at 70°C, mix cells with the liquid matrix and gently spread the cell–agarose mixture (~20 μL) onto microscope slides using a microtip. Use covered surface slides for cell adhesion. For spreading the cells, use surface tension to move agarose mixture throughout the slide. Cover gently this thin layer of agarose/cell mixture with a perfusion chamber (CoverWell™).
3.2. Osmium Staining
Properties: Osmium tetroxide binds to unsaturated lipids, and is reduced by organic materials to elemental osmium, an easily visible and permanent black substance (Fig. 9.1a). Reduction of osmium by thiocarbohydrazide highly enhances lipid labeling.
Prepare slides with samples using a cytocentrifuge (see Section 3.1).
Fix samples, while still moist, with paraformaldehyde or formalin for 10 min. Refer to Note 2 for cell fixation and Note 6 for “moist cells”.
Rinse slides in distilled water.
Circumscribe the adhered cells with liquid blocker pen to facilitate the staining procedure.
Stain the adhered cells by adding one drop of 0.1 M cacodylate buffer and one drop of 1.5% osmium tetroxide for 30 min. Refer to Note 1 for osmium manipulation.
Rinse slides in distilled water.
Immerse in thiocarbohydrazide solution for 5 min at room temperature.
Rinse the adhered cells twice with distilled water.
Re-stain by adding one drop of 0.1 M cacodylate buffer and one drop of 1.5% osmium tetroxide for 3 min.
Rinse slides in distilled water.
Let the slides dry.
Mount with aqueous mounting medium.
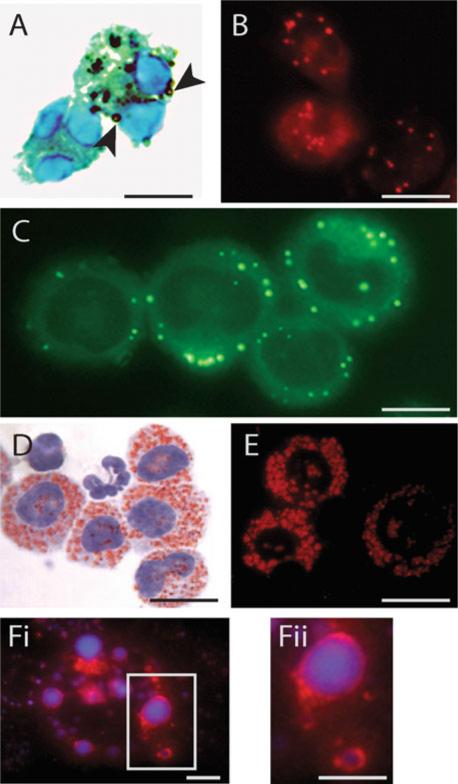
Fig. 9.1.
Lipid bodies within leukocytes imaged by light microscopy after staining with osmium (a), Nile red (b), BODIPY (c), oil red O (ORO) (d, e) or double labeled with 1-pyrenedodecanoic acid (P 96) and anti-adipose differentiation-related protein (ADRP) (f). Lipid bodies appear as round, dark (a, arrowheads), fluorescent red (b, e) or green (c) organelles distributed throughout the cytoplasm. ORO-stained lipid bodies appear as round red organelles at both bright field (d) and fluorescence (e) microscopy, while the nucleus is imaged in light blue after counterstaining with hematoxylin (d). In f, merged images show P 96-labeled lipid bodies in blue at UV filter and ADRP immunolabeling as a ring in the periphery of the lipid body. In a, cells were counterstained with Hema 3® kit (Fisher Scientific). a and b show eosinophils isolated from the blood of normal human donors and stimulated with eotaxin (a) or calcium ionophore (b) as before (20, 21). c and d show murine peritoneal leukocytes and macrophages, respectively. Bars, 6 μm (A, B, D, E and F), 10 μm (c).
Alternatively, osmium staining can be performed on agarose preparations.
Spread a mixture of cells/agarose onto a microscope slide and cover with a perfusion chamber (see Section 3.1).
Carefully pipet 400 μL of the fixative (2% paraformaldehyde) over sample through the chamber access port, ensuring that chamber area is uniformly saturated and let for 10 min.
Rinse slides in distilled water through the chamber access port.
Rinse slides in 0.1 M cacodylate buffer through the chamber access port.
Stain by adding 1.5% osmium tetroxide through the chamber access port for 30 min. Refer to Note 1 for osmium manipulation.
Rinse slides in 0.1 M cacodylate buffer through the chamber access port.
Carefully remove the chamber.
Immerse in thiocarbohydrazide for 5 min at room temperature.
Rinse slides in 0.1 M cacodylate buffer.
Re-stain with 1.5% osmium tetroxide for 3 min.
Let the slides dry.
Mount with aqueous mounting medium.
3.3. Nile Red Staining
Properties: Nile red is intensely fluorescent, and can serve as a sensitive stain for the detection of cytoplasmic lipid bodies (11, 12).
Incubate a cell suspension (1.0 × 106 cells/mL) with a working solution of Nile red (see Section 2.2, Item 1) for 5 min at room temperature and protected from light. Cells are incubated in a test tube.
Centrifuge (120 g/5 min) and resuspend in HBSS–/– or PBS to wash cells. Repeat this step once.
Cytospin onto slides using 100 μL of cell suspension at 18–23 g for 5 min.
Fix with paraformaldehyde or formalin (see Note 2 and Section 2.1, Item 3, for fixative preparation) for 5 min at room temperature.
Wash twice in HBSS–/– or PBS.
Mount while wet using anti-fading mounting medium. Keep slides in the dark (see Notes 3 and 7).
Alternatively, Nile Red staining can be done on agarose preparations.
Spread a mixture of cells/agarose onto a microscope slide and cover with a perfusion chamber (see Section 3.1).
Carefully pipet 400 μL of the fixative (2% paraformaldehyde) over sample through the chamber access port, ensuring that chamber area is uniformly saturated and let for 5 min. This step can be performed after incubation with Nile Red. Refer to Note 8 for Nile red staining in fixed/unfixed cells.
Wash twice with HBSS–/– (2 × 400 μL) adding buffer through the chamber access port.
Incubate with a working solution of Nile Red (400 μL) for 5–10 min at room temperature. Keep slides protected from light (see Note 3).
Wash twice with HBSS–/–.
Carefully remove the chamber.
Mount while moist with HBSS–/– or with anti-fading mounting medium after drying. Keep slides in the dark (see Note 3).
3.4. Oil Red O Staining
Properties: Oil red O belongs to the polyazo group of dyes which also includes the Sudan series of dyes. The principle of the lipid staining is based on the physical properties of the dye that preferentially divide into lipid-rich compartments. Oil red O staining can be readily visualized in both bright field and fluorescent microscopy (Fig. 9.1d, f) (13).
For slide preparation, cytocentrifuge 100 μL of a sample cell suspension, at 18–23 g for 5 min.
Fix cells with 3.7% formalin in HBSS–/– (see Section 2.1, Item 3).
Wash twice in distilled water.
Place slides in absolute propylene glycol for 5 min.
Stain in 0.5% oil red O solution (see Section 2.4) for 10 min in the incubator at 60°C.
Rinse cells in 85% propylene glycol solution for 5 min.
Wash twice in distilled water.
Counterstain with hematoxylin solution for 30 s (see Note 9).
Wash thoroughly in tap water.
Mount with aqueous mounting medium.
3.5. BODIPY Staining
Properties: BODIPY® lipid probe is an effective dye for staining neutral lipids and, for this reason it is very efficient for lipid body staining (Fig. 9.1c). The fluorescence quantum yield of the BODIPY dyes is not diminished in water and this method can be used in conjugation with immunofluorescence (see Note 10 and Chapter 11).
Incubate the cell suspension with 1 μM BODIPY for 1 h at 37°C. Cells are incubated in a test tube inside a water bath.
Pellet the cells (120 g/5 min) and resuspend in HBSS–/– or PBS to wash cells. Repeat this step once.
Cytospin onto slides using 100 μL of cell suspension at 18–23 g for 5 min.
Fix with paraformaldehyde or formalin in HBSS–/– or PBS for 5–10 min (see Section 2.1, Item 3, for fixative preparation).
Wash twice in HBSS–/– or PBS.
Mount while wet using anti-fading mounting medium. Slides are stored at room temperature in the dark until analysis (see Notes 3 and 7).
Alternatively, the BODIPY staining can be done using an agarose preparation.
Spread a mixture of cells/agarose onto a microscope slide and cover with a perfusion chamber (see Section 3.1).
Wash twice with HBSS–/– (2× 400 μL) adding buffer through the chamber access port.
Carefully pipet 400 μL of BODIPY solution over sample through the chamber access port, ensuring that chamber area is uniformly saturated. Place slides on a tray atop hydrated pad. Place tray in humidified incubator (37°C, 5% CO2) for 1 h.
Wash twice with HBSS–/– (2× 400 μL) adding buffer through the chamber access port.
Pipet 400 μL of the fixative (2% paraformaldehyde) over sample through the chamber access port, ensuring that chamber area is uniformly saturated and let for 5–10 min.
Wash twice in HBSS–/– or PBS.
Carefully remove the chamber.
Mount while wet with HBSS–/– or anti-fading mounting medium. Keep slides protected from light (see Note 3).
3.6. 1-Pyrenedodecanoic Acid Staining
Properties: 1-Pyrenedodecanoic acid (P-96) is a fluorescent fatty acid analog with the environment sensitive pyrene attached to the terminal carbon atom that is furthest from the carboxylate moiety. P96 is readily incorporated into lipid bodies and P96 fluorescent-labeled lipid bodies are visualized under the UV (excitation/emission 340/376 nm) (Fig. 9.1e) (14, 15).
Incubate a cell suspension with a working solution of P96 (see Section 2.5, Item 1). for 1 h at room temperature and protected from light (see Note 3).
Pellet the cells (120 g/5 min) and resuspend in HBSS–/– or PBS to wash cells. Repeat this step once.
Cytospin onto slides using 100 μL of cell suspension at 18–23 g for 5 min.
Fix with 3.7% formalin in HBSS–/– or PBS for 5 min at room temperature (see Section 2.1, Item 3, for fixative preparation).
Wash twice in distilled water.
Mount while wet using anti-fading medium for fluorescence microscopy (see Note 7).
3.7. ADRP Staining
Properties: Adipose differentiation-related protein (ADRP) is a structural protein of lipid bodies considered essential for lipid storage and metabolism (reviewed in (16)). ADRP is ubiquitously associated with cytoplasmic lipid bodies in different types of cells and is described as a specific protein marker for lipid bodies (Fig. 9.1f) (17–19). This method can be conjugated with immuno-labeling for other proteins (see Note 11).
For slide preparation, cytocentrifuge 100 μL of a sample cell suspension, at 18–23 g for 5 min.
Fix slides with 3.7% formalin in HBSS–/– or PBS (see Section 2.1, Item 3, for fixative preparation).
Wash once in HBSS–/– or PBS.
Permeabilize the cells with 0.1% Triton X-100 in HBSS–/– for 10 min.
Circumscribe the adhered cells with liquid blocker pen.
- For human cells incubate with mouse anti-human ADRP at dilution of 1:20 (2.5 μg/mL, final concentration) for 1 h at room temperature.
- For mouse, rat, human or bovine cells incubate with guinea pig anti-human ADRP polyclonal antibody at dilution of 1:300 (final dilution) for 1 h at room temperature.
Wash three times in HBSS–/– or PBS.
Incubate with fluorescent-labeled secondary antibody for 1 h room temperature.
Wash three times in HBSS–/– or PBS.
Mount in mounting medium for fluorescence microscopy (see Note 7).
3.8. Lipid Body Analysis and Quantification
Lipid body analysis is performed on a bright field (osmium and oil red O staining) or fluorescence microscope (oil red O and fluorescent probes) at 1000×. For example, analyses and image acquisition can be obtained using an Olympus BX-FLA fluorescence microscope equipped with a Plan Apo 100 × 1.4 Ph3 objective (Olympus) and CoolSNAP-Pro CF digital camera in conjunction with Image Pro Plus® software (Media Cybernetics). The detection of lipid bodies using different techniques will appear as round dark (osmium staining, Fig. 9.1a) or fluorescent red (Nile red or Oil red O) (Fig. 9.1b and e, respectively) or green (BODIPY) (Fig. 9.1c) organelles. Of note, Nile red can be observed through both green (fluorescein) and red (rhodamine) channels and Oil red O can be also observed at bright field microscopy (Fig. 9.1d). ADRP immuno-labeling has a characteristic ring-shape appearance as this protein localizes preferentially at the periphery of the lipid body (Fig. 9.1f). Lipid bodies are usually enumerated using a 100× objective lens in 50 consecutively scanned cells (10).
Alternatively, lipid bodies can be quantified by the measurement of oil red O (ORO) or BODIPY fluorescent area. The measurement of the area of lipid bodies is obtained with a 60 objective (at least four fields per slide). Images are transformed into black and white pictures and analyzed with the Image 2D software (GE Healthcare). Spots are determined by automatic spot detection and the total area of fluorescent lipid bodies is obtained for each field and divided by the number of cells in the respective field.
Acknowledgments
Supported by CNPq and FAPEMIG (Brazil) to RCNM; CNPq, FAPERJ and PRONEX (Brazil) to PTB and NIH grants (USA) AI020241, AI051645 and AI022571 to PFW. We acknowledge Clarissa M. Maya-Monteiro for the Fig. 9.1c used in this chapter.
Footnotes
Osmium tetroxide is volatile and its fumes are very toxic (causes severe irritation to eyes, skin and respiratory tract). Thus, any manipulation involving this chemical must be performed in a fume hood and wearing gloves.
Fixation of cells before osmium, Nile Red or BODIPY staining can be done using either 2% paraformaldehyde or 3.7% formalin.
When staining with fluorescent probes such as Nile red, P 96 and BODIPY, fluorescence is usually not stable for a long period and fluorescence bleaching will occur after a certain time. Keep the cell preparations in the dark to avoid fluorescence loss.
For some type of cells, the P96 staining can have better results using HBSS+/+.
In general, cells kept in agarose show better morphology compared with cells from cytospin preparations because cells are kept in a hydrated system. In addition, shape changes, a feature of activated leukocytes, can be observed when cells are embedded in an agarose matrix. On the other hand, cytospin slides are fast prepared. In both cytospin and agarose preparations, lipid bodies are well preserved and can be easily detected.
It is very important to keep cells moist during the staining with osmium. Dried cells will appear with very bad morphology.
For fluorescence microscopy, it is important to use a mounting medium that prevents rapid loss of fluorescence during microscopic examination and retains its anti-fading ability during long-term storage.
Staining with Nile red can be carried out on either fixed or unfixed cells with no apparent difference in distribution or intensity of fluorescence.
Counterstaining with hematoxylin is important to show nuclear aspects. This step is not mandatory if visualization of nuclei is not necessary.
BODIPY staining can be combined with an immunofluorescence protocol (for example, ADRP immuno-labeling). In this case, BODIPY can be mixed with the secondary antibody (Chapter 10).
Immuno-labeling for ADRP can be conjugated with immuno-labeling for other proteins by simultaneous incubation with two primary antibodies (raised in distinct hosts) followed by incubation with two secondary antibodies (with distinct ranges of excitation/emission).
References
- 1.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 2.Bozza PT, Melo RCN, Bandeira-Melo C. Leukocyte lipid bodies regulation and function: contribution to allergy and host defense. Pharmacol Ther. 2007;113:30–49. doi: 10.1016/j.pharmthera.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Dvorak AM, Dvorak HF, Peters SP, Shulman ES, MacGlashan DW, Jr., Pyne K, Harvey VS, Galli SJ, Lichtenstein LM. Lipid bodies: cytoplasmic organelles important to arachidonate metabolism in macrophages and mast cells. J Immunol. 1983;131:2965–2976. [PubMed] [Google Scholar]
- 4.Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- 5.Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 6.Weller PF, Monahan-Earley RA, Dvorak HF, Dvorak AM. Cytoplasmic lipid bodies of human eosinophils. Subcellular isolation and analysis of arachi-donate incorporation. Am J Pathol. 1991;138:141–148. [PMC free article] [PubMed] [Google Scholar]
- 7.Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Wan HC, Melo RC, Jin Z, Dvorak AM, Weller PF. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J. 2007;21:167–178. doi: 10.1096/fj.06-6711com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bozza PT, Magalhaes K, Weller PF. Leukocyte lipid bodies–biogenesis and functions in inflammation. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbalip.2009.01.005. doi:10.1016/j.bbalip_2009_01_005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melo RCN, Sabban A, Weller PF. Leukocyte lipid bodies: inflammation-related organelles are rapidly detected by wet scanning electron microscopy. J Lipid Res. 2006;47:2589–2594. doi: 10.1194/jlr.D600028-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Greenspan P, Mayer EP, Fowler SD. Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol. 1985;100:965–973. doi: 10.1083/jcb.100.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukumoto S, Fujimoto T. Deformation of lipid droplets in fixed samples. Histochem Cell Biol. 2002;118:423–428. doi: 10.1007/s00418-002-0462-7. [DOI] [PubMed] [Google Scholar]
- 13.Koopman R, Schaart G, Hesselink MK. Optimisation of oil red O staining permits combination with immunofluorescence and automated quantification of lipids. Histochem Cell Biol. 2001;116:63–68. doi: 10.1007/s004180100297. [DOI] [PubMed] [Google Scholar]
- 14.Radom J, Salvayre R, Maret A, Negre A, Douste-Blazy L. Metabolism of 1-pyrenedecanoic acid and accumulation of neutral fluorescent lipids in cultured fibroblasts of multisystemic lipid storage myopathy. Biochim Biophys Acta. 1987;920:131–139. doi: 10.1016/0005-2760(87)90252-9. [DOI] [PubMed] [Google Scholar]
- 15.Yu W, Bozza PT, Tzizik DM, Gray JP, Cassara J, Dvorak AM, Weller PF. Co-compartmentalization of MAP kinases and cytosolic phospholipase A2 at cytoplasmic arachidonate-rich lipid bodies. Am J Pathol. 1998;152:759–769. [PMC free article] [PubMed] [Google Scholar]
- 16.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: Stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007;48:2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ, Londos C. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J Lipid Res. 1997;38:2249–2263. [PubMed] [Google Scholar]
- 18.Heid HW, Moll R, Schwetlick I, Rackwitz HR, Keenan TW. Adipophilin is a specific marker of lipid accumulation in diverse cell types and diseases. Cell Tissue Res. 1998;294:309–321. doi: 10.1007/s004410051181. [DOI] [PubMed] [Google Scholar]
- 19.D'Avila H, Melo RCN, Parreira GG, Werneck-Barroso E, Castro-Faria-Neto HC, Bozza PT. Mycobacterium bovis bacillus Calmette-Guerin induces TLR2-mediated formation of lipid bodies: intracellular domains for eicosanoid synthesis in vivo. J Immunol. 2006;176:3087–3097. doi: 10.4049/jimmunol.176.5.3087. [DOI] [PubMed] [Google Scholar]
- 20.Melo RCN, Perez SAC, Spencer LA, Dvorak AM, Weller PF. Intragranular vesiculotubular compartments are involved in piecemeal degranulation by activated human eosinophils. Traffic. 2005;6:866–879. doi: 10.1111/j.1600-0854.2005.00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandeira-Melo C, Perez SAC, Melo RCN, Ghiran I, Weller PF. EliCell assay for the detection of released cytokines from eosinophils. J Immunol Methods. 2003;276:227–237. doi: 10.1016/s0022-1759(03)00076-0. [DOI] [PubMed] [Google Scholar]