Abstract
BTZ043, a tuberculosis drug candidate with nanomolar whole-cell activity, targets the DprE1 enzyme of the essential decaprenylphosphoryl-β-D-ribofuranose-2′-epimerase thus blocking biosynthesis of arabinans, vital cell-wall components of mycobacteria. Crystal structures of DprE1, in its native form and in complex with BTZ043, unambiguously reveal formation of a semimercaptal adduct between the drug and an active-site cysteine, as well as contacts to a neighbouring catalytic lysine residue. Kinetic studies confirm BTZ043 as a mechanism-based, covalent inhibitor. This explains the exquisite potency of BTZ043, which, when fluorescently labelled, localizes DprE1 at the poles of growing bacteria. Menaquinone can reoxidize the FAD cofactor in DprE1 and may be the natural electron acceptor for this reaction in the cell. Our structural and kinetic analysis provides both insight into a critical epimerization reaction and a platform for structure-based design of improved inhibitors. Surprisingly, given the colossal tuberculosis burden globally, BTZ043 is the only new drug candidate to have been co-crystallized with its target.
INTRODUCTION
Tuberculosis (TB) represents a formidable challenge to global health and results from infection with the air-borne pathogen Mycobacterium tuberculosis. In recent decades, synergy with the HIV/AIDS pandemic, widespread social instability and poverty have all contributed to a resurgence of TB that increasingly manifests in multidrug-resistant (MDR) and extensively drug-resistant (XDR) forms (1, 2). Faced with the specter of untreatable disease, intensive efforts have been made to discover new drugs to replace the existing TB treatment that was developed in the 1960s. New TB drugs should be highly potent in order to reduce therapy duration and inhibit novel targets to ensure activity against both the various drug-susceptible and drug-resistant strains of M. tuberculosis in circulation.
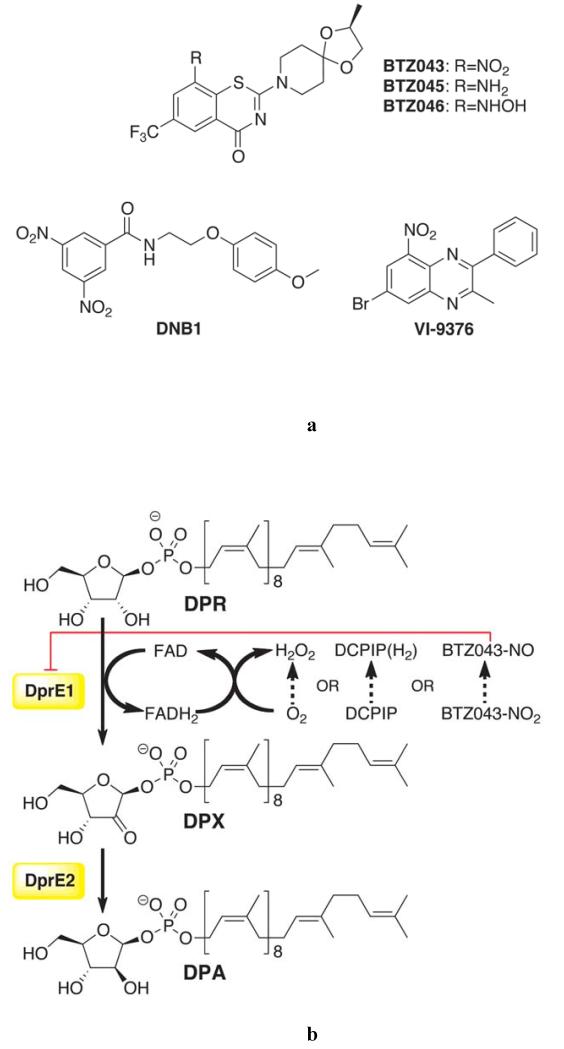
For the first time in 50 years, a portfolio of promising new antitubercular agents is on the horizon (3, 4). Among these, the benzothiazinones (BTZs) represent a class of nitroaromatic molecules that kill M. tuberculosis cells in vitro, ex vivo, and in animal models of the disease (5). BTZ043, currently in the late stages of preclinical development (4), exhibits a minimal inhibitory concentration (MIC) of 1 ng/mL against M. tuberculosis, making it significantly more potent than any of the currently used drugs (Fig. 1A). The cellular target of BTZs is the DprE1 component of the mycobacterial decaprenylphosphoryl-β-D-ribofuranose-2′-epimerase encoded by the neighboring dprE1/dprE2 genes (5), which are essential for growth of M. tuberculosis and Mycobacteriums megmatis (6, 7). Concerted expression of DprE1 and DprE2 is required to carry out the epimerization reaction of decaprenylphosphoryl-β-D-ribofuranose (DPR) into decaprenylphosphoryl-β-D-arabinose (DPA) (Fig. 1B) (8). DPA is the sole precursor for the synthesis of the arabinan moiety of the mycobacterial cell wall, whose unique composition renders mycobacteria insensitive to a number of antibiotics (9). DprE1 from most actinobacteria is susceptible to BTZ043 (5). Furthermore, all M. tuberculosis clinical isolates tested so far, from drug sensitive-, MDR- and XDR-TB cases, were susceptible to BTZ043, making this drug a promising candidate for the treatment of all forms of tuberculosis and possibly for other mycobacterial diseases, such as leprosy (10). More recently, two other families of less potent antitubercular compounds, namely the dinitrobenzamides such as DNB1 (MIC 0.072 μg/mL) and benzoquinoxalines such as VI-9376 (MIC 1 μg/mL) (Fig. 1A), were also found to target DPR epimerization and several other scaffolds targeting this function are being developed (11, 12).
Fig. 1.
Inhibitors and enzymatic activity of DprE1.(A) Structures of antitubercular compound families that target DprE1. BTZ043 (MIC 1 ng/mL) is in late preclinical development (5). Reduced BTZ043 analogues BTZ045 (amino) and BTZ046 (hydroxylamino) show MIC values >500-fold higher than that of BTZ043. DNB1 represents the dinitrobenzamide family of inhibitors (11) (MIC 0.072 μg/mL or 0.02 μM). VI-9376, a benzoquinoxaline, was also reported to target DprE1 (12) (MIC 1 μg/mL or 2.9 μM). (B) Epimerization reaction on the 2′ hydroxyl group of DPR, catalyzed by the mycobacterial DprE1/DprE2. DPR is converted into DPA, an essential precursor for the synthesis of the arabinan moiety of the mycobacterial cell wall (9). DprE1 catalyzes the first step through a FAD-dependent process that requires an electron acceptor for enzyme turnover, which] in vitro can be either molecular oxygen, DCPIP or menaquinone (MQ), as described in this report. BTZ043 was suggested to be converted into a nitroso derivative by DprE1 reduced flavin cofactor (13).
The epimerization of DPR to DPA takes place in two sequential oxidation-reduction reactions: firstly, DprE1 oxidizes DPR to decaprenylphosphoryl-2-keto-β-D-erythro-pentofuranose (DPX), which is then reduced by DprE2 to DPA (Fig. 1B) (13). Genetic analyses of resistant mutants restricted the molecular target of BTZs (as well as that of DNB1 and VI-94376,Fig. 1A) to the DprE1 component of the epimerase (5, 14). In particular, point mutations at a cysteine residue(Cys387 in M. tuberculosis, Cys394 in M. smegmatis)drastically increased the MIC of BTZ043, by up to 10,000-fold, highlighting the potential of DprE1 as a truly selective drug target (15). Mass spectrometric analysis of M. tuberculosis DprE1 over-expressed in M. smegmatis cells treated with BTZ043 demonstrated that the inhibitor is a pro-drug, which is activated inside the cell to a nitroso derivative that covalently reacts with a cysteine residue on the target protein (16). The covalent nature of BTZ043 inhibition was further confirmed in vitro using purified recombinant M. smegmatis DprE1 (13).
DprE1, a 51 kDa protein highly conserved among mycobacteria, shows 83% sequence identity between the M. tuberculosis and M. smegmatis orthologs (Fig. S1). DprE1 shares moderate sequence similarity to flavoenzymes of the vanillyl-alcohol oxidase class (17) and, on this basis, it was predicted to act as a decaprenylphosphoryl-β-D-ribofuranose-2′-oxidoreductase through an FAD-dependent mechanism. Here, we disclose the crystal structure of M. smegmatis DprE1 in its native form and in complex with the BTZ043 inhibitor, revealing the mechanism for covalent inhibition. In addition, we report the biochemical analysis of both wild-type and mutant forms of the protein, and used a fluorescent BTZ analogue to probe the subcellular localization of DprE1.
RESULTS
DprE1 structure reveals a flavoenzyme two-domain topology
Numerous attempts to produce M. tuberculosis DprE1 were made, using multiple constructs and expression systems, but these yielded insoluble or inactive enzyme (see Supplementary Methods and Fig. S2). Consequently, we focused on M. smegmatis DprE1 that was produced in soluble form with good yields from a pET SUMO construct (His6–SUMO tag) or from a pET32b construct (thioredoxin–His6 tag). Cleavage of the protein tags by specific proteases and subsequent purification afforded DprE1 of high purity (Fig. S3), which was used for crystallization trials and activity assays.
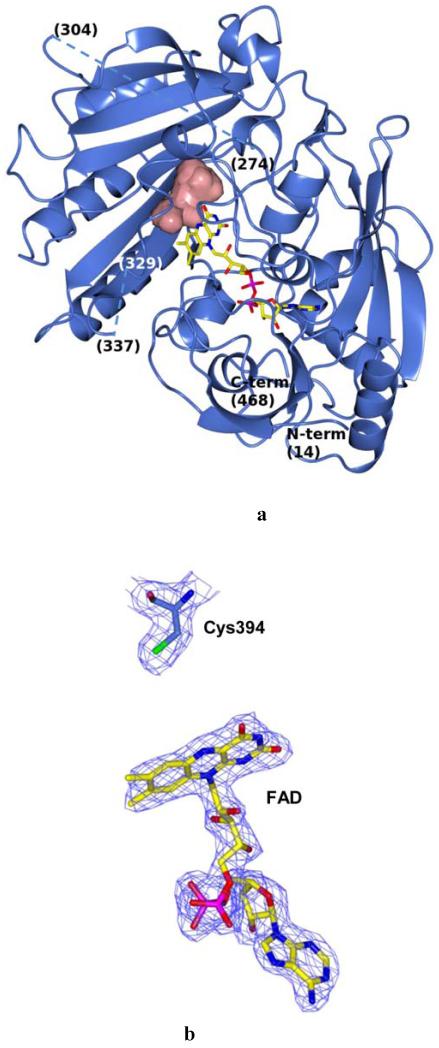
The crystal structure of native M. smegmatis DprE1 in complex with its FAD cofactor was solved at 2.1 Å resolution (Table 1, Fig. 2A). Molecular replacement and energy-optimized rebuilding procedures by the Phaser and Rosetta programs were used to obtain an initial DprE1 model starting from the coordinates of cytokinin dehydrogenase (PDB code 1w1o) (18). Despite the low homology of the starting model (17% sequence identity), the highquality X-ray data (Table 1) combined with powerful computational algorithms produced an excellent electron density map (Fig. 2B). This yielded an almost complete initial DprE1 model which, upon further manual building and refinement, gave the final structure of DprE1. The structure obtained of the 468 amino acid DprE1 protein does not include the first 13 residues and two other segments (residues 275-303 and 330-336), which are not visible in the electron density probably because they are either disordered or adopt multiple conformations (Fig. 2A). SDS-PAGE analysis on dissolved crystals using silver staining ruled out possible proteolytic events.
Table 1.
Data collection and refinement statistics for DprE1 structures
| Native | BTZ043 complex | |
|---|---|---|
| Data collection | ||
| Space group | P3221 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 64.1, 64.1, 191.2 | 39.92, 64.31, 202.84 |
| α, β, γ (°) | 90.00, 90.00, 120.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 60.00-2.10 | 50.00-2.62 |
| R meas | 5.7 (46.6) | 13.8 (68.3) |
| I/ σI | 14.7 (3.0) | 11.6 (2.5) |
| Completeness (%) | 99.9 (99.9) | 98.60 (91.3) |
| Redundancy | 4.9 (4.6) | 4.80 (4.3) |
| Refinement | ||
| Resolution (Å) | 60.00-2.10 | 50.00-2.62 |
| No. reflections | 26,106 | 15,505 |
| Rwork / Rfree | 0.20/0.27 | 0.20/0.24 |
| No. atoms | ||
| Protein | 3229 | 3229 |
| Ligand/ion | 53 | 87 |
| Water | 95 | 88 |
| B-factors | ||
| Protein | 45.4 | 35.7 |
| Ligand/ion | 32.7 | 34.2 |
| Water | 43.6 | 29.9 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.015 | 0.006 |
| Bond angles (°) | 2.031 | 1.108 |
One crystal was used for each structure. Values in parentheses are for highest-resolution shell.
Fig. 2.
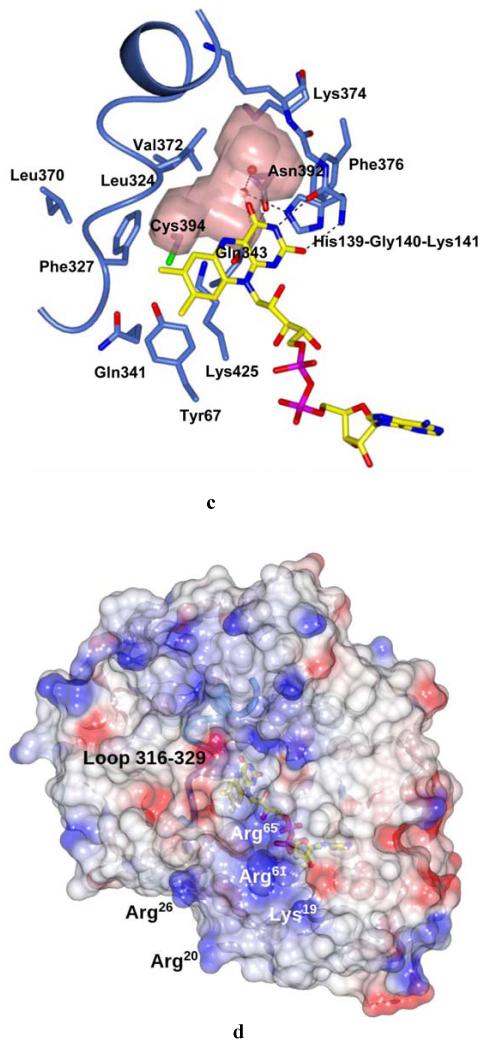
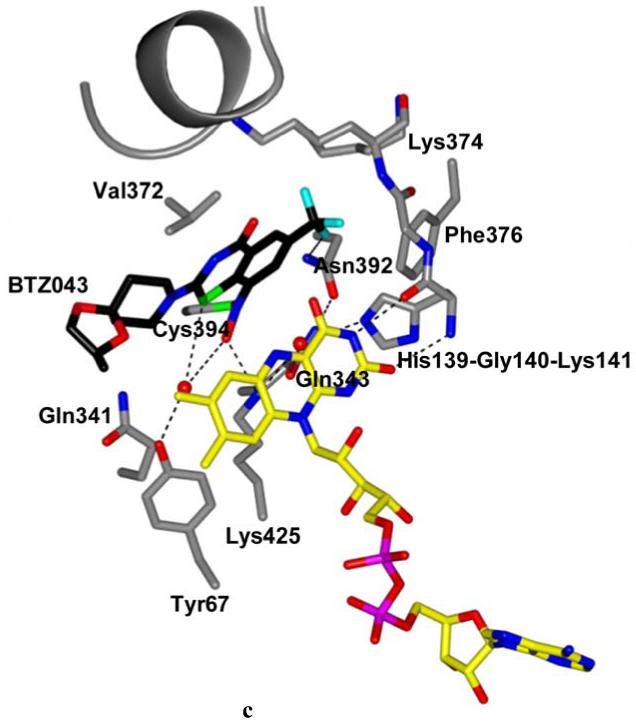
Crystal structure of M. smegmatis DprE1. (A) Light blue ribbon diagram of the DprE1 overall structure with labeled N- and C-termini. Disordered regions are indicated by dashed lines (numbers in parentheses correspond to the residues that flank the disordered parts). The FAD cofactor is represented in yellow with nitrogen, oxygen, and phosphorous atoms colored in blue, red, and magenta, respectively. The active site cavity is shown as pink surface. (B) Refined 2mFo-DFc electron density map (contoured at 1.2σ) for the FAD cofactor and the Cys394 known to be the target of covalent modification by BTZ043. For clarity, electron density contours further than 3Å from atoms in the figure have been omitted. Orientation and color code are as in Fig. 2A, except for protein carbon atoms, which are in light blue; sulfur atom is in green. (C) Close-up view of the DprE1 active site. Molecule orientation and color code are as in Fig. 2B.Water molecules are drawn as red spheres. Black dashed lines indicate H-bonds. The loop formed by residues 316-329 is shown, which shields the active site cavity from outside. (D) Electrostatic surface potential of DprE1 showing positively- and negatively-charged areas in blue and red, respectively. The FAD cofactor is visible in semi-transparency (color code as in Fig. 2B). Loop 316-329 is also visible in semi-transparency.
The DprE1 structure features the common two-domain topology (Fig. 2A) observed in other flavoenzymes such as alditol oxidase and cytokinin dehydrogenase (rmsd values calculated for the 345 Cαatoms of the aligned residues are 2.1 and 2.5 Å, respectively) (18, 19). The isoalloxazine ring of the non-covalently bound FAD lies at the interface between the cofactor-binding domain and the substrate-binding domain, where the two disordered regions are also located. The prosthetic group borders the active site cavity of about 130 Å3, which includes the critical Cys394 (Cys387 in M. tuberculosis DprE1, Figs. 2, B and C) (13). Interestingly, despite the structural and functional divergence, the DprE1 active site shares similarity with that of alditol oxidase (19). In particular, His139, Gly140, Gln341, and Lys425 (Fig. 2C) are conserved, whereas Tyr67, Gln343 and Lys374 are replaced by relatively conserved residues (Phe, Glu and Arg, respectively). This similarity reflects the common chemical activity of the two enzymes, as both oxidize a C-OH group in a sugar substrate. The active site cavity is shielded by a loop formed by residues 316-329, which reside on the protein surface and are followed by one of the two disordered regions of the structure (Fig. 2, A and C). The protein surface located close to this loop and belonging to the FAD-binding domain is sprinkled with a cluster of positively-charged residues (Fig. 2D), which might be involved in interactions with the cell membrane.
To assess the main differences between the DprE1 proteins of M. smegmatis and M. tuberculosis, we generated a homology model of the latter based on the M. smegmatis DprE1 structure using the SWISS-MODEL interface (20-22). Given the high sequence identity between the two DprE1 proteins (83%) the model shows conservation of all residues in the active site, with more significant differences observed on the protein surface.
The DprE1 complex with BTZ043 shows a covalent adduct with Cys394
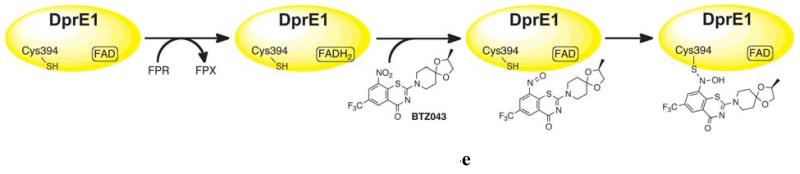
As discussed in the next sections, BTZ043 is a mechanism-based covalent inhibitor, which requires the enzymatic activity of the protein to reduce BTZ043 to the cysteine-reacting nitrosoanalogue. Consequently, the DprE1-BTZ043 adduct had to be generated prior to crystallization trials. This was achieved by incubating DprE1 with BTZ043 and FPR (farnesylphosphoryl-β-D-ribofuranose), an analogue of DPR with a shorter polyprenyl chain, which was shown to be a good enzyme substrate (13). Adduct formation was confirmed by mass spectrometry (Fig. S4). Crystals of the DprE1-BTZ043 complex were obtained in conditions different from those that yielded crystals of the native protein. Inclusion of the ionic liquid tetrabutylphosphonium bromide in the crystallization condition proved crucial to generate good quality crystals of the BTZ043-labeled protein that diffracted to 2.6 Å (Table 1).
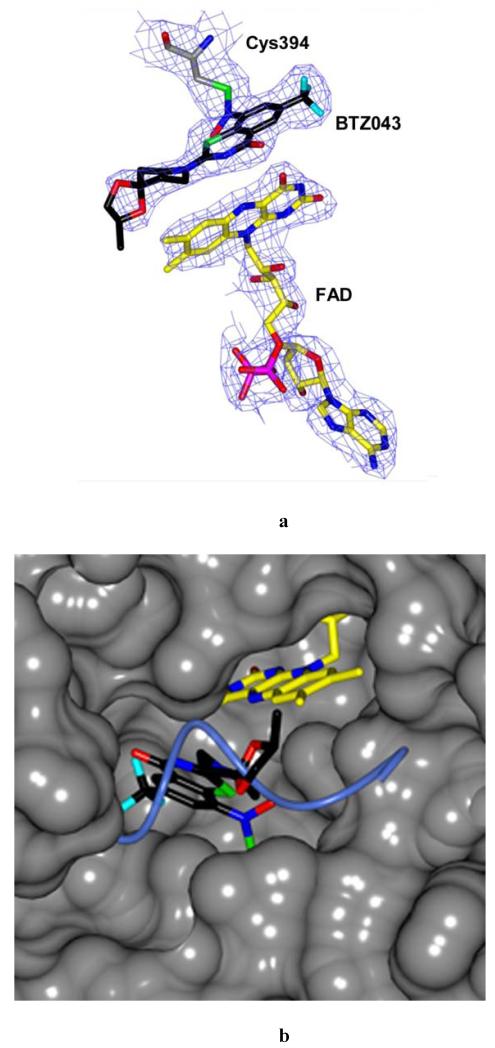
The overall structure of the DprE1-BTZ043 adduct is very similar to that of the native protein (rmsd of 0.285 Å for 346 Cαcarbons), with the FAD cofactor bound exactly in the same conformation. The electron density map was of very good quality and unambiguously showed the presence of a covalent adduct between BTZ043 and Cys394 (Fig. 3, A and B). The covalent adduct causes Cys394 to adopt a rotamer conformation different from that observed in the native structure. BTZ043 binds in front of the FAD isoalloxazine ring with an angle of approximately 45° between the ring systems of the two molecules (Fig. 3A). The loop(residues 324-329) blocking the active site cavity in the native structure (Fig. 2C) is partly disordered in the BTZ043-bound protein (Fig. 3C). Superposition to the unbound protein clearly shows that movement of the loop is required to accommodate the BTZ043 molecule (Fig. 3B) and, most likely, also to bind the DPR substrate.
Fig. 3.
Structure of DprE1 in complex with BTZ043. (A) Refined 2mFo-DFc electron density map provides clear evidence of the formation of the semimercaptal adduct between the reduced BTZ043 form and Cys394 of DprE1. For clarity, electron density contours further than 3Å from atoms in the figure have been omitted. The protein molecule is oriented as the native structure in Figs.2A,B. Color code is as in Fig. 2B except for BTZ043 carbon atoms which are black, with fluorine atoms in cyan. (B) Surface representation showing the DprE1 active site with BTZ043 bound to Cys394 in front of the FAD cofactor. Color code is as in Fig.3A. The structure is rotated approximately 90° around an axis perpendicular to the plane of the figure with respect to the orientation in Figure 3A. Superposition with the native structure shows that a portion of the flexible loop shielding the active site cavity (drawn in light blue as in Fig. 2C) is displaced by the BTZ043 molecule. (C) BTZ043 binding to DprE1 and interactions with active site residues. Structure orientation is the same as in Figure 2C. Color code is as in Figure 3A. Water molecules are represented as red spheres. Dashed lines represent H-bonds. The visible portion (residues 316-323) of the cavity loop is shown in grey.
Comparison with the native structure (Fig. 2C) shows that two water molecules occupying the inner part of the cavity are displaced to a different position in front of the flavin ring, with one of them bridging a network of hydrogen bonds between the inhibitor and Tyr67.Moreover, a key interaction is observed between the semimercaptal hydroxyl group and the side chain of Lys425 (Fig. 3C). The CF3 group of BTZ043 is well placed in a small pocket lined by His139, Gly140, Lys141, Lys374, Phe376 and interacts with the amide of Asn392 (Fig. 3C). Notably, no major interactions are observed with other parts of BTZ043, except for a weak hydrophobic interaction between the sidechain of Leu 370 and the piperidine ring of BTZ043. The spirocyclic moiety of BTZ043 is located at the protein surface and lacks full electron density, namely to account for its methyl group (Fig. 3A). As indicated by the mass spectrometry data, the protein used for crystallization was intact and contained the full BTZ043 semimercaptal adduct (Fig.S4), suggesting that the partially undefined electron density is due to disorder and/or multiple conformations, rather than to degradation.
DprE1: an oxidase or a dehydrogenase?
The oxidative reaction catalyzed by DprE1 at the 2′-hydroxyl group of the DPR substrate implies that its FAD cofactor undergoes a reductive half-reaction generating reduced flavin (FADH2), which has to be re-oxidized by an electron acceptor to start a new catalytic cycle (Fig. 1B). We verified that DprE1 can use molecular oxygen as electron acceptor using a horseradish peroxidase-coupled Amplex Red assay that probes for hydrogen peroxide production due to flavin re-oxidation by oxygen (23), in the presence of the substrate FPR. We also found that DprE1 activity increases by about 30% when excess FAD is present, suggesting that a significant fraction of the protein molecules is in the apo form. This fact is in agreement with our observation that DprE1 tends to lose FAD during purification. Initial kinetic data were obtained at 25°C and the initial velocity versus substrate concentration plot resulted in a sigmoidal curve that was best fitted with the Hill equation (Table 2). This was unexpected because DprE1 is not predicted to be an allosteric enzyme, and gel-filtration chromatography indicated that the protein is monomeric, ruling out activation mechanisms due to oligomerization. The sigmoidal shape of the steady-state kinetics is less pronounced when experiments are performed at 37°C with essentially unchanged kinetic constants.
Table 2.
Steady-state kinetics parameters of M. smegmatis DprE1 enzymes.
| HRP-coupled assaya,b |
DCPIP assaya,d |
|||||
|---|---|---|---|---|---|---|
|
kcat (min−1) |
Km (mM) |
kcat / Km ( min−1 mM−1) |
kcat (min−1) |
Km (mM) |
kcat / Km ( min−1 mM−1) |
|
| Wild type |
3.1 ± 0.4 | 0.19 ± 0.02 | 16.3 ± 2.7 | 12.7 ± 0.6 | 0.11 ± 0.01 | 115.5 ± 10.9 |
| C394G | n.d.c | n.d.c | n.d.c | 2.9 ± 0.2 | 0.36 ± 0.03 | 8.0 ± 0.9 |
| Q343A | n.d.c | n.d.c | n.d.c | 3.2 ± 0.1 | 0.29 ± 0.02 | 11.0 ± 1.3 |
| K425A | not active | not active | ||||
All assays were performed at 25 °C (at 37 °C for progress curves analysis, Fig. 4C) in 20 mM glycylglycine pH 8.5, as this pH value yielded the best kinetic parameters for enzyme activity. No effect of ionic strength up to 250 mM NaCl was observed.
By using the HRP-coupled assay the Michaelis-Menten curve showed a sigmoidal shape with a Hill coefficient of 2.0, which might be related to a low reactivity of DprE1with molecular oxygen (see text). The substrate concentration at half of the maximum velocity is reported as Km (rather than in the more rigorous form s0.5).
n.d., not determined.
The kinetic constants determined for the DCPIP as substrate of the reduced enzyme are Km= 4.2 ± 0.3 μM and kcat= 11.1 ± 0.1 min−1.
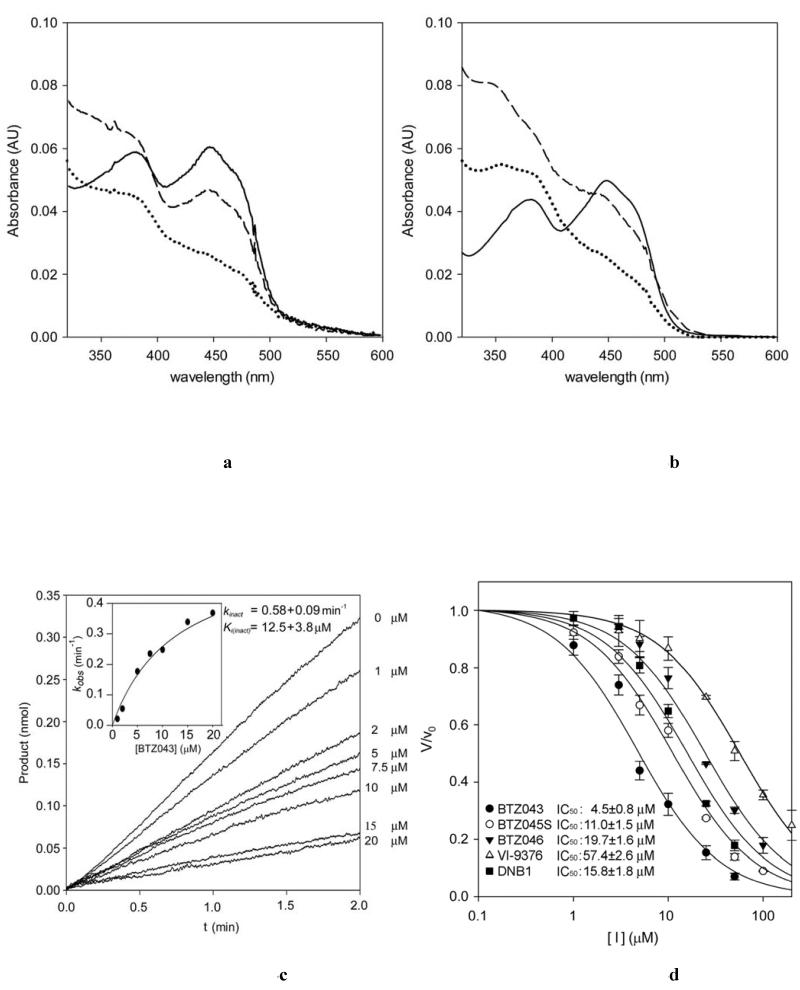
We then probed other known electron acceptors to assess whether these could replace molecular oxygen in re-oxidizing the reduced FAD in DprE1. We observed that 2,6-dichlorophenolindophenol (DCPIP) (24) is an efficient electron acceptor substrate for DprE1, showing a 4-fold increase in the value of kcat compared to that measured for oxygen (Table 2). Furthermore, using DCPIP, the steady-state data could be fitted to the Michaelis-Menten equation without any indication of sigmoidal behavior. These findings indicate that DprE1 is unlikely to be a true oxidase but rather a dehydrogenase that reacts poorly with oxygen and likely uses an alternative electron acceptor in the bacterium such as menaquinone. We then tested whether this electron acceptor, present in mycobacterial membranes, could reoxidize the DprE1-bound FADH2 following its reduction by exposure to light and deazaflavin. On monitoring the UV/V is absorbance spectrum, we observed full reduction of FAD to FADH2, which was promptly converted to its re-oxidized form upon addition of menaquinone (Fig. 2A). Therefore menaquinone could indeed be the physiological substrate for regenerating the oxidized form of DprE1 following each catalytic cycle.
Site-directed mutagenesis identifies Lys 425 as a key residue for DprE1 activity
Following analysis of the active site architecture and the DprE1 residues that interact with BTZ043, we generated three active-site mutants, namely Cys394Gly, Gln343Ala, and Lys425Ala (Table 2) and characterized them by activity studies, using the DCPIP functional assay. Firstly, replacement of Cys394 with Gly hadan evident effect on activity by causing a14-fold decrease in the catalytic efficiency (kcat/Km). Similarly to the Cys394Gly variant, the Gln343Ala mutant is also enzymatically active, but displayed a significant decrease in catalytic efficiency (10-fold), indicating that this residue also plays a role in the active site architecture and enzyme catalysis (Table 2).
Lys425 is homologous to a Lys residue (Lys375) present in the active site of alditol oxidase that is directly involved in catalysis, where it was proposed to activate the sugar substrate by interacting with the hydroxyl group oxidized by the enzyme (19). Strikingly, in DprE1 the Lys425Ala mutant was completely inactive (FAD cofactor was normally incorporated as indicated by its spectral properties), suggesting that Lys425 plays the same role in substrate binding and catalysis as in alditol oxidase. This is consistent with its position in front of the flavin (Fig. 2C) and therefore this residue can be predicted to interact with the 2′-OH group of the substrate, the site of oxidation of DPR by DprE1 (Fig. 1B).
Characterization of DprE1 inhibition by BTZ043
With the DCPIP assay described above, we were able to perform an in-depth analysis of DprE1 inhibition by BTZ043. To verify that DprE1 is responsible for the activation of BTZ043 through its reduction to the nitroso analogue (13, 16), we probed the ability of BTZ043 to re-oxidize protein-bound FADH2 as described above for menaquinone. We verified that following full reduction of FAD to FADH2 as described, this was rapidly re-oxidized upon addition of BTZ043 (Fig. 4B). Enzyme re-oxidation was not observed with the analogues of BTZ043 (BTZ045 and BTZ046)lacking a nitro group. This is consistent with previous results (13), unequivocally demonstrating that BTZ043 is a suicide inhibitor, activated in mycobacteria by reacting with reduced DprE1 and then forming the semimercaptal covalent adduct with Cys394, as illustrated in Fig. 4E. We also performed a kinetic study of DprE1 inactivation by analysis of progress curves at different BTZ043 concentrations (Fig. 4C). We used oxygen as electron-accepting substrate instead of DCPIP as the latter is known to interact with Cys residues over a certain timeframe (25) and could interfere with the formation of the BTZ043-Cys394 adduct. The assays were performed at 37°C to reduce the sigmoidal behavior of the progress curves, which could then be fitted to the single exponential equation (Fig. 4C). The Ki(inact) and kinact for the inhibition of DprE1 by BTZ043 were measured to be 12.5 μM and 0.58 min−1, indicating that covalent in activation of DprE1 by BTZ043 occurs very slowly.
Fig. 4.
DprE1 inhibition by BTZs, DNB1 and VI-9376. (A) UV/Vis absorbance spectrum showing the anaerobic DprE1 flavin re-oxidation carried out by menaquinone. Continuous, dotted and dashed lines correspond to oxidized, photoreduced and BTZ043-reoxidized DprE1, respectively. (B) UV/Vis absorbance spectrum showing the anaerobic DprE1 flavin re-oxidation carried out by BTZ043. Continuous, dotted and dashed lines correspond to oxidized, photoreduced and BTZ043-reoxidized DprE1, respectively. (C) Steady-state progress curves obtained for the inactivation of DprE1 by BTZ043 at different concentrations (reported on the right of each curve) using the peroxidase/Amplex Red coupled assay. The standard reaction mixture contained 20 mM glycylglycine pH 8.5, 150 μM FPR, 0.3μM DprE1, 50 μM Amplex Red, 0.35 μM horseradish peroxidase and the assay was performed at 37°C. Progress curves were fit to the single exponential equation for slow-binding inhibitors: product = v0[1 − exp(-kobst)]/kobs, where v0 is the initial velocity for each curve. The kobs values were then analyzed by the method of Kitz and Wilson to yield kinact and Ki(inact)(see inset) (41). (D) DprE1 initial velocities measured by the DCPIP assay plotted versus inhibitor concentration. In each measurement the protein sample (3 μM) was incubated with 100 μM FPR and variable concentrations of the inhibitor (0-50 or 0-200μM depending on the inhibitor). The IC50 values were determined by fitting the data as described in the online methods. (E) Mechanism of DprE1 inhibition by BTZ043.
Remarkably, the non-covalent BTZ045 and BTZ046 inhibitors exhibit IC50 values that are not drastically different from that of BTZ043; 4.5 μM for BTZ043, 11.0 μM for BTZ045, and 19.7 μM for BTZ046 (measured by the DCPIP assay with a seven-minute incubation time for the enzyme-inhibitor complex; Fig. 4D). Nevertheless, their different mechanisms of inhibition translate into widely different antibacterial activities as discussed below.
Dinitrobenzamides and nitrobenzoquinoxalines are also suicide inhibitors
The dinitrobenzamides (DNBs) and the nitro-benzoquinoxaline VI-9376, initially discovered in whole-cell screens for antitubercular activity, also target DprE1 (11, 12). To investigate the mechanism of inhibition by these compounds, we attempted co-crystallization with DprE1, using the procedure described above for BTZ043. However, despite considerable efforts, high diffraction grade crystals have not yet been obtained. Therefore, we took advantage of the DprE1 crystal structure and performed docking studies with DNB1 and VI-9376 to generate models of the complexes formed between DprE1 and these compounds. The docking results suggest that both compounds indeed fit nicely in the BTZ043 binding site and make the key interactions observed for BTZ043 (Fig. S5). The weaker inhibitory activity of VI-9376 might be due to its small size and the limited number of functional groups, therefore exhibiting only weak interactions with the active site. DNB1, being a more flexible molecule, was seen to adopt a bent conformation in the docked pose, exploring space available in the active site that was not occupied by BTZ043.
We further characterized the DprE1 inhibition by these compounds through enzyme activity studies and mass-spectrometry analysis. Mass spectrometry confirmed that DNB1 (DNB selected for this study) and VI-9376 also form a covalent adduct with DprE1 (Fig. S4). Enzyme labeling reactions were performed in parallel and in the same conditions for BTZ043, DNB1 and VI-9376. Whereas for BTZ043 full labeling of DprE1 was observed, for DNB1 and VI-9376 only partial labeling was achieved (67% and 53%, respectively). Furthermore, the masses observed for the DNB1- and VI-9376-DprE1 adducts were different from those expected based solely on the formation of the semimercaptal product, indicating secondary structural modifications. Finally, the IC50 values determined for DNB1 (15.8 μM) and VI-9376 (57.4 μM) were 3.5- and 12.8-fold higher, respectively, than that determined for BTZ043 (Fig. 4D). Collectively, these data show that DNB1 and VI-9376 are also suicide inhibitors of DprE1 and reflect the weaker antimycobacterial activities of these two compounds, compared to that of BTZ043.
Fluorescently-labeled BTZ localizes DprE1 in the poles of live M. tuberculosis cells
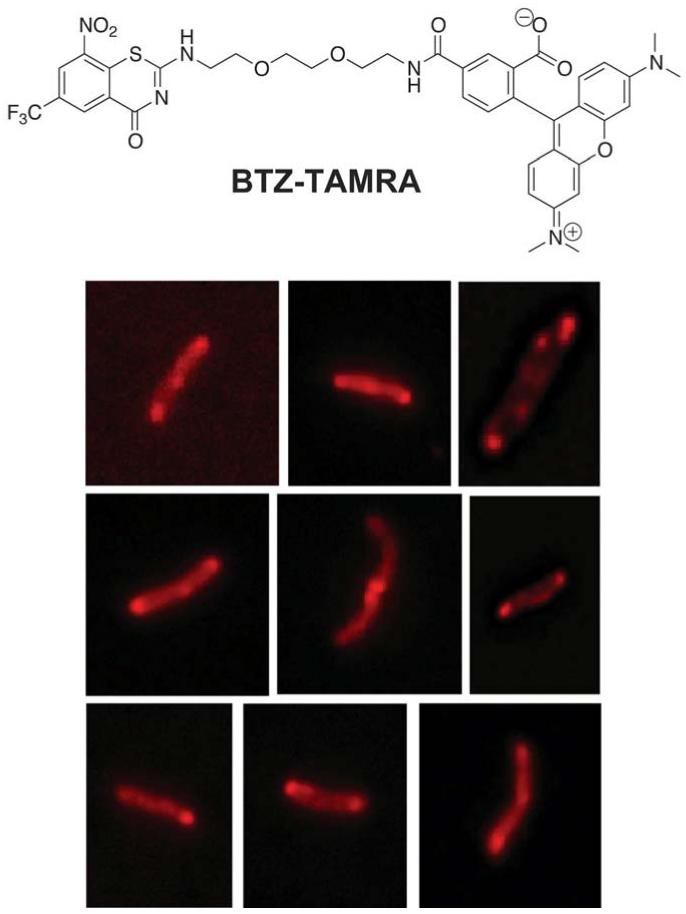
Given that BTZ043covalently binds to recombinant DprE1 very efficiently, we wondered whether a labeled BTZ could be used in live mycobacteria to probe the subcellular localization of this protein and consequently gain further insight into the cidality of this family of compounds. This could provide valuable information regarding uptake of DprE1 inhibitors by mycobacteria and indicate whether the protein is localized in the cytosol or the cell membranes.
We synthesized a fluorescent BTZ043 analogue labeled with TAMRA (BTZ-TAMRA; Fig. 5 and Fig. S6; synthesis described in the Supplementary Materials). To avoid clashes of the fluorophore with the active site and allow efficient binding, TAMRA was connected directly to the benzothiazinone ring through a flexible tri-PEG linker. This compound was tested for antimycobacterial activity and it still exhibited appreciable efficacy (6 μg/mL), taking into account the substantial structural differences with BTZ043, namely the presence of the bulky TAMRA fluorophore. M. tuberculosis was incubated with BTZ-TAMRA, at three different concentrations around the MIC value, and images taken at different time points by fluorescence microscopy. The kinetic analysis clearly showed that BTZ-TAMRA is able to enter the cells quickly and gives rise to highly fluorescent spots localized at the poles of the bacteria there by defining the subcellular localization of DprE1 (Fig. 5).
Fig. 5.
Subcellular localization of DprE1 in M. tuberculosis, probed by BTZ-TAMRA, a fluorescently-labeled BTZ analogue. Exponential M. tuberculosis cultures were exposed to BTZ-TAMRA and observed by fluorescence microscopy at different time points. Images were obtained with excitation and emission wavelengths of 542 and 585 nm, respectively. Higher fluorescence intensity is observed at the poles of the cells.
DISCUSSION
Isoniazid (INH) and ethambutol, two of the four first-line tuberculosis drugs, inhibit cell wall biosynthesis, which remains an attractive target for drug development (26). However, replacement of ethambutol, a weak drug inhibiting the last step in the DPA biosynthetic pathway, by a more potent compound has been envisioned and a possible candidate is BTZ043 (Fig. 1A), which is now in late-stage pre-clinical development. BTZ043 acts two steps upstream of ethambutol by preventing formation of DPA (Fig. 1B), the sole donor of arabinose for biosynthesis of the essential cell wall arabinan components. DprE1 has been widely acclaimed as a novel and tractable target for TB drug development and is one of the few to have been pharmacologically validated. Here, through the structural and functional characterization of DprE1 and its inhibition by BTZ043 and other nitro-compounds, we present a substantial advance in understanding the mechanism of action at the atomic level.
The major difference between the native and BTZ043-bound DprE1 structures resides in a loop segment (residues 323-329) that restricts access to the active site in the native protein (Fig. 2, A and C). Superposition of the two structures clearly shows that binding of BTZ043 and most likely of the DPR substrate to the active site requires a conformational change of this loop (Fig. 3B), which might then serve as a switch between active and inactive forms of the enzyme. DprE1 forms a membrane-associated complex with DprE2 (8) thus the conformational change might take place upon formation of the complex, leading to DprE1 activation. Another disordered loop (residues 275-303) and the disordered N-terminus of the protein (Fig. 2A) are also candidates for protein-protein and protein-membrane interactions. The observed cluster of positively-charged residues at the surface of the protein could provide additional interactions between DprE1 and the membrane surface (Fig. 2D). Further support for this hypothesis comes from the fact that DPR, the natural substrate of DprE1, contains a long hydrophobic decaprenyl chain, and is embedded in the cell membrane. DprE1 is likely a dehydrogenase, and not an oxidase, as it uses organic electron acceptors more efficiently than molecular oxygen to regenerate its FAD cofactor. In particular, our biochemical data indicate that the membrane-embedded menaquinone may represent the natural electron acceptor for reoxidation of the FAD cofactor of DprE1 in mycobacteria.
Subcellular localization studies with the fluorescent BTZ-TAMRA provide a clear indication that DprE1 is located at the poles of mycobacterial cells, demonstrating that this is the site where arabinan biosynthesis occurs (Fig. 5), as was previously reported for peptidoglycan and mycolic acids (27). Synthesis of all three major cell wall components is thus spatially coordinated.
DprE1 oxidative activity and the concomitant FAD reduction are essential for activation of BTZ043 and effective inhibition of the enzyme through the formation of a covalent adduct. Residues that make key interactions with BTZ043, in particular Cys394 and Lys425, are also critical for full enzyme activity. Notably, the mutation of Lys425 led to complete loss of DprE1 activity, showing that this residue plays a key role in the oxidation of the 2′-hydroxyl group of DPR. On the other hand, despite losing significant activity, the Cys394Gly enzyme remained functional, indicating that this residue is not strictly needed for catalysis. This is fully consistent with the viability of the naturally BTZ043-resistant mycobacteria, M. aurum and M. avium where Cys394 has been replaced by Ser or Ala in DprE1, respectively, and with the spontaneous resistant mutants of M. tuberculosis, M. smegmatis or M. bovis BCG bearing Gly or Ser in this position (5). Decreased DprE1 activity may account for a slight defect in growth rate observed for the Cys mutants in M. tuberculosis, M. bovis BCG and M. smegmatis(our unpublished results).
The inactivation of DprE1 by BTZ043 is time-dependent, whereas inhibition by its reduced amino and hydroxylamino analogues (BTZ045 and BTZ046, respectively) is not. However, the respective IC50 values are similar ranging from 4.5 to 20 μM (Fig. 4D) indicating that formation of the initial enzyme-inhibitor complex is likely to be the rate-limiting step in the inhibition process. Hence, the key difference between BTZ043 and its reduced forms lies in the steps that follow binding to the reduced enzyme: whereas BTZ043 can be reduced to the nitroso analogue and immediately react with the adjacent Cys394 there by irreversibly inactivating the enzyme, BTZ045 and BTZ046 cannot undergo this reaction. This explains the striking difference observed in their antitubercular activity (5) and the loss of potency of BTZ043 against the Cys394 mutants: being unable to form a covalent adduct with Cys394, BTZ043 becomes a weak inhibitor of DprE1, just like BTZ045 and BTZ046 with the wild type enzyme. These inhibition studies also highlight a potential limitation of target-based screens using DprE1 since inhibitors like BTZ046 will be found that may not prove effective in killing mycobacteria.
The DNBs and the nitro-benzoquinoxalines, two other families of compounds known to target DprE1 (11, 12), are also mechanism-based irreversible inhibitors. The observed slower labeling of DprE1 by DNB1 and VI-9376 detected by mass spectrometry indicates that these are either reduced less efficiently to their nitroso analogue or, that once generated, this intermediate does not react with Cys394 as promptly as the nitroso-BTZ043. These observations and the higher IC50 values measured for DNB1 and VI-9376 are in agreement with their weaker antimycobacterial activity, when compared to BTZ043.
The structural and enzymatic characterization of DprE1 here described are crucial to deepen our understanding of this target and the mode of action of BTZ043. This is an important development for a drug that is on track to enter clinical studies (4) and for which no resistant M. tuberculosis clinical isolates have been found so far, even among MDR- and XDR-TB cases (10). Nevertheless, our structural data will underpin the development of other drugs, for instance through structure-based design, and enable rational exploration of the active site of the enzyme. Alternative inhibitor scaffolds for DprE1 could then be found, including non-covalent inhibitors, which might be less susceptible to the development of resistance due to mutation of the active site cysteine targeted by BTZ. The potential of this approach is highlighted by the docking studies presented here, in particular for DNB1 that partly occupies a pocket adjacent to the BTZ043 binding site (Fig. S5).
To our knowledge, this is only the second structure of a tuberculosis drug in complex with its target, after that of the INH-NADH adduct in the enoyl-(Acyl-Carrier-Protein) reductase, InhA (28). This is paradoxical given the importance of tuberculosis as a global health problem and reflects in part both the dearth of good targets and the difficult nature of mycobacterial drug discovery.
MATERIALS AND METHODS
Protein expression and purification
All chemicals were purchased from Sigma-Aldrich, unless otherwise stated. M. smegmatisdpre1 (mc2155 strain, msmeg_6382 gene) was cloned into a modified version of pET32b vector (Novagen) containing a PreScission protease cleavage site between the tag and DprE1 (13). All point mutants were generated using the QuickChange (Stratagene) PCR-based method, on the pET32b construct. Mutant enzymes were purified with yields comparable to the wild-type. The proteins expressed from the pET32b construct were produced and purified using a modified protocol (Supplementary Methods). Improvement in protein yield was obtained by re-cloning the gene in the pET SUMO vector (Invitrogen) both in the full-length form of the DprE1 protein and as a N-terminal truncated construct lacking the first 5 amino acids (DprE1-Δ6,Fig.S1), which were predicted to be disordered by bioinformatics analysis. Recombinant proteins were expressed fused to a N-terminal His6-SUMO tag in E. coli BL21-DE3 in LB medium at 25°C for 5 hours (after induction with 1 mM isopropyl-β-D-1-thiogalactopyranoside). Cells were harvested by centrifugation and resuspended in 50 mM sodium phosphate pH 8.0, 300 mM NaCl, 5% (v/v) glycerol, 1 mM phenylmethanesulfonyl fluoride (20 g of cells/100 mL buffer with 2 complete EDTA-free protease inhibitors cocktail tablets, Roche). Cell disruption was carried out by Emulsiflex C3 (Avestin) and the extract was obtained by centrifuging at 70,000g for 45 minutes at 4°C. Protein purification was carried out on an Akta Purifier FPLC system. The cell extract was loaded onto a HisTrap (1 mL, GE Healthcare) and the protein eluted with 100 mM imidazole. The eluted fractions were yellow, indicating that the purified protein incorporated the FAD cofactor. Tag cleavage was achieved by overnight incubation with 0.3 mg of SUMO protease dialyzed against resuspension buffer, followed by a second HisTrap purification step to remove both tag and SUMO protease. After this step the protein tends to lose the FAD cofactor and the sample was therefore incubated with 100 μM FAD for 1 h prior to the following purification steps, namely ion-exchange chromatography using a Mono Q column (GE Healthcare) followed by gel filtration (Superdex 200 HR 10/300, GE Healthcare) in 25 mM potassium phosphate buffer pH 7.2, 5% glycerol. Sample purity was checked by SDS-PAGE and protein concentration was evaluated by UV/Vis absorbance spectrum. The extinction coefficient at 449 nm (related to the FAD cofactor) was experimentally determined to be 10300 M−1 cm−1 for the full-length protein and 13530 M−1 cm−1 for the DprE1-Δ6 construct. The Abs280/Abs449 ratio was used to evaluate the fraction of holoenzyme (i.e. containing FAD) with respect to total protein.
Activity and inhibition studies
The enzyme activity of DprE1 was spectrophotometrically determined using farnesylphosphoryl-β-D-ribofuranose (FPR) as substrate and 2,6-dichlorophenolindophenol (DCPIP) as electron acceptor. FPR was synthesized as previously reported (29). Enzyme assays were performed on a Varian Cary 100 UV-visible spectrophotometer equipped with temperature-controlled cell holder (T = 25 °C), measuring the decrease in absorbance at 600 nm of DCPIP (ε= 19100 M−1 cm−1). The standard reaction mixture contained 20 mM glycylglycine buffer pH 8.5, 500 μM FPR, 50 μM DCPIP, and the reaction was started by adding the enzyme solution (final concentration 0.3μM). Alternatively, the reactivity with molecular oxygen was determined measuring the H2O2 formation by the horseradish peroxidase/Amplex Red coupled assay, following the formation of resorufin at 560 nm (ε = 54000 M−1 cm−1) at 25 °C (or 37 °C for the progress curve analysis). The standard reaction mixture contained 20 mM glycylglycine pH 8.5, 500 μM FPR, 50 μM Amplex Red, 0.35 μM horseradish peroxidase. Steady-state kinetics studies were performed by assaying the activity at different FPR concentrations (10-500 μM). All measurements were performed in triplicate. The kinetic constants, Km and kcat, were determined fitting the data to the Hill equation using Origin 8 software.
For inhibition studies, DprE1 (3 μM) was incubated for 7 minutes with 100 μM FPR in the presence of different inhibitor concentrations (0-100 μM for BTZs and DNB1 inhibitors; 0-200 μM for VI-9376; all inhibitors were dissolved in 100% DMSO whose concentration was kept constant in the incubation mixture). Then the enzyme activity was measured using the DCPIP assay with a final protein concentration of 0.3 μM (see above). The IC50 values were obtained by plotting the initial velocities with the following equation:
where A[I] is the enzyme activity at inhibitor concentration [I] and A[0] is the enzyme activity without inhibitor.
The ability of menaquinone and BTZs to reoxidize the flavin of DprE1 was determined as follows: DprE1 solution (10 μM) was made anaerobic in a rubber septum-sealed cuvette (Hellma) by alternating cycles of vacuum with flushing of O2-free argon. The enzyme was then photoreduced by light irradiation in the presence of 0.1 μM 5-deazaflavin, and the photoreaction was followed spectrophotometrically. BTZ043 or menaquinone(10 μM) was then added and the flavin re-oxidation monitored spectrophotometrically.
Crystallization and structure determination
Two crystal forms of DprE1 were obtained with the recombinant protein produced from the pET32b and the pET SUMO constructs. Crystals of the native protein were obtained with the DprE1-Δ6 pET SUMO construct (Fig. S1) by the hanging-drop vapour diffusion method at 20°C. Experiments were set up by mixing 1 μl of the protein sample (3 mg/mL in 25 mM potassium phosphate buffer pH 7.2, 5% glycerol, 100 μM FAD) with an equal volume of the reservoir solution containing 30% PEG4000, 100 mM sodium citrate pH 5.6, 200 mM ammonium acetate. Yellow crystals grew in approximately 4-5 weeks and were transferred to a cryo-protectant solution (reservoir solution with 15% glycerol) prior to flash-cooling in liquid nitrogen. X-ray data were collected at the X06SA beamline of the Swiss Light Synchrotron (SLS, Villigen) and at the ID14-EH1 beamline of the European Synchrotron Radiation Facility (ESRF, Grenoble). Data processing and scaling were carried out by MOSFLM (30) and the CCP4 package (31) (Table 1). The DprE1 crystal structure was solved by molecular replacement with Phaser (32), using as a model an ensemble constructed from six distant homologues: PDB entry 2vfr (20% sequence identity), 2i0k (15%), 3js8 (15%), 2exr (17%), 2bvf (17%) and 1w1o (17%). Selection and sequence alignment of the homologues was performed with HHPRED (33). The sequence alignment was used to trim off non-identical side chains in Sculptor (34), and the program Ensembler (http://www.phenix-online.org/documentation/ensembler.htm) was used to superimpose the six structures and trim non-conserved surface loops. All 6 individual models from the ensemble were used for automated rebuilding using phenix.mr_rosetta (35), and the best results were obtained starting from cytokinin dehydrogenase (1w1o), which gave a model with a Free R factor of 33%. The model was then partly manually rebuilt in Coot (36) and refined by REFMAC5 (37). Cavity calculations were carried out with VOIDOO (38). Structural illustrations were produced using CCP4mg (39).
Crystals of full-length DprE1 (fl-DprE1, Fig. S1) with BTZ043 were obtained with protein produced from the pET32b construct. The protein (approximately 20 μM) was incubated for 2h at 37°C in the presence of 25 μM FAD, 50 μM BTZ043 and 100 μM FPR, in 20 mM Tris pH 7.5, 100 mM NaCl and 1 mM MgCl2. The protein was concentrated to approximately 10 mg/mL on an Amicon centrifugal device (10,000 MWCO, Millipore). Crystals were obtained by the hanging-drop vapor diffusion method at 18°C. Experiments were set up by mixing 0.5 μl of the protein sample with 1 μl of the reservoir solution containing 11% PEG300, 90 mM MES pH 5.9, 5% tetrabutylphosphonium bromide (Hampton Research). Yellow crystals grew in approximately 2-3 days and were transferred to a cryo-protectant(reservoir solution with 25% glycerol) prior to flash-cooling in liquid nitrogen. X-ray data were collected at the X06DA beamline of the Swiss Light Synchrotron (SLS, Villigen). Data processing and scaling were carried out in XDS (40) (Table 1). The DprE1-BTZ043 complex crystal structure was solved by molecular replacement using the native DprE1 structure as model. Manual adjustments of the model were made in COOT (36), followed by refinement using REFMAC5 (37), part of the CCP4i program suite (31). In the refined structures all residues fall in the allowed regions of the Ramachandran plot.
Subcellular localization studies with BTZ-TAMRA
BTZ-TAMRA was synthesized as described in the Supplementary Materials. Exponential cultures of M. tuberculosis grown in 7H9 medium were exposed to different concentrations of BTZ-TAMRA (1-12 μg/ml) for periods of 24-48 hours. A few microliters of culture were placed between two coverslips and cells were observed by fluorescence microscopy using a personal Delta Vision inverted microscope (Applied Precision, Washington) equipped with a 100X objective (Olympus Plan Semi Apochromat, 1.3 numerical aperture). Cells were illuminated using Insight SSI solid state illumination with excitation wavelengths 475/28 (FITC), 542/27 (RD-TR-PE) and emission filters 512/18 for FITC and 585/29 for RD-TR-PE. Exposure times varied from 0.05 s to 0.2 s. Images were captured using a CoolSnap HQ2 camera and SoftWorx software (Applied Precision, Washington), and processed for publishing using Adobe Illustrator.
One Sentence Summary: The crystal structure of DprE1 benzothiazinone BTZ043, a potent antitubercular drug.
Supplementary Material
Acknowledgements
We thank Vadim Makarov, Priscille Brodin and Gyorgy Keri for kindly providing inhibitors, Laure Menin for technical assistance with mass spectrometry, Avram Liav for advice on the synthesis of FPR, Kai Johnsson for comments on the manuscript, Roberto Orrù for technical assistance and helpful discussions, the Protein Crystallography Core Facility of EPFL, the European Synchrotron Radiation Facility and the Swiss Light Source for beam time and excellent support during X-ray data collection.
Funding:Supported by the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 260872, and by Fondazione Cariplo (N° 2008.3148). J.N. is the recipient of an International Incoming Marie Curie fellowship (252802 – DPRETB) from the European Commission. F.P. is a Swiss National Science Foundation MHV Post-doctoral Fellow. R.J.R. is funded by a Principal Research Fellowship from the Wellcome Trust (082961/Z/07/Z).
Footnotes
Competing interests: All authors declare no competing financial interests except STC who holds patents related to this work.
Data and materials availability: Coordinates and structure factors have been deposited in the Protein Data Bank with access codes 4AUT for native DprE1 and 4F4Q for the DprE1-BTZ043 complex.
References
- 1.Velayati AA, Farnia P, Masjedi MR, Ibrahim TA, Tabarsi P, Haroun RZ, Kuan HO, Ghanavi J, Varahram M. Totally drug-resistant tuberculosis strains: evidence of adaptation at the cellular level. European Respiratory Journal. 2009;34:1202. doi: 10.1183/09031936.00081909. [DOI] [PubMed] [Google Scholar]
- 2.Zignol M, van Gemert W, Falzon D, Sismanidis C, Glaziou P, Floyd K, Raviglione M. Surveillance of anti-tuberculosis drug resistance in the world: an updated analysis, 2007-2010. Bulletin of the World Health Organization. 2012;90:111. doi: 10.2471/BLT.11.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole ST, Riccardi G. New tuberculosis drugs on the horizon. Current Opinion in Microbiology. 2011;14:570. doi: 10.1016/j.mib.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 4.Lienhardt C, Raviglione M, Spigelman M, Hafner R, Jaramillo E, Hoelscher M, Zumla A, Gheuens J. New Drugs for the Treatment of Tuberculosis: Needs, Challenges, Promise, and Prospects for the Future. Journal of Infectious Diseases. 2012;205:S241. doi: 10.1093/infdis/jis034. [DOI] [PubMed] [Google Scholar]
- 5.Makarov V, Manina G, Mikusova K, Moellmann U, Ryabova O, Saint-Joanis B, Dhar N, Pasca MR, Buroni S, Lucarelli AP, Milano A, De Rossi E, Belanova M, Bobovska A, Dianiskova P, Kordulakova J, Sala C, Fullam E, Schneider P, McKinney JD, Brodin P, Christophe T, Waddell S, Butcher P, Albrethsen J, Rosenkrands I, Brosch R, Nandi V, Bharath S, Gaonkar S, Shandil RK, Balasubramanian V, Balganesh T, Tyagi S, Grosset J, Riccardi G, Cole ST. Benzothiazinones Kill Mycobacterium tuberculosis by Blocking Arabinan Synthesis. Science. 2009;324:801. doi: 10.1126/science.1171583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crellin PK, Brammananth R, Coppel RL. Decaprenylphosphoryl-beta-D-Ribose 2′-Epimerase, the Target of Benzothiazinones and Dinitrobenzamides, Is an Essential Enzyme in Mycobacterium smegmatis. Plos One. 2011;6 doi: 10.1371/journal.pone.0016869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12989. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikusova K, Huang HR, Yagi T, Holsters M, Vereecke D, D’Haeze W, Scherman MS, Brennan PJ, McNeil MR, Crick DC. Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. Journal of Bacteriology. 2005;187:8020. doi: 10.1128/JB.187.23.8020-8025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolucka BA. Biosynthesis of D-arabinose in mycobacteria - a novel bacterial pathway with implications for antimycobacterial therapy. Febs Journal. 2008;275:2691. doi: 10.1111/j.1742-4658.2008.06395.x. [DOI] [PubMed] [Google Scholar]
- 10.Pasca MR, Degiacomi G, Ribeiro A. L. d. J. L., Zara F, De Mori P, Heym B, Mirrione M, Brerra R, Pagani L, Pucillo L, Troupioti P, Makarov V, Cole ST, Riccardi G. Clinical Isolates of Mycobacterium tuberculosis in Four European Hospitals Are Uniformly Susceptible to Benzothiazinones. Antimicrobial Agents and Chemotherapy. 2010;54:1616. doi: 10.1128/AAC.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christophe T, Jackson M, Jeon HK, Fenistein D, Contreras-Dominguez M, Kim J, Genovesio A, Carralot J-P, Ewann F, Kim EH, Lee SY, Kang S, Seo MJ, Park EJ, Skovierova H, Pham H, Riccardi G, Nam JY, Marsollier L, Kempf M, Joly-Guillou M-L, Oh T, Shin WK, No Z, Nehrbass U, Brosch R, Cole ST, Brodin P. High Content Screening Identifies Decaprenyl-Phosphoribose 2′ Epimerase as a Target for Intracellular Antimycobacterial Inhibitors. Plos Pathogens. 2009;5 doi: 10.1371/journal.ppat.1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magnet S, Hartkoorn RC, Szekely R, Pato J, Triccas JA, Schneider P, Szantai-Kis C, Orfi L, Chambon M, Banfi D, Bueno M, Turcatti G, Keri G, Cole ST. Leads for antitubercular compounds from kinase inhibitor library screens. Tuberculosis. 2010;90:354. doi: 10.1016/j.tube.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Trefzer C, S’Kovierova H, Buroni S, Bobovska A, Nenci S, Molteni E, Pojer F, Pasca MR, Makarov V, Cole ST, Riccardi G, Mikusova K, Johnsson K. Benzothiazinones Are Suicide Inhibitors of Mycobacterial Decaprenylphosphoryl-beta-D-ribofuranose 2 ’-Oxidase DprE1. Journal of the American Chemical Society. 2012;134:912. doi: 10.1021/ja211042r. [DOI] [PubMed] [Google Scholar]
- 14.Ribeiro A. L. d. J. L., Degiacomi G, Ewann F, Buroni S, Incandela ML, Chiarelli LR, Mori G, Kim J, Contreras-Dominguez M, Park Y-S, Han S-J, Brodin P, Valentini G, Rizzi M, Riccardi G, Pasca MR. Analogous Mechanisms of Resistance to Benzothiazinones and Dinitrobenzamides in Mycobacterium smegmatis. Plos One. 2011;6 doi: 10.1371/journal.pone.0026675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manina G, Pasca MR, Buroni S, De Rossi E, Riccardi G. Decaprenylphosphoryl-beta-D-Ribose 2 ’-Epimerase from Mycobacterium tuberculosis is a Magic Drug Target. Current Medicinal Chemistry. 2010;17:3099. doi: 10.2174/092986710791959693. [DOI] [PubMed] [Google Scholar]
- 16.Trefzer C, Rengifo-Gonzalez M, Hinner MJ, Schneider P, Makarov V, Cole ST, Johnsson K. Benzothiazinones: Prodrugs That Covalently Modify the Decaprenylphosphoryl-beta-D-ribose 2 ’-epimerase DprE1 of Mycobacterium tuberculosis. Journal of the American Chemical Society. 2010;132:13663. doi: 10.1021/ja106357w. [DOI] [PubMed] [Google Scholar]
- 17.Fraaije MW, Van Berkel WJH, Benen JAE, Visser J, Mattevi A. A novel oxidoreductase family sharing a conserved FAD-binding domain. Trends in Biochemical Sciences. 1998;23:206. doi: 10.1016/s0968-0004(98)01210-9. [DOI] [PubMed] [Google Scholar]
- 18.Malito E, Coda A, Bilyeu KD, Fraaije MW, Mattevi A. Structures of Michaelis and product complexes of plant cytokinin dehydrogenase: Implications for flavoenzyme catalysis. Journal of Molecular Biology. 2004;341:1237. doi: 10.1016/j.jmb.2004.06.083. [DOI] [PubMed] [Google Scholar]
- 19.Forneris F, Heuts DPHM, Delvecchio M, Rovida S, Fraaije MW, Mattevi A. Structural analysis of the catalytic mechanism and stereo selectivity in Streptomyces coelicolor alditol oxidase. Biochemistry. 2008;47:978. doi: 10.1021/bi701886t. [DOI] [PubMed] [Google Scholar]
- 20.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 21.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 22.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Research. 2003;31:3381. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith TA, Baker JH. In: Progress in polyamine reseacrh. Novel biochemical, pharmacological and clinical aspects. Zappia AEPV, editor. Plenum Press; NY: 1988. pp. 573–589. [Google Scholar]
- 24.Vanderjagt DJ, Garry PJ, Hunt WC. Ascorbate in plasma as measured by liquid-chromatography and by dichlorophenolindophenol colorimetry. Clinical Chemistry. 1986;32:1004. [PubMed] [Google Scholar]
- 25.Coffey DS, Hellerma L. Interaction of 2,6-dichloroindophenol and protein sulfhydryl groups. Biochimica Et Biophysica Acta. 1965;100:98. doi: 10.1016/0304-4165(65)90431-9. [DOI] [PubMed] [Google Scholar]
- 26.Tahlan K, Boshoff HI. New cell wall biosynthesis inhibitors under active development for tuberculosis. Drugs of the Future. 2009;34:739. [Google Scholar]
- 27.Thanky NR, Young DB, Robertson BD. Unusual features of the cell cycle in mycobacteria: Polar-restricted growth and the snapping-model of cell division. Tuberculosis. 2007;87:231. doi: 10.1016/j.tube.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Rozwarski DA, Grant GA, Barton DHR, Jacobs WR, Sacchettini JC. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science. 1998;279:98. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 29.Liav A, Swiezewska E, Ciepichal E, Brennan PJ. Stereoselectivity in the synthesis of polyprenylphosphoryl beta-D-ribofuranoses. Tetrahedron Letters. 2006;47:8781. [Google Scholar]
- 30.Leslie AGW. Integration of macromolecular diffraction data. Acta Crystallographica Section D-Biological Crystallography. 1999;55:1696. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 31.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallographica Section D-Biological Crystallography. 2011;67:235. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallographica Section D-Biological Crystallography. 2007;63:32. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Research. 2005;33:W244. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunkoczi G, Read RJ. Improvement of molecular-replacement models with Sculptor. Acta Crystallographica Section D-Biological Crystallography. 2011;67:303. doi: 10.1107/S0907444910051218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D. F. Terwilliger TC, Read RJ, Baker D, Bunkóczi G, Adams PD, Grosse-Kunstleve RW, Afonine PV, Echols N. phenix.mr_rosetta: molecular replacement and model rebuilding with Phenix and Rosetta. J Struct Funct Genomics. 2012 doi: 10.1007/s10969-012-9129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D-Biological Crystallography. 2004;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D-Biological Crystallography. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallographica Section D-Biological Crystallography. 1994;50:178. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 39.McNicholas S, Potterton E, Wilson KS, Noble MEM. Presenting your structures: the CCP4 mg molecular-graphics software. Acta Crystallographica Section D-Biological Crystallography. 2011;67:386. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch W. XDS. Acta Crystallographica Section D-Biological Crystallography. 2010;66:125. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Copeland RA. Enzymes: A practical introduction to structure, mechanism and data analysis. ed. 2nd John Wiley & Sons Inc.; New York, NY: 2000. [Google Scholar]
- 42.Jones G, Willett P, Glen RC. Molecular recognition of receptor-sites using a genetic algorithm with a description of desolvation. Journal of Molecular Biology. 1995;245:43. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]
- 43.Makarov VA. Application: WO2011132070. 2011
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.