Abstract
In Drosophila, Hedgehog (Hh) signal transduction has been shown to require a multiprotein complex (Hedgehog signaling complex (HSC)), which includes the Kinesin-related protein Costal2 (Cos2), the serine/threonine protein kinase Fused (Fu), and the transcription factor Cubitus interruptus (Ci). We present evidence that a biologically relevant fraction of the HSC is found in association with cellular membranes. We demonstrate that Cos2 is capable of tethering an exogenous protein to vesicular membranes and that Cos2 association with membranes is Hh-sensitive. In addition, we demonstrate that Cos2 associates with membranes in cells that lack the transmembrane protein Smoothened (Smo) through a domain of Cos2 distinct from its recently characterized Smo binding domain. We suggest that an Hh-regulated membrane binding activity of Cos2 is part of the mechanism by which Cos2 contributes to Hh signaling. We propose a model in which there are two distinct HSCs with discrete subcellular localizations and activities: one is endosome-associated and facilitates production of a repressor form of Ci (HSC-R), and one is Smo-associated and promotes Ci activation (HSC-A). In response to Hh and through interaction with Cos2, Smo mediates both inhibition of the endosome-associated HSC-R and activation of HSC-A at the plasma membrane.
The Hedgehog (Hh)1 signaling pathway is conserved from invertebrates to humans where it is required for differentiation and growth of a diverse array of cell types during development (1). Consequently misregulation of the Hh pathway causes congenital defects and oncogenesis (2-12). We are only just beginning to understand the mechanism of Hh signal transduction. In Drosophila, the secreted protein Hh binds to the 12-transmembrane protein Patched (Ptc) (13-19), which, upon binding Hh, no longer inhibits the activity of the 7-transmembrane protein Smoothened (Smo) (20-26). Smo then activates the downstream components of the pathway, which include the Kinesin-related protein (KRP) Costal2 (Cos2), the serine/threonine protein kinase Fused (Fu), and the transcription factor Cubitus interruptus (Ci) (27-42). Fu, Cos2, and Ci form the core of a multiprotein complex (HSC) that is required for Hh signal transduction (43-46). The HSC has been found to associate with microtubules (MTs) and to do so in an Hh-sensitive manner, suggesting that a Kinesin-like activity of Cos2, regulated MT binding, is relevant to Hh signal transduction (37, 43).
In the absence of Hh, Cos2 and Fu are required to maintain the proteolytic conversion of Ci to Ci75, an active repressor of some Hh target genes (33, 47, 48). It has been postulated that Cos2 tethers the HSC to MT to provide for Ci75 production as well as to provide cytoplasmic retention of full-length Ci (42, 43, 49, 50). In the presence of Hh, the production of Ci75 is halted, resulting in an increased amount of full-length Ci that equilibrates between the nucleus and the cytosol (46, 48). Stabilization of Ci in its full-length form is not sufficient to stimulate maximal transcription of Hh target genes (41, 49). Additional activation steps, which also require Cos2 and Fu, are necessary to activate the full-length form of Ci (33, 47, 49-55). It is currently unknown how Cos2 contributes to the stabilization and activation of full-length Ci. Recently it has been shown that Cos2 can bind to Smo and that this interaction is important for Ci activation (56, 57).
Kinesins have been shown to play a role in several signaling pathways, providing required subcellular localization to membrane-associated complexes of signaling components (58, 59). KRPs generally contain a conserved motor domain (which includes an MT binding domain and an ATPase domain) and an extracatalytic tail domain (60). KRPs localize signaling components to specific membrane-bound vesicles and do so in a regulated manner (61-65). Here we investigate the role of Cos2 in targeting the HSC to various subcellular compartments in a manner consistent with it being a KRP. We demonstrate that the HSC can exist as a membrane-associated complex whose localization is altered by Hh. We demonstrate that Cos2 and Fu associate with vesicular and plasma membranes and that Cos2 is able to provide this localization to an exogenous protein. Finally we provide evidence that the bulk of Cos2 associates with membranes in a manner independent of Smo. We conclude that Cos2 has a membrane tethering domain that is distinct from its Smo association domain and from its putative MT binding domain. We suggest a model in which an HSC tethered directly to membranes by Cos2 (HSC-R) provides for production of Ci75, while an HSC bound to Smo through Cos2 (HSC-A) provides for Ci activation. The differential targeting of these complexes may provide the subcellular localization required for differential processing of Ci into a transcriptional activator or a transcriptional repressor.
EXPERIMENTAL PROCEDURES
Preparation of Cell Lysates
Drosophila embryos and S2 cells were hypotonically lysed as described previously (56). Low speed supernatants were generated by centrifugation at 5,000 or 2,000 × g for 20 min and then centrifuged for 1 h at 100,000 × g. The resulting high speed supernatant consisted of the cytosolic fraction and was termed “S2.” The high speed pellet, “P2,” consisted of total cellular membranes and was washed by homogenization with a tight fitting glass Dounce in 0.15 m NaCl HLB (50 mm β-glycerophosphate, 10 mm NaF, 1 mm EGTA, 1 mm dithiothreitol, protease inhibitor cocktail (pH 7.6). This suspension was centrifuged at 100,000 × g for 1 h, yielding “S3” and “P3.” In this manner the high speed pellet P3 was sequentially extracted in 0.5 m NaCl, 0.75 m NaCl, 1 m NaCl, and finally 1% Nonidet P-40 HLB. Each extraction was normalized to total protein, separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted. The 0.5 m NaCl (“S4”) extraction was also used to immunoprecipitate HSC members. The immunoprecipitates were separated by SDS-PAGE and immunoblotted.
Equilibrium Density Centrifugation
The P2 from S2 cells was washed by homogenization with a Dounce in TNE (150 mm NaCl, 100 mm Tris, 0.20 mm EGTA, pH 7.4) and centrifuging at 100,000 × g for 1 h. This pellet, P3, was resuspended in 1.4 m sucrose TNE (3-ml final volume). This was overlaid in a centrifuge tube with 1.22 m sucrose TNE and 0.1 m sucrose TNE. After centrifugation for 18 h at 112,000 × g, fractions were collected starting from the top of the centrifuge tube.
Antibodies
Anti-Fu was FuH (31), anti-Cos2 was 5D6 (33), anti-Ci was 2A1 (40), and anti-Suppressor of fused (Su(fu)) is described in Ref. 46. Anti-Tubulin was β512 from Sigma, mouse IgG1 was from Zymed Laboratories Inc., anti-Kinesin was Kin01 from Cytoskeleton. Anti-HA was HA.11 or anti-HA-488; both were from Covance. Anti-Fasciclin was F5H7, a kind gift from Dr. M. Hortsch (66). Anti-Rab11 was a kind gift from Dr. R. Cohen. Anti-Cadherin was DCAD1, a kind gift from Dr. T. Uemura (67). Secondary antibodies for immunoblots, from Jackson Immunoresearch Laboratories, were conjugated to horseradish peroxidase. Secondary antibodies, from Molecular Probes, for immunofluorescence were anti-rabbit-546 and anti-rat-546.
Electron Microscopy
Following equilibrium density centrifugation, fractions containing the vesicular and pellet fractions were diluted 1:3 in TNE, centrifuged at 100,000 × g, and then resuspended in TNE. These resuspensions were mixed with an equal volume of 5% glutaraldehyde in wash buffer (100 mm sodium cacodylate, pH 7.4) and allowed to fix for 5 min followed by centrifugation at 100,000 × g for 0.5 h. The pellet was fixed an additional 30 min in 2.5% glutaraldehyde in wash buffer, washed in wash buffer, treated with 1% OsO4 for 30 min, washed, dehydrated, and embedded in Epon resin. 85-nm sections were made using a Reichert Ultra-cut E. Sections were collected on copper grids, stained with uranyl acetate and lead citrate, and then examined using a JEOL 1230 JEM transmission electron microscope at 80 kV.
Reporter Assays
Transfections were carried out using Cellfectin transfection reagent according to the manufacturer’s instructions (Invitrogen) and according to procedures described previously (56). ptcΔ136-Luc 35 was used as the reporter construct for Hh target gene transcription (41).
Smo dsRNA
We prepared dsRNA using the primers and method previously shown to reduce Smo protein levels in Drosophila cells (77, 78). We treated S2 and Cl8 cells as described previously (77). Briefly we treated cells with dsRNA during transfection with appropriate plasmids using Cellfectin. Cells were lysed or prepared for immunofluorescence 48 h post-treatment.
Immunofluorescence of S2 Cells
S2 cells were prepared for indirect immunofluorescence and detection of eGFP fluorescence as described previously (56). Confocal images were collected using the LSM-510 confocal laser scanning microscope (Zeiss) and processed using LSM Image Browser software (Zeiss) and Adobe Photoshop Version 6.0. Non-confocal images were taken with an Orca-ER, black and white cooled CCD camera attached to a Zeiss Axioplan Imaging 2 microscope. These images were deconvolved using AutoDeblur software (Version 8, AutoQuant) and processed using Photoshop Version 6.0.
DNA Constructs
Various Cos2 truncation mutants were generated by PCR amplification of the appropriate regions using Cos2 cDNA as a template (37). PCR oligonucleotides were designed with BglII restriction sites flanking the amplified Cos2 fragment. These PCR products were then ligated in-frame into a pAct plasmid that provides for an amino-terminal triple HA epitope tag or a triple-HA/eGFP addition. The pDA-Flag-HhN expression vector was a kind gift from Dr. R. Fukunaga (32).
RESULTS
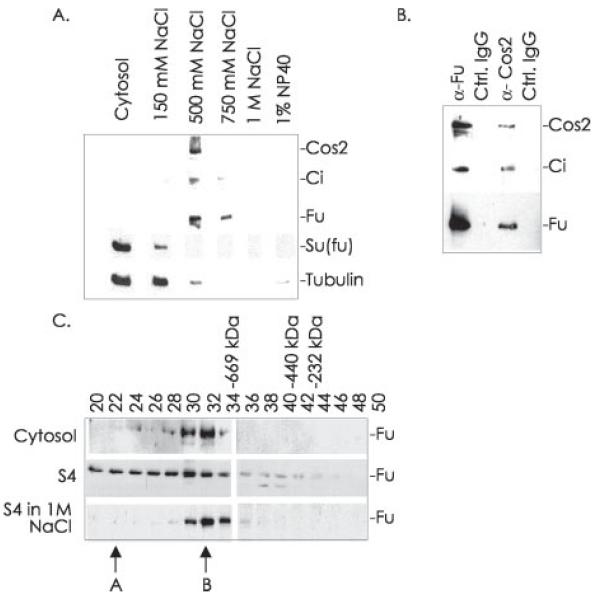
We have previously demonstrated that the HSC associates with MTs in an Hh-sensitive manner (43). As Kinesin family members are known to associate with membrane-bound vesicular cargo, we hypothesized that Cos2 might also associate with membranes. While a large fraction of the HSC, isolated from Drosophila embryonic extracts, is cytosolic, a significant percentage is also found in a 100,000 × g membrane-enriched pellet (Fig. 1A). The HSC is highly enriched in the membrane-containing pellet compared with the cytosol when both are normalized to protein. To determine the nature of the HSC membrane association we isolated a 100,000 × g membrane-enriched pellet from postnuclear Drosophila embryo lysate and sequentially extracted it with a variety of buffers. The vast majority of membrane-associated Fu, Cos2, and Ci was extracted with a buffer containing 0.50–0.75 m NaCl, leaving residual amounts of these proteins in the membrane pellet. Su(fu) is a protein of largely unknown function that we have previously shown to be a part of a tetrameric HSC that does not enrich on MTs (46). Su(fu) does not enrich in the high speed pellet and appears to be predominantly cytosolic. We conclude that the HSC is peripherally associated with cellular membranes.
Fig. 1. The HSC associates with membranes.
A, following hypotonic lysis and fractionation of Drosophila embryos, total cellular membranes were sequentially extracted by homogenization with a Dounce in lysis buffer containing the indicated concentrations of NaCl or finally 1% Nonidet P-40 (NP40). The cytosol and sequential extractions were normalized to protein, separated by SDS-PAGE, and immunoblotted for the indicated proteins. B, total embryo membranes were washed in lysis buffer containing 0.15 m NaCl and then extracted with lysis buffer containing 0.5 m NaCl (producing the S4 fraction). This salt extraction was immunoprecipitated with antibodies to Fu or Cos2 or with rabbit IgG or mouse IgG1 as controls (Ctrl.). The immunoprecipitates were separated by SDS-PAGE and immunoblotted for the indicated proteins. C, the cytosolic fraction (top panel) and S4 extraction of embryo cellular membranes (middle and bottom panels) were fractionated by size on a Superose 6 column. The size exclusion column was equilibrated in lysis buffer containing 0.15 m NaCl (top panel), 0.5 m NaCl (middle panel), or 1 m NaCl (bottom panel). Every other fraction was immunoblotted for Fu. Fractions where standard size markers elute are indicated. Peaks A and B of Fu elution are referred to in the text.
We washed a 100,000 × g membrane pellet from Drosophila embryos with 0.15 m NaCl and then extracted it with a buffer containing 0.5 m NaCl (the S4 fraction). To determine whether the components of the HSC found on cellular membranes are associated with each other, we immunoprecipitated from this extract using Fu antiserum, Cos2 monoclonal antibody, or control IgGs. The immunoprecipitates were separated by SDS-PAGE and then immunoblotted with antibodies to components of the HSC (Fig. 1B). Since antibodies to Fu or Cos2 were able to co-immunoprecipitate Fu, Cos2, and Ci, we conclude that the components of the HSC are complexed together on cellular membranes.
We have demonstrated in previous work that Fu is found in two major complexes, which elute as peaks A and B of Fu immunoreactivity, when cytosolic Drosophila extracts are fractionated by size exclusion chromatography (see Ref. 43 and Fig. 1C, top panel, which is a short exposure of the cytosolic fractions showing only peak B.) Ci is only associated with peak A, the largest Fu-containing complex, which is thus identified as the intact HSC (43). To determine whether the intact HSC is membrane-associated, we fractionated the 0.5 m extraction of cellular membranes (S4) by size exclusion chromatography (Fig. 1C, middle panel). The bulk of Fu in the S4 elutes earlier than peak B, consistent with membrane-associated Fu being a part of a complex larger than that in peak B. Additionally Fu that elutes in the earlier fractions continues to specifically co-immunoprecipitate Cos2 and Ci (data not shown), indicating that the larger Fu-containing complex is the intact HSC. When the S4 was fractionated by size exclusion chromatography in buffer containing 1 m NaCl the bulk of Fu shifted to peak B (Fig. 1C, bottom panel). This suggests a precursor/product relationship between the complexes in which the intact HSC in peak A breaks down to form the smaller complex in peak B. We conclude that the physiologically relevant complex of Fu, Cos2, and Ci, the intact HSC, enriches on cellular membranes.
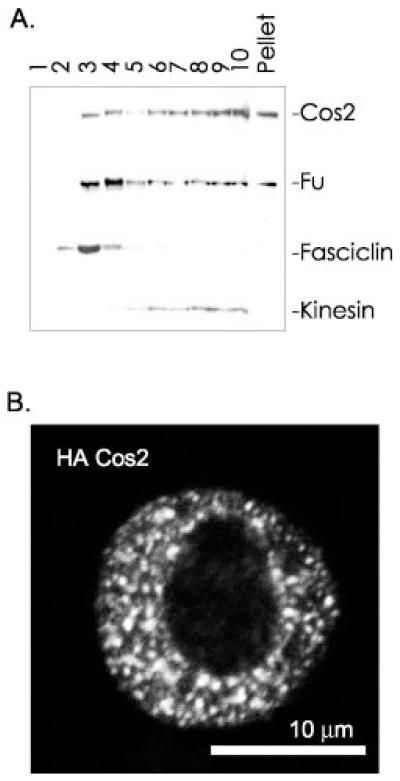
To begin to characterize the membranes with which the HSC associates, we subjected total cellular membranes to equilibrium density centrifugation. We overlaid washed total membranes, P3, with a discontinuous sucrose gradient designed to separate plasma membranes from vesicular membranes (68). After centrifugation, fractions were taken from the top of the tube and analyzed by SDS-PAGE and immunoblot (Fig. 2A). While some Cos2 fractionated with the plasma membrane marker Fasciclin (fractions 2–4), the bulk of Cos2 is associated with vesicular membranes in a manner analogous to Kinesin (fractions 6–10). Fu fractionates in largely the same manner as Cos2. Therefore, the HSC exhibits a second characteristic of a Kinesin-targeted complex; in addition to binding MTs, it associates with membrane vesicles (61, 63). As we hypothesize that the HSC is targeted by the KRP Cos2, we used immunofluorescence microscopy to determine whether the localization of Cos2 in cells would be similar to that reported for various Kinesins (63). We examined the distribution of HA-tagged Cos2 expressed in S2 cells using confocal microscopy (Fig. 2B). HA-Cos2 localized to discrete cytoplasmic puncta, consistent with localization to membrane-bound vesicles. Thus, our immunofluorescence microscopy results are consistent with our biochemical analysis. We conclude that Cos2 localizes to vesicular and plasma membranes.
Fig. 2. Cos2 associates with vesicles and plasma membrane.
A, total membranes prepared from S2 cells were washed in 0.15 m NaCl, then resuspended in 1.4 m sucrose, and overlaid with 1.22 m sucrose and then with 0.1 m sucrose. Following equilibrium density centrifugation (112,000 × g for 18 h), fractions were taken from the top of the tube. Each fraction was separated by SDS-PAGE and immunoblotted. Approximately 30% of Kinesin associates with vesicular membranes and is used here as a vesicular membrane marker (fractions 6–10) (61, 63). Fasciclin is a glycosylphosphatidylinositol-anchored protein and is used here as a plasma membrane marker (fractions 2–4) (66). The apparent accumulation of Fu in the plasma membrane to a greater degree than Cos2 is not consistently observed. B, S2 cells were transfected with HA-Cos2, then fixed, and stained with anti-HA-Alexa 488 and with 4,6-diamidino-2-phenylindole (not shown) to visualize DNA followed by confocal microscopy.
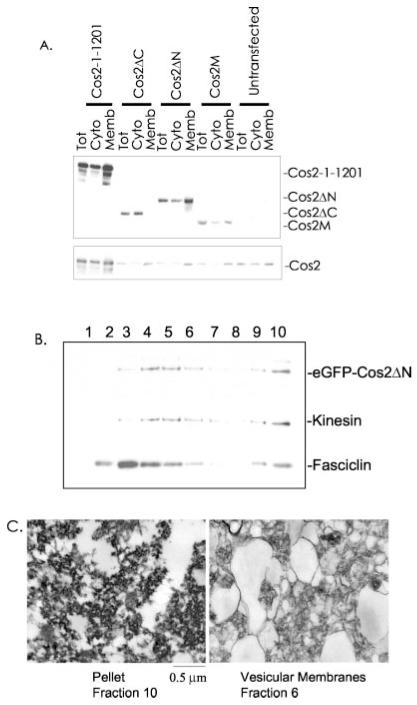
To identify the domain of Cos2 responsible for membrane association, we transfected S2 cells with a series of aminoterminal HA-tagged Cos2 constructs: full-length Cos2 (residues 1–1201); Cos2ΔC (residues 1–500), which contains the putative MT binding domain; Cos2M (residues 500–1000); and Cos2ΔN (residues 500–1201) (60, 69, 70). Postnuclear hypotonic lysates were centrifuged at 100,000 × g. High speed supernatants and high speed pellets were then immunoblotted following normalization to volume (Fig. 3A, upper panel). Approximately 60–70% of full-length Cos2, Cos2M, and Cos2ΔN associated with cellular membranes. However, Cos2ΔC, which contains the putative MT binding domain, was unable to associate with membranes. These results localized the membrane binding domain of Cos2 between amino acids 500 and 1000, which is distinct from the MT binding domain. Interestingly the percentage of endogenous Cos2 associated with cellular membranes remained unchanged in the presence of the various Cos2 truncation mutants (Fig. 3A, bottom panel). This suggests that Cos2 may be binding a non-protein component of membranes directly as the overexpressed membrane binding domain of Cos2 did not appear to compete with endogenous Cos2 for membrane binding. Alternatively Cos2 may associate with an abundant membrane protein.
Fig. 3. A central region of Cos2 contains a membrane binding domain.
A, S2 cells were transfected with expression vectors containing HA-tagged Cos2 (residues 1–1201), Cos2ΔC, Cos2ΔN, or Cos2M. Two days post-transfection, cells were lysed in hypotonic lysis buffer. Total lysate (Tot), postnuclear cytosol (Cyto), and total membrane (Memb) fractions were normalized to volume, separated by SDS-PAGE, and immunoblotted with α-HA antibody (top panel). An immunoblot of endogenous Cos2 is also shown (bottom panel) to demonstrate that endogenous Cos2 is not displaced by overexpressed Cos2. B, S2 cells transfected with eGFP-Cos2ΔN were lysed, and total membranes were separated by equilibrium density centrifugation. eGFP-Cos2ΔN fractionates in a manner similar to endogenous Cos2. C, electron microscopy of the vesicular membrane fraction (fraction 6) compared with the pellet (fraction 10) of the sucrose gradient in B.
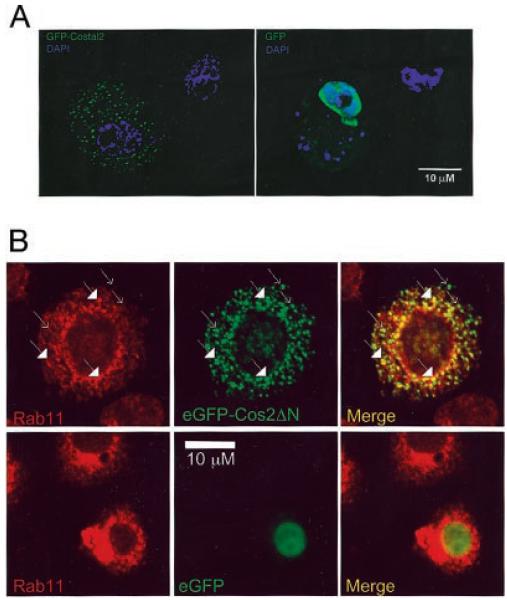
To demonstrate that Cos2ΔN, when overexpressed, was actually associating with cellular membranes, we expressed a similar construct, eGFP-Cos2ΔN, in S2 cells and separated total membranes into vesicular and plasma membrane fractions by equilibrium density centrifugation (Fig. 3B). The overexpressed eGFP-Cos2ΔN fractionates with vesicular and plasma membrane marker proteins in a manner similar to endogenous Cos2 (compare with Fig. 2A). To verify that we were enriching for vesicular membranes in our analysis, we prepared a vesicular fraction and the pellet fraction for electron microscopy (Fig. 3C). We observed vesicular membrane structures in the fraction in which Cos2 and Kinesin fractionate, while the pellet fraction contained few such vesicles. This result also suggests that a piece of Cos2 was able to tether an exogenous protein, eGFP, to membranes. To verify this result, we compared the distribution of eGFP fluorescence in S2 cells transfected with eGFP or with eGFP-Cos2ΔN. By fluorescent light microscopy we observed that eGFP-Cos2ΔN localizes in a manner similar to full-length Cos2 in S2 cells (Fig. 4A). That is, eGFP fused to Cos2ΔN, which does not contain the Cos2 MT binding domain, localizes to punctate cytoplasmic structures, consistent with its being targeted to membrane-bound vesicles. In contrast, eGFP itself is diffuse and mostly nuclear when expressed in S2 cells. Thus, the MT binding domain of Cos2 is not necessary to provide for the Kinesin-like staining pattern of Cos2. Additionally our results suggest that the proposed role of Cos2 in tethering associated proteins (like Ci) out of the nucleus requires the Cos2 membrane binding domain, not the MT binding domain (37, 43, 49). Indeed, upon transfection of S2 cells with HA-Cos2-ΔC, which contains only the putative motor domain of Cos2, we observed a diffuse, mostly nuclear distribution similar to eGFP alone (data not shown). Therefore, we conclude that Cos2ΔN contains a domain that is capable of tethering exogenous proteins to various membrane vesicles.
Fig. 4. The Cos2 membrane binding domain can tether an exogenous protein to vesicles.
A, S2 cells expressing eGFP-Cos2ΔN or eGFP were fixed, stained with 4,6-diamidino-2-phenylindole (DAPI) (blue) and observed by fluorescence microscopy. The distribution of eGFP-Cos2ΔN is cytoplasmic and punctate similar to full-length Cos2 (see Fig. 2B). However, eGFP is predominantly nuclear with a diffuse cytoplasmic pattern. B, S2 cells were transfected with eGFP-Cos2ΔN, fixed, and stained with α-Rab11 (red) and observed by confocal microscopy. Rab11 is a marker for early/recycling endosomes (73). Plain arrows indicate eGFP-Cos2ΔN signal alone; filled arrows indicate yellow puncta where co-localization of Rab11 and Cos2 signals occurs.
We next asked whether Cos2 is associated with endosomal membrane vesicles since both Ptc and Smo cycle through the endosomal pathway (71, 72). Rab11 is a marker for some early endosomes and for recycling endosomes (73), the latter of which play an important role in the recycling of membrane proteins back to the plasma membrane. To determine whether the punctate signal of eGFP-Cos2ΔN is at least partly due to association with endosomal vesicles, we measured the co-localization of Cos2 with Rab11 (Fig. 4B). In S2 cells, ~54% of eGFP-Cos2ΔN co-localized with Rab11-containing vesicles. The co-localization of eGFP-Cos2ΔN with Rab11-associated vesicles is especially pronounced in perinuclear regions of the cell, which are rich in recycling endosomes. There was less than 0.5% co-localization between wild type eGFP and Rab11-containing vesicles. These results verified that the punctate staining pattern of eGFP-Cos2ΔN is due to its association with membrane vesicles. Additionally these results suggest that a portion of Cos2 localizes to recycling endosomes.
It has been suggested that the ability of Kinesin to associate with MTs in vivo is regulated by its association with vesicular cargo (74-76). In the absence of cargo, Kinesin adopts a conformation unfavorable for association with MTs under physiological conditions. Upon cargo binding, Kinesin is stabilized into a conformation capable of binding MTs. Such a mechanism would prevent MT-stimulated ATPase activity of the motor domain in the absence of cargo. By analogy, we hypothesized that the Hh-regulated MT binding of the HSC we have reported previously (43) is mediated in vivo by differential association of Cos2 with a vesicular cargo. That is, in response to Hh, Cos2 may adopt a conformation that inhibits its association with membranes and therefore decreases its affinity for MTs. Indeed we found substantially less Fu and Cos2 associated with membranes isolated from Hh-stimulated S2 cells than from unstimulated S2 cells (Fig. 5A). Instead a greater percentage of Fu and Cos2 fractionated with the cytosol in response to Hh. Thus, the association of the HSC with membranes can be regulated by exposure to Hh. This suggests that differential affinity of Cos2 for membranes may provide a mechanism of Hh signal regulation.
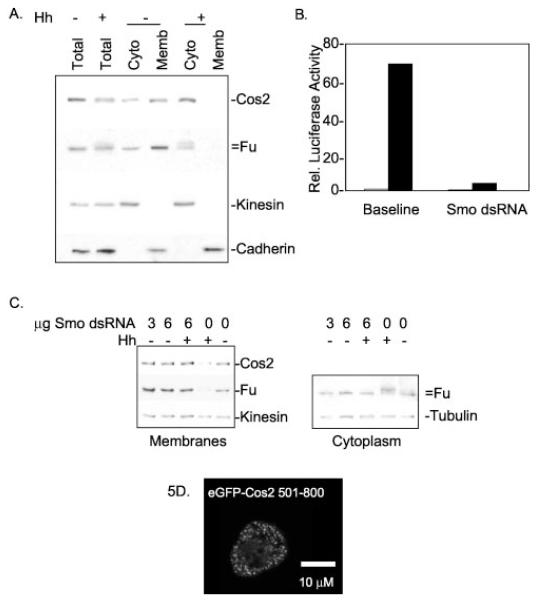
Fig. 5. The HSC association with membranes is Hh-sensitive and independent of Smo.
A, S2 cells were transfected with Hh or a control vector. Two days post-transfection, cells were lysed in hypotonic lysis buffer and fractionated as described under “Experimental Procedures.” 0.5× total lysate, 1× postnuclear cytosol (Cyto), and 1× total membrane (Memb) samples were normalized to volume, separated by SDS-PAGE, and immunoblotted. Kinesin and Cadherin serve as positive controls for soluble and membrane-associated proteins, respectively. Kinesin is generally about 70% soluble upon hypotonic lysis, while the remainder associates with vesicles (61, 63). Cos2 and Fu are more cytoplasmic in Hh-treated cells. B, Cl8 cells were transfected with a ptc-Luciferase reporter construct and treated with or without dsRNA homologous to Smo in the presence or absence of Hh. Relative luciferase activity was measured post-lysis and demonstrates reduced Hh-activated transcription in cells treated with Smo dsRNA. White bars indicate −Hh; black bars indicate +Hh. C, S2 cells were transfected with Hh or a control vector with or without the addition of Smo dsRNA. Two days post-transfection, cells were hypotonically lysed and fractionated into postnuclear cytosol and total cellular membranes. Left panel, total cellular membranes separated by SDS-PAGE and immunoblotted for Fu, Cos2, and Kinesin. Fu and Cos2 continue to fractionate with membranes despite Smo dsRNA treatment. Right panel, postnuclear cytosols are immunoblotted for Fu and Tubulin. This panel demonstrates that the Hh-induced phosphorylation of Fu does not occur in cells treated with Smo dsRNA, verifying depletion of Smo in these cells. D, S2 cells were transfected with eGFP-Cos2-(501–800), which expresses a fusion protein that contains the membrane binding domain of Cos2 but not the Smo association domain. Confocal microscopy reveals that eGFP-Cos2-(501–800) distribution is similar to full-length Cos2; eGFP-Cos2-(501–800) is both cytoplasmic and punctate. Thus Cos2 does not require a Smo association domain to tether eGFP to vesicles. Rel., relative.
Recently it has been shown that Cos2 associates with the 7-transmembrane protein Smo (56, 57). However, we propose that Cos2 is also able to bind membranes itself as Smo protein levels are likely too low to provide for membrane association of overexpressed Cos2. To verify that Cos2 is able to bind membranes independently of Smo, we used an established method to deplete tissue culture cells of Smo protein using RNA interference (77, 78). We then demonstrated, using a ptc-Luciferase reporter assay (77), that Smo dsRNA treatment reduces Hh-stimulated Ci activation by 94% (Fig. 5B). Additionally we have shown that this dsRNA treatment will greatly reduce the expression of a Myc-tagged Smo construct when detected by indirect immunofluorescence (data not shown). To determine the distribution of Cos2 in cells that lack Smo, we used this same Smo dsRNA to treat S2 cells in the presence or absence of Hh. Upon lysis and fractionation by centrifugation, we found that Cos2 and Fu continue to associate with cellular membranes in cells in which Smo levels are depleted due to RNA interference (Fig. 5C, left panel). Interestingly Smo is required for Cos2 to be released from membranes. Therefore, Smo provides an activity that increases the solubility of the HSC due to Hh. As an internal control for Smo depletion, we compared the state of Fu phosphorylation in the dsRNA-treated cells to the control cells. A shift in Fu mobility, upon SDS-PAGE, is due to Hh-dependent hyperphosphorylation and has been shown to be Smo-dependent (31, 43, 79). In cells treated with Smo dsRNA Fu is not hyperphosphorylated in the presence of Hh but is hyperphosphorylated in the Hh-exposed controls cells (Fig. 5C, right panel). This observation verifies that Smo is depleted in dsRNA-treated cells. To further test the hypothesis that Cos2 contains a domain that associates with membranes independently of Smo, we constructed a plasmid expressing eGFP-Cos2-(501–800). This fusion protein contains the Cos2 membrane binding domain but lacks the domain of Cos2 (amino acids 900–1201) that associates with Smo (57). Using confocal microscopy, we observed that eGFP-Cos2-(501–800) expressed in S2 cells localizes to cytoplasmic puncta similar to full-length Cos2 (Fig. 5D). Thus, we conclude that eGFP can be tethered to vesicular membranes by a Cos2 membrane binding domain. Taken together, these results suggest that Cos2 binds membranes in a Smo-independent manner.
DISCUSSION
We have presented evidence that a subset of the HSC is enriched on cellular membranes. We demonstrate that the HSC component Cos2 is capable of tethering an exogenous protein to various cellular membranes through a domain that maps between amino acids 501 and 800 of Cos2. We show that Cos2 continues to associate with cellular membranes in the absence of Smo. We demonstrate that the Cos2 membrane binding domain is distinct from the MT binding domain and from the recently identified Smo interaction domain. In contrast to the association of Cos2 with Smo, we also show that the association of Cos2 with membranes is Hh-sensitive. Thus, the ability of Cos2 to associate with cellular membranes appears to be distinct from its ability to associate with Smo. We propose that the ability of Cos2 to associate with cellular membranes, through direct binding or through association with the membrane protein Smo, may underlie its ability to function as both a positive and negative regulator of the Hh pathway.
Cos2 Tethers the HSC to Membranes
It has been proposed that the MT association of Cos2, through its predicted aminoterminal MT binding domain, is required to tether Ci out of the nucleus. This model assumed that Cos2 was working as an ATP-regulated scaffolding protein. However, the membrane binding domain of Cos2 now appears sufficient to tether an exogenous protein, eGFP, out of the nucleus. This result suggests that Ci may actually be tethered out of the nucleus through vesicular association, not MT association. It was initially unclear whether Cos2 would have the ability to act as a true Kinesin as an arginine highly conserved in the motor domain of other KRPs is missing in Cos2. However, it now appears that Cos2 has additional similarities to KRPs beyond MT binding; it is capable of associating with membrane vesicles in a regulated manner. Thus, the MT binding domain of Cos2 may play an active role in transporting the HSC, while the Cos2 membrane binding domain plays the role of tethering Ci out of the nucleus.
The amount of membrane-associated Cos2 and Fu decreases in response to Hh, resulting in a subsequent increase of Cos2 and Fu in the cytoplasmic fraction. Thus, Cos2 appears to redistribute the HSC in response to Hh, separating it from the membrane trafficking system. It has been proposed that in vivo the ability of KRPs to associate with MTs is regulated by association with membrane vesicles. Therefore, the differential MT binding we observe in the HSC, in S2 cell versus Hh-stimulated S2 cell lysates, may be a reflection of a lack of vesicular binding in Hh-stimulated cells. Although no vesicles should have been present in the high speed supernatants used for our MT binding assays, the Cos2 in these lysates could have maintained their conformation after lysis such that the soluble Cos2 retains its ability to bind MT in the unstimulated cell lysates. Thus, it is likely that this Hh-regulated redistribution of the HSC is provided by Cos2 acting as a KRP. Release of Cos2 from membranes would limit the HSC to soluble locations and leave it unable to be trafficked or to be activated by Smo. This lack of trafficking might affect Ci processing, which may occur at various intracellular sites; see below for a suggested model.
It was recently suggested that Fu and Cos2 are degraded in response to Hh signaling (57). In these experiments, diminished amounts of Fu and Cos2 were detected after Hh treatment of cells or by visualizing Fu and Cos2 in various tissues by indirect immunofluorescence microscopy. Our results do not rule out a role for Cos2 and Fu degradation in the regulation of the Hh pathway. However, it is apparent that degradation alone cannot explain our results as upon Hh signaling Fu and Cos2 levels increase in the cytosolic fractions as well as decrease in the membrane fractions (see Fig. 5, A and B). In Ruel et al. (57), see Fig. 3A; there is also an increase in the cytoplasmic fraction of the HSC in response to Hh, supporting the proposal that degradation of membrane-associated Fu and Cos2 cannot solely account for the redistribution of the HSC after Hh treatment. We conclude, from both these and our data, that release of Cos2 and Fu from membranes may play a role in regulation of the Hh pathway. Thus, in response to Hh, destabilization of Fu and Cos2 may occur subsequent to HSC redistribution and may not be relevant to initial Hh signaling events. There are many examples of signaling systems that have both acute and chronic desensitization systems (80-82). In this case, redistribution of Cos2 and Fu to the cytoplasmic fraction of the cell may be an acute down-regulation mechanism, while decreased Cos2 and Fu stability may represent a more permanent way to limit their activity.
Cos2 Associates with Cellular Membranes through Two Distinct Domains
If Cos2 associates with membranes through a protein intermediate, one might expect the binding of Cos2 to that protein would be saturable. However, when we overexpress Cos2 in cells we still observe vesicular association. Additionally in the presence of overexpressed Cos2 the percentage of endogenous Cos2 on cellular membranes is unchanged. Thus, we speculate that Cos2 may associate directly with an abundant, perhaps lipid, component of membranes. Additionally we recently reported that Cos2 associates directly with the 7-transmembrane protein Smo and that this association is necessary for Ci activation. Thus, Cos2 appears to associate with cellular membranes through two different mechanisms encoded by distinct domains of Cos2. Through its membrane binding domain, amino acids 501–800, Cos2 associates directly with cellular membranes, and through its Smo interaction domain, amino acids 900–1201, Cos2 associates indirectly with membranes through Smo. Additionally the affinities of the two domains of Cos2 for their targets are regulated in different manners. The association of Cos2 with Smo did not appear to be affected by Hh treatment in both S2 cells treated with Hh and in embryos engineered to be chronically in the Hh off state (ptc-overexpressing) or in the Hh on state (hh-overexpressing) (56). This is in contrast to the Cos2 membrane association reported here in which the bulk of Cos2 leaves the membrane in response to Hh. One explanation for how these two different interactions could play a role in Hh signaling is that there are two distinct forms of the HSC. One HSC may be tethered directly to membranes through Cos2, and one may be bound to Smo. In response to Hh, Cos2 would release the bulk of the HSC from endosomal vesicles, disrupting Ci75 production. Simultaneously the HSC tethered to Smo would be redistributed to the plasma membrane, allowing Ci to be activated (see below).
Previously, based on quantitative immunoprecipitation results, we estimated that only 3–5% of Cos2 associated with Smo. This result is consistent with our new report as the small percentage of Cos2 remaining on membranes after Hh treatment could be Cos2 in association with Smo. However, we had previously suggested, based on the percentage of Smo and Cos2 that co-localized by immunofluorescence microscopy, that as much as 50% of Cos2 and Smo may associate. This co-localization in various imaginal disc cells did not appear to vary with the position of the cell in the wing imaginal disc. This is consistent with the hypothesis that the Smo-Cos2 association is not affected by Hh. However, the high degree of Cos2 co-localization with Smo across the wing disc is seemingly inconsistent with the proposal that the bulk of Cos2 leaves cellular membranes in response to Hh. While it is unclear what the reasons behind this discrepancy are, we suggest that the resolution of our immunofluorescence studies may not be sufficient to detect alteration of Cos2 distribution due to Hh. Alternatively the pool of Cos2 that becomes soluble due to acute Hh signal reception may be degraded and undetectable in the chronic Hh reception state of the late third instar wing disc.
Multiple Signaling Complexes
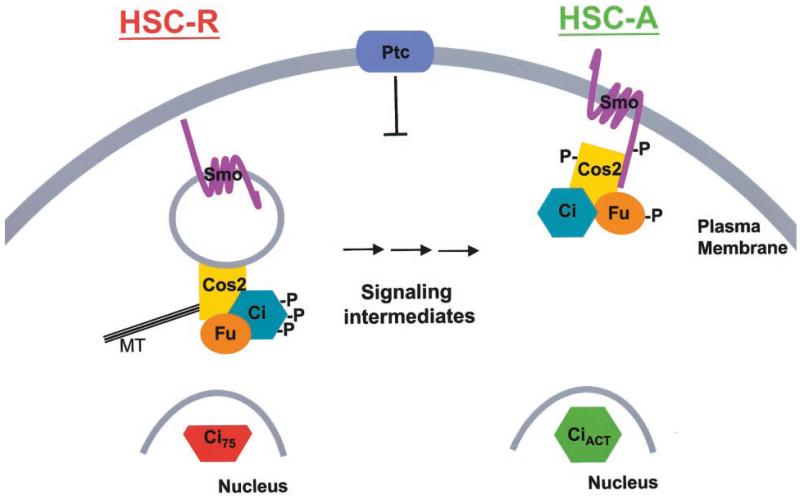
We suggest a model in which there are two HSCs, one involved in converting Ci into Ci75 (HSC-R) and one involved in converting Ci into an activator (HSC-A) (see Fig. 6). HSC-R associates with endosomes through Cos2, which may associate directly with membranes. HSC-A is also tethered to membranes through Cos2, but in this case Cos2 associates with membranes indirectly through the transmembrane protein Smo. In the absence of Hh, the Hh off state, HSC-R potentiates Ci processing into Ci75. In the presence of Hh, the HSC-R begins to release from vesicular membranes. This release of HSC-R from endosomal membranes, which would require initial Smo activity, attenuates Ci processing to Ci75, thus activating low Hh responses. Additionally Hh promotes Smo relocalization to the plasma membrane where it appears to be activated (72, 83). The Kinesin-like properties of Cos2 and its direct association with Smo might facilitate this relocalization. The fraction of the HSC that is associated with activated Smo becomes HSC-A, which involves hyperphosphorylated Fu and Cos2 (31, 42, 48, 50), allowing conversion of Ci into an activated form (CiAct) in a manner that requires localization to the plasma membrane (54). Thus, in response to Hh only a small amount of Ci would become activated, while the bulk of the HSC would accumulate in the cytoplasm. This model is consistent with the lack of correlation between Ci levels and target gene activation (41, 42, 50-52, 55, 84, 85).
Fig. 6. Model.
See text for details.
This model is based on four main observations. 1) Smo association with Cos2 does not change appreciably in response to Hh, while apparently simultaneously the majority of Cos2 and Fu is released from endosomal membranes. 2) SmoC, a membrane-targeted truncation of the intracellular carboxyl-terminal tail of Smo, inhibits Hh-induced Ci activation but also activates the low Hh responses thought to proceed through blocking Ci75 production (56, 86). 3) Genetic interactions between HSC components suggest the function of at least two complexes (85, 87). 4) Hh target gene activation is not correlated with blockage of Ci75 production (41, 42). A cytosolic HSC, HSC-C, whose association with MTs is regulated by Hh has been described previously (37, 43, 46). Although the relationship between the various HSCs is not yet clear, we speculate that HSC-C may represent a signaling intermediate between HSC-R and HSC-A, or it may be an end point where HSC-R accumulates, after Hh exposure, to be inactivated and perhaps degraded. Alternatively HSC-C may represent components of a non-membrane-associated HSC that are able to reassemble in the presence of the high concentrations of MTs formed in various in vitro MT binding assays. Like other KRPs, Cos2 may cycle on and off membranes and MT always generating a cytosolic pool of HSC. In the absence of Hh, HSC-C may be in a conformation capable of associating with MTs. In the presence of Hh, HSC-C, containing hyperphosphorylated Cos2, may be in a conformation with a weaker affinity for both MTs and membranes.
Thus, between the Hh off state, in which the endosome-associated HSC-R is the primary form of the HSC, and the completely activated state, in which the plasma membrane-localized Smo-associated HSC-A is the primary form of the HSC, there may be many intermediate forms. This model is consistent with the requirements of a signaling system that translates a morphogen gradient into distinct biological outcomes. Direct association between Smo and Cos2 may allow Smo to directly activate HSC-A and inhibit HSC-R. This testable model will focus future investigations on the dynamic equilibrium of Smo with various forms of the HSC.
Acknowledgments
We thank Dr. M. Hortsch (University of Michigan) for Fasciclin antibody (66), Dr. T. Uemura (Kyoto University, Kyoto, Japan) for Cadherin antibody (67), Dr. R. Fukunaga (Osaka University, Osaka, Japan) for the HhN expression vector (32), Dr. R. Cohen (University of Kansas, Lawrence, KS) for Rab11 antibody (73), and Dr. P. Beachy (The Johns Hopkins University) for the ptc reporter construct (41). We thank the University of Cincinnati Department of Cell Biology Microscopy Core for expert assistance. We thank Dr. J. Hooper for many helpful discussions and insights. We thank C. Nasrallah for expert technical assistance.
Footnotes
This work was supported by National Institutes of Health Grant CA82628 (to D. J. R.) and National Institutes of Health Training Grant 5-T32 ES07250 (to S. K. O. and M. A.).
The abbreviations used are: Hh, Hedgehog; HSC, Hedgehog signaling complex; Cos2, Costal2; Fu, Fused; Su(fu), Suppressor of fused; Ci, Cubitus interruptus; Ci75, a repressor form of Ci; CiAct, an activated form of Ci; Ptc, Patched; Smo, Smoothened; MT, microtubule; KRP, Kinesin-related protein; HA, hemagglutinin; ds, double-stranded; eGFP, enhanced green fluorescent protein.
REFERENCES
- 1.Ingham PW, McMahon AP. Genes Dev. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell HF, Donis-Keller H, Helms C, Hing AV, Heng HH, Koop B, Martindale D, Rommens JM, Tsui LC, Scherer SW. Nat. Genet. 1996;14:353–356. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- 3.Wechsler-Reya R, Scott MP. Annu. Rev. Neurosci. 2001;24:385–428. doi: 10.1146/annurev.neuro.24.1.385. [DOI] [PubMed] [Google Scholar]
- 4.Xie J, Johnson RL, Zhang X, Bare JW, Waldman FM, Cogen PH, Menon AG, Warren RS, Chen LC, Scott MP, Epstein EH., Jr. Cancer Res. 1997;57:2369–2372. [PubMed] [Google Scholar]
- 5.Unden AB, Zaphiropoulos PG, Bruce K, Toftgard R, Stahle-Backdahl M. Cancer Res. 1997;57:2336–2340. [PubMed] [Google Scholar]
- 6.Unden AB, Holmberg E, Lundh-Rozell B, Stahle-Backdahl M, Zaphiropoulos PG, Toftgard R, Vorechovsky I. Cancer Res. 1996;56:4562–4565. [PubMed] [Google Scholar]
- 7.Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, Negus K, Smyth I, Pressman C, Leffell DJ, Gerrard B, Goldstein AM, Dean M, Toftgard R, Chenevix-Trench G, Wainwright B, Bale AE. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 8.Chidambaram A, Goldstein AM, Gailani MR, Gerrard B, Bale SJ, DiGiovanna JJ, Bale AE, Dean M. Cancer Res. 1996;56:4599–4601. [PubMed] [Google Scholar]
- 9.Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH, Jr., Scott MP. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 10.Reifenberger J, Wolter M, Weber RG, Megahed M, Ruzicka T, Lichter P, Reifenberger G. Cancer Res. 1998;58:1798–1803. [PubMed] [Google Scholar]
- 11.Dahmane N, Lee J, Robins P, Heller P, Ruiz i Altaba A. Nature. 1997;389:876–881. doi: 10.1038/39918. [DOI] [PubMed] [Google Scholar]
- 12.Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, Jr., de Sauvage FJ. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 13.Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Nature. 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 14.Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, Noll M, Hooper JE, de Sauvage F, Rosenthal A. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Struhl G. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- 16.Nakano Y, Guerrero I, Hidalgo A, Taylor A, Whittle JR, Ingham PW. Nature. 1989;341:508–513. doi: 10.1038/341508a0. [DOI] [PubMed] [Google Scholar]
- 17.Hooper JE, Scott MP. Cell. 1989;59:751–765. doi: 10.1016/0092-8674(89)90021-4. [DOI] [PubMed] [Google Scholar]
- 18.Ingham PW, Taylor AM, Nakano Y. Nature. 1991;353:184–187. doi: 10.1038/353184a0. [DOI] [PubMed] [Google Scholar]
- 19.Fuse N, Maiti T, Wang B, Porter JA, Hall TM, Leahy DJ, Beachy PA. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10992–10999. doi: 10.1073/pnas.96.20.10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE. Cell. 1996;86:221–232. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- 21.van den Heuvel M, Ingham PW. Nature. 1996;382:547–551. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- 22.Quirk J, van den Heuvel M, Henrique D, Marigo V, Jones TA, Tabin C, Ingham PW. Cold Spring Harb. Symp. Quant. Biol. 1997;62:217–226. [PubMed] [Google Scholar]
- 23.Ingham PW, Nystedt S, Nakano Y, Brown W, Stark D, van den Heuvel M, Taylor AM. Curr. Biol. 2000;10:1315–1318. doi: 10.1016/s0960-9822(00)00755-7. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Struhl G. Development. 1998;125:4943–4948. doi: 10.1242/dev.125.24.4943. [DOI] [PubMed] [Google Scholar]
- 25.Strutt H, Thomas C, Nakano Y, Stark D, Neave B, Taylor AM, Ingham PW. Curr. Biol. 2001;11:608–613. doi: 10.1016/s0960-9822(01)00179-8. [DOI] [PubMed] [Google Scholar]
- 26.Taipale J, Cooper MK, Maiti T, Beachy PA. Nature. 2002;418:892–897. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- 27.Monnier V, Dussillol F, Alves G, Lamour-Isnard C, Plessis A. Curr. Biol. 1998;8:583–586. doi: 10.1016/s0960-9822(98)70227-1. [DOI] [PubMed] [Google Scholar]
- 28.Forbes AJ, Nakano Y, Taylor AM, Ingham PW. Dev. Suppl. 1993:115–124. [PubMed] [Google Scholar]
- 29.Mariol MC, Preat T, Limbourg-Bouchon B. Mol. Cell. Biol. 1987;7:3244–3251. doi: 10.1128/mcb.7.9.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preat T, Therond P, Lamour-Isnard C, Limbourg-Bouchon B, Tricoire H, Erk I, Mariol MC, Busson D. Nature. 1990;347:87–89. doi: 10.1038/347087a0. [DOI] [PubMed] [Google Scholar]
- 31.Therond PP, Knight JD, Kornberg TB, Bishop JM. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4224–4228. doi: 10.1073/pnas.93.9.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukumoto T, Watanabe-Fukunaga R, Fujisawa K, Nagata S, Fukunaga R. J. Biol. Chem. 2001;276:38441–38448. doi: 10.1074/jbc.M105871200. [DOI] [PubMed] [Google Scholar]
- 33.Ascano M, Jr., Nybakken KE, Sosinski J, Stegman MA, Robbins DJ. Mol. Cell. Biol. 2002;22:1555–1566. doi: 10.1128/mcb.22.5.1555-1566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grau Y, Simpson P. Dev. Biol. 1987;122:186–200. doi: 10.1016/0012-1606(87)90344-7. [DOI] [PubMed] [Google Scholar]
- 35.Simpson P, Grau Y. Dev. Biol. 1987;122:201–209. doi: 10.1016/0012-1606(87)90345-9. [DOI] [PubMed] [Google Scholar]
- 36.Ho K. Doctoral dissertation. Stanford University; Stanford, CA: 2002. Regulation of Hh Target Gene Transcription by the Kinesin-Related Protein Costal2. [Google Scholar]
- 37.Sisson JC, Ho KS, Suyama K, Scott MP. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 38.Orenic TV, Slusarski DC, Kroll KL, Holmgren RA. Genes Dev. 1990;4:1053–1067. doi: 10.1101/gad.4.6.1053. [DOI] [PubMed] [Google Scholar]
- 39.Motzny CK, Holmgren R. Mech. Dev. 1995;52:137–150. doi: 10.1016/0925-4773(95)00397-j. [DOI] [PubMed] [Google Scholar]
- 40.Slusarski DC, Motzny CK, Holmgren R. Genetics. 1995;139:229–240. doi: 10.1093/genetics/139.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen CH, von Kessler DP, Park W, Wang B, Ma Y, Beachy PA. Cell. 1999;98:305–316. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]
- 42.Wang QT, Holmgren RA. Development. 2000;127:3131–3139. doi: 10.1242/dev.127.14.3131. [DOI] [PubMed] [Google Scholar]
- 43.Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 44.Monnier V, Ho KS, Sanial M, Scott MP, Plessis A. BMC Dev. Biol. 2002;2:4. doi: 10.1186/1471-213X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nybakken KE, Turck CW, Robbins DJ, Bishop JM. J. Biol. Chem. 2002;277:24638–24647. doi: 10.1074/jbc.M110730200. [DOI] [PubMed] [Google Scholar]
- 46.Stegman MA, Vallance JE, Elangovan G, Sosinski J, Cheng Y, Robbins DJ. J. Biol. Chem. 2000;275:21809–21812. doi: 10.1074/jbc.C000043200. [DOI] [PubMed] [Google Scholar]
- 47.Lefers MA, Wang QT, Holmgren RA. Dev. Biol. 2001;236:411–420. doi: 10.1006/dbio.2001.0345. [DOI] [PubMed] [Google Scholar]
- 48.Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 49.Wang G, Amanai K, Wang B, Jiang J. Genes Dev. 2000;14:2893–2905. doi: 10.1101/gad.843900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang QT, Holmgren RA. Development. 1999;126:5097–5106. doi: 10.1242/dev.126.22.5097. [DOI] [PubMed] [Google Scholar]
- 51.Methot N, Basler K. Development. 2000;127:4001–4010. doi: 10.1242/dev.127.18.4001. [DOI] [PubMed] [Google Scholar]
- 52.Wang G, Wang B, Jiang J. Genes Dev. 1999;13:2828–2837. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallet A, Angelats C, Kerridge S, Therond PP. Development. 2000;127:5509–5522. doi: 10.1242/dev.127.24.5509. [DOI] [PubMed] [Google Scholar]
- 54.Ohlmeyer JT, Kalderon D. Nature. 1998;396:749–753. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- 55.Chen Y, Cardinaux JR, Goodman RH, Smolik SM. Development. 1999;126:3607–3616. doi: 10.1242/dev.126.16.3607. [DOI] [PubMed] [Google Scholar]
- 56.Ogden SK, Ascano M, Stegman MA, Suber LM, Hooper JE, Robbins DJ. Curr. Biol. 2003;13:1998–2003. doi: 10.1016/j.cub.2003.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Therond PP. Nat. Cell Biol. 2003;5:907–913. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- 58.Verhey KJ, Meyer D, Deehan R, Blenis J, Schnapp BJ, Rapoport TA, Margolis B. J. Cell Biol. 2001;152:959–970. doi: 10.1083/jcb.152.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mok H, Shin H, Kim S, Lee JR, Yoon J, Kim E. J. Neurosci. 2002;22:5253–5258. doi: 10.1523/JNEUROSCI.22-13-05253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldstein LS. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6999–7003. doi: 10.1073/pnas.111145298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hollenbeck PJ. J. Cell Biol. 1989;108:2335–2342. doi: 10.1083/jcb.108.6.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hirokawa N, Sato-Yoshitake R, Kobayashi N, Pfister KK, Bloom GS, Brady ST. J. Cell Biol. 1991;114:295–302. doi: 10.1083/jcb.114.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pfister KK, Wagner MC, Stenoien DL, Brady ST, Bloom GS. J. Cell Biol. 1989;108:1453–1463. doi: 10.1083/jcb.108.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamal A, Goldstein LS. Curr. Opin. Cell Biol. 2002;14:63–68. doi: 10.1016/s0955-0674(01)00295-2. [DOI] [PubMed] [Google Scholar]
- 65.Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elkins T, Hortsch M, Bieber AJ, Snow PM, Goodman CS. J. Cell Biol. 1990;110:1825–1832. doi: 10.1083/jcb.110.5.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oda H, Uemura T, Harada Y, Iwai Y, Takeichi M. Dev. Biol. 1994;165:716–726. doi: 10.1006/dbio.1994.1287. [DOI] [PubMed] [Google Scholar]
- 68.Rietveld A, Neutz S, Simons K, Eaton S. J. Biol. Chem. 1999;274 doi: 10.1074/jbc.274.17.12049. [DOI] [PubMed] [Google Scholar]
- 69.Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Nature. 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Nature. 1996;380:555–559. doi: 10.1038/380555a0. [DOI] [PubMed] [Google Scholar]
- 71.Incardona JP, Gruenberg J, Roelink H. Curr. Biol. 2002;12:983–995. doi: 10.1016/s0960-9822(02)00895-3. [DOI] [PubMed] [Google Scholar]
- 72.Denef N, Neubuser D, Perez L, Cohen SM. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- 73.Dollar G, Struckhoff E, Michaud J, Cohen RS. Development. 2002;129:517–526. doi: 10.1242/dev.129.2.517. [DOI] [PubMed] [Google Scholar]
- 74.Coy DL, Hancock WO, Wagenbach M, Howard J. Nat. Cell Biol. 1999;1:288–292. doi: 10.1038/13001. [DOI] [PubMed] [Google Scholar]
- 75.Hackney DD, Stock MF. Nat. Cell Biol. 2000;2:257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 76.Stock MF, Guerrero J, Cobb B, Eggers CT, Huang TG, Li X, Hackney DD. J. Biol. Chem. 1999;274:14617–14623. doi: 10.1074/jbc.274.21.14617. [DOI] [PubMed] [Google Scholar]
- 77.Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Science. 2003;299:2039–2045. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- 78.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramirez-Weber FA, Casso DJ, Aza-Blanc P, Tabata T, Kornberg TB. Mol. Cell. 2000;6:479–485. doi: 10.1016/s1097-2765(00)00046-0. [DOI] [PubMed] [Google Scholar]
- 80.Hardman JG, Limbird LE. In: Goodman & Gilman’s the Pharmacological Basis of Therapeutics. 10th Gilman AG, editor. McGraw-Hill Medical Publishing Division; New York: 2001. [Google Scholar]
- 81.Sorkin A, Von Zastrow M. Nat. Rev. Mol. Cell. Biol. 2002;3:600–614. doi: 10.1038/nrm883. [DOI] [PubMed] [Google Scholar]
- 82.Wiley HS, Burke PM. Traffic. 2001;2:12–18. doi: 10.1034/j.1600-0854.2001.020103.x. [DOI] [PubMed] [Google Scholar]
- 83.Zhu AJ, Zheng L, Suyama K, Scott MP. Genes. Dev. 2003;17:1240–1252. doi: 10.1101/gad.1080803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Methot N, Basler K. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- 85.Alves G, Limbourg-Bouchon B, Tricoire H, Brissard-Zahraoui J, Lamour-Isnard C, Busson D. Mech. Dev. 1998;78:17–31. doi: 10.1016/s0925-4773(98)00130-0. [DOI] [PubMed] [Google Scholar]
- 86.Hooper JE. Development. 2003;130:3951–3963. doi: 10.1242/dev.00594. [DOI] [PubMed] [Google Scholar]
- 87.Pham A, Therond P, Alves G, Tournier FB, Busson D, Lamour-Isnard C, Bouchon BL, Preat T, Tricoire H. Genetics. 1995;140:587–598. doi: 10.1093/genetics/140.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]