Abstract
The microRNA-induced silencing complex (miRISC) protein TNRC6 (also called GW182) uses dispersed tryptophan-containing repeats in unstructured regions to recruit the CCR4–NOT nuclease complex leading to mRNA deadenylation and inhibition of translation initiation according to new research.
Research on microRNAs (miRNAs) and the mechanism by which they repress their target mRNAs constitutes one of the frontiers in the study of post-transcriptional gene regulation. miRNAs are 21–23 nucleotide (nt) short, conserved, noncoding RNAs, which are expressed in a cell type–specific manner and can accumulate to tens of thousands of copies per cell. miRNAs guide the miRISC to partially complementary binding sites of target mRNAs1, leading to their destabilization and/or translational repression. Well-expressed miRNAs typically target hundreds of different mRNAs2,3 and consequently participate in many physiological and developmental processes4-7. Though significant progress has been made in miRNA target identification, the precise molecular mechanism by which target RNA inhibition is achieved is still a source of scientific debate8. The series of molecular events triggered by the recruitment of TNRC6/GW182 proteins to miRNA target sites and leading to mRNA destabilization and/or translational repression have now been further dissected in three recent studies, two of which are appearing in this issue9,10 and one elsewhere11.
In cells, mature miRNAs are bound by Argonaute (also called EIF2C1–EIF2C14 or Ago1–Ago4) proteins12, which present a 6- to 8-nt 5′ terminal segment, known as a ‘seed’, in a solvent-exposed helical geometry for target RNA recognition13. Upon recognition of seed-complementary target RNAs, miRISC undergoes conformational transitions dependent on the extent of complementarity14 and additional protein components, including TNRC6 proteins, join the miRISC–target RNA complex. In instances where the miRNA is nearly perfectly complementary to the target and bound by Ago proteins with catalytically active RNase H domains, the target RNA is directly cleaved, in a process known as RNA interference (RNAi). However, most animal miRNA-binding sites show only seed complementarity2,3, thus implicating other mechanisms of target RNA repression. How this regulation is mediated in molecular terms has been intensely debated over the past years8, and different mechanisms have been proposed ranging from predominant translational repression of various kinds to almost exclusive alterations of mRNA stability.
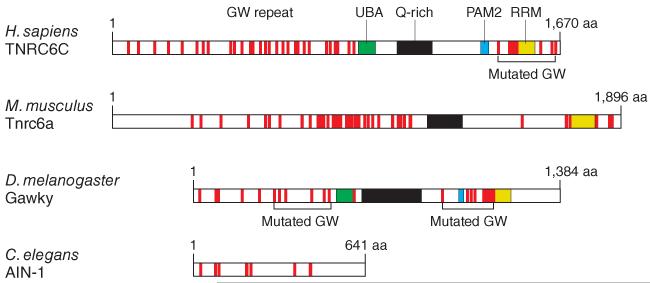
The new molecular link tying miRISC to mRNA processing and translational regulation was discovered in 2005 when proteins of the TNRC6 family, which are concentrated in mRNA-containing cytoplasmic speckles (P bodies or GW bodies), were shown to participate in RNAi by direct binding to Ago proteins15. TNRC6 proteins were first identified as antigens for autoimmune antisera from a patient with motor and sensory neuropathy16. Humans and mice contain three highly similar family members, TNRC6A–TNRC6C, whereas Drosophila melanogaster has one, Gawky (also called GW182), and nematodes have two, AIN-1 and AIN-2, with truncations at their C termini (Fig. 1)17. All TNRC6 proteins are characterized by stretches of glycinetryptophan (GW and WG) repeats across the entire protein, whereas the C-terminal region of the mammalian and fly homologs contains additional domains: a glutamine-rich domain, a ubiquitin-binding domain (UBA), a PAM2 motif and a C-terminal RRM domain.
Figure 1.
Domain structure of representative members of the TNRC6 protein family. Structurally or functionally defined domains are indicated; red lines show the location of the (G/S/T)W(G/S/T) repeat elements, referred to as GW repeats. GW repeats mutated in ref. 9 are shown.
Tethering assays have proven to be an invaluable tool in the dissection of the contribution of the individual protein domains of TNRC6 in RNA silencing. In such assays, the protein or domain of interest is fused to the 22-amino-acid-long bacteriophage lambda N peptide, which specifically binds with high affinity to repeated B-Box stem-loop structures introduced at defined sites into coexpressed reporter mRNAs18-20. Other biochemical tricks widely used to characterize TNRC6 proteins included pulldown experiments with GST-tagged protein fragments followed by mass spectrometric analysis, western blotting to identify specific protein interactors or rescue assays to document the importance of TNRC6 mutations in genuine miRNA-mediated repression. The three new studies9-11 made extensive use of these approaches testing fulllength TNRC6 proteins, their domains and mutants thereof, providing a detailed map of molecular interactions with protein partners and of the regulatory consequences of complex formation using reporter and natural target mRNAs (Fig. 2). Similar results were obtained from studies of TNRC6 in human and Drosophila systems and underscore the evolutionary conservation of these molecular interactions and processes.
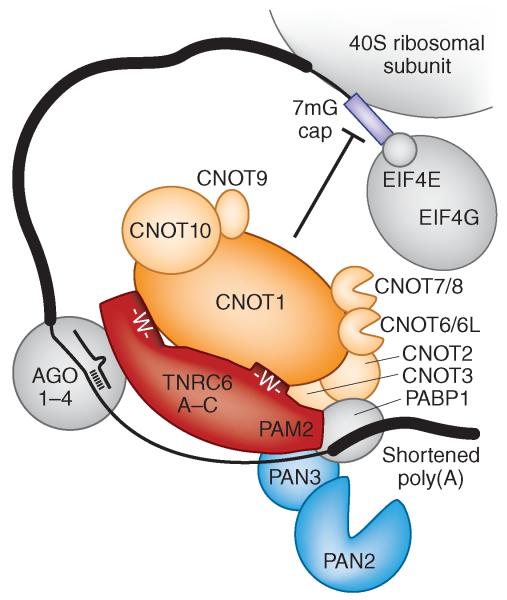
Figure 2.
Model of TNRC6 interaction with the CCR4–NOT and PAN2–PAN3 deadenylase complexes in humans. Protein sizes are drawn to scale. GW repeats in the N and C termini of human TNRC6 proteins are indicated and bind CNOT1. The TNRC6 N-terminal region also interacts with Ago proteins. The PAM2 domain of TNRC6 associates with PAN3 and PABP1. Recruitment of CCR4–NOT by GW repeats and PAN2–PAN3 by the PAM2 motif or flanking GW repeats causes deadenylation of target mRNAs and translation inhibition. In fly, TNRC6 corresponds to Gawky, and CNOT proteins are termed NOT proteins except for CNOT6, CNOT7 and CNOT9, which are named Twin, Pop2 and CAF40, respectively. Human CNOT6L is a paralog of CNOT6, and CNOT8 is a paralog of CNOT7, and each family is represented as single-copy genes in fly.
All three studies demonstrate that TNRC6 proteins directly recruit CNOT1, which is a component of the cytoplasmic CCR4–NOT deadenylase complex and acts as a scaffold for its assembly. The components of the CCR4–NOT complex are deeply conserved in eukaryotes, and the nuclear and cytoplasmically distributed complex has been implicated in many aspects of mRNA and protein expression, including transcription initiation, elongation, mRNA degradation, ubiquitination and protein modification. Among these various functions, regulation of mRNA abundance rates by deadenylation-triggered degradation is of greatest importance21-23. Indeed, CCR4–CNOT recruitment by tethering assays in the absence of miRNA or Ago proteins confers deadenylation9,10 and reporter mRNA repression9-11.
It was shown that GW repeats, or more generally tryptophan residues flanked by aliphatic side chain amino acids such as glycine, serine or threonine, located in unstructured regions in the C-terminal domain of TNRC6 (Gawky), recruit CNOT1 (Not1) and thereby the CCR4–NOT complex in an additive manner9. GW repeats in the N-terminal region of Gawky, which is also known to bind to Ago protein, also recruited CCR4–NOT, as did the transfer of GW repeats to a heterologous yeast protein, demonstrating that GW repeats were sufficient to bind the CCR4–NOT complex9.
Furthermore, independent of the CCR4–NOT interaction, the C-terminal domain of TNRC6 proteins also recruited the PAN2–PAN3 poly(A) tail-trimming nuclease complex. The mechanism of PAN2–PAN3 recruitment remains uncertain, with either PAM2 (ref. 10) or GW-containing regions flanking the RRM domain9,11 being identified as main interactors. The real surprise in these studies was that CCR4–NOT recruited to reporter mRNAs lacking poly(A) tails was still repressive, leading to the conclusion that the CCR4–NOT complex also acts as translational repressor during translation initiation9,11 and independently of its deadenylase activity. Finally, the recruitment of deadenylase and 3′-to-5′ exonuclease–containing complexes to miRNA targets provides an additional rationale for why miRNA binding sites located in the 3′ untranslated region (UTR)—that is, proximal to the mRNA 3′ end—show stronger mRNA destabilizing effects than binding sites situated in the coding regions2,3. Given the simplicity of the GW motif, it is conceivable that other mRNA-interacting factors also take advantage of CCR4–NOT recruitment to confer posttranscriptional gene repression.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/nsmb/.
References
- 1.Lim LP, et al. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK, Burge CB, Bartel DP. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hafner M, et al. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambros V. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 5.Ventura A, Jacks T. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Nat. Rev. Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 7.Liu N, Olson EN. Dev. Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huntzinger E, Izaurralde E. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 9.Chekulaeva M, et al. Nat. Struct. Mol. Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabian MR, et al. Nat. Struct. Mol. Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 11.Braun JE, Huntzinger E, Fauser M, Izaurralde E. Mol. Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Hutvagner G, Simard MJ. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, et al. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eulalio A, Behm-Ansmant I, Izaurralde E. Nat. Rev. Mol. Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 16.Eystathioy T, et al. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider MD, et al. J. Cell Biol. 2006;174:349–358. doi: 10.1083/jcb.200512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Gregorio E, Baron J, Preiss T, Hentze MW. RNA. 2001;7:106–113. doi: 10.1017/s1355838201000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 20.Pillai RS, Artus CG, Filipowicz W. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashita A, et al. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 22.Tucker M, et al. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, et al. EMBO J. 2010;29:2566–2576. doi: 10.1038/emboj.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]