Abstract
The Hedgehog (Hh) signal transduction pathway plays critical instructional roles during development. Activating mutations in human Hh signaling components predispose to a variety of tumor types, and have been observed in sporadic tumors occurring in a wide range of organs. Multiple insights into the regulation of Hh signaling have been achieved through studies using Drosophila melanogaster as a model organism. In Drosophila, regulation of the transcription factor Cubitus interruptus (Ci) is the ultimate target of the Hh pathway. Ci is regulated through communication of the membrane proteins Patched (Ptc) and Smoothened (Smo) to the intracellular Hedgehog Signaling Complex (HSC) in response to a graded concentration of Hh ligand. The HSC consists of the Kinesin Related Protein, Costal2 (Cos2), the serine-threonine protein kinase, Fused (Fu) and Ci. In the absence of Hh stimulation, the HSC is involved in processing of Ci to a truncated repressor protein. In response to Hh binding to Ptc, processing of Ci is blocked to allow for accumulation of full-length Ci activator protein(s). Differential concentrations of Hh ligand stimulate production of Ci transcriptional activators of varying strength, which facilitate activation of distinct subsets of target genes. The mechanism(s) by which Ptc and Smo communicate with the HSC in response to differential ligand concentrations to regulate Ci function are not yet fully elucidated. Here, we review what is known about regulation of individual Hh signaling components, concentrating on the mechanisms by which the Hh signal is propagated through Smo to the HSC.
Keywords: Hedgehog, Signaling, Signal transduction, Cancer, Development, Regulation
1. Introduction
The Hedgehog (Hh) family of morphogens plays important instructional roles in the development of numerous metazoan structures (reviewed in [1]). The Hh ligands, Sonic, Indian and Desert Hh in vertebrates and Hh in Drosophila, signal through binding to the membrane receptor Patched (Ptc) [2-4], to reverse the Ptc-mediated inhibition of signaling by the trans-membrane protein Smoothened (Smo) [5]. This allows Smo to activate the intracellular signaling components, resulting in stabilization of down-stream transcriptional activator(s) and activation of target genes [3,5-7]. Transcription activation is facilitated through the Gli family of transcription factors in vertebrates and Cubitus interruptus (Ci) in Drosophila (reviewed in [1]).
Hh signaling can initiate cellular growth, division, lineage specification, axon guidance and function as a survival factor (reviewed in [8,9]). Given this range of biological functions, it is not surprising that mutations in components of the Hh pathway are associated with both developmental defects and tumor progression (reviewed in [9]). Developmental disorders including holoprosencephaly, Greig cephalopolysyndactyly syndrome and Pallister–Hall syndrome result from mutations in SHH and GLI genes, while Gorlin’s syndrome is associated with PTC mutation (reviewed in [9,10]). Disruption of PTC, which functions as a negative regulator of the pathway, is implicated in cancer development in both inherited and sporadic cancers. Mutations in PTC and/or SMO trigger inappropriate activation of the Hh pathway, and have been identified in tumor types including basal cell carcinoma, rhabdomyosarcoma and medulloblastoma (reviewed in [10]). Recent studies also implicate activated hedgehog signaling as a mediating factor in small-cell lung cancer, pancreatic cancer and various digestive tract tumors [11-13]. Thus, an understanding of the functional components of this pathway, and the processes by which they are regulated, is critical in order to elucidate treatments for these various human pathologies.
Numerous advances in understanding Hh signaling have come from studies involving the fruit fly Drosophila melanogaster. In flies, Hh signaling is necessary for proper patterning of the embryo and multiple adult structures (reviewed in [1]). In particular, the adult wing affords a unique read-out of Hh signaling: proper patterning of wing veins, intervein space and wing bristles develop based on cellular position within a Hh morphogen gradient that forms across the larval wing imaginal disc, Fig. 1 (reviewed in [14,15]). The wing disc can be divided into anterior (A) and posterior (P) compartments based primarily on established gene expression patterns and relative intercellular affinities [16-20]. Cells in the P compartment produce and release Hh ligand, but do not respond to it, as they do not express Ptc or Ci [18,21]. Hh diffuses into the adjacent A compartment in a graded manner, where it acts on Ptc-expressing A cells (reviewed in [1,15]). Cells near the A/P compartment boundary receive the highest level of Hh stimulation, while cells further from the border receive lower levels of stimulation. Each group of cells transduces the Hh signal via Smo-mediated communication to intracellular signaling components [22]. These components include the kinesin-like protein Costal-2 (Cos2) [23], the serine-threonine protein kinase Fused (Fu) [24], Ci [25] and the novel protein, Suppressor of Fused (Su(fu)) [26] (Fig. 2). The sex-determination master switch Sex-lethal (Sxl) has also been demonstrated to be in association with Hh signaling components [27,28]. However, a direct role for Sxl in the regulation of Hh pathway target gene has not been described.
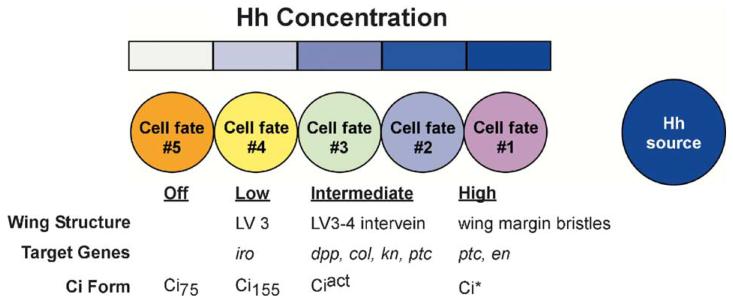
Fig. 1.
Differing levels of Hedgehog stimulation specify distinct cell fates. Hh diffusing into a field of cells specifies differing cell fates in cells near and far from the Hh source. In the Drosophila wing imaginal disc, the level of Hh stimulation controls target gene activation by regulating the relative amounts of Ci transcriptional repressors (Ci75) and activators (Ciact and Ci*). Ci transcription factors then control expression of specific subsets of genes to establish cell fate for development of specific adult structures. iro: iroquois; col: collier; kn: knot; ptc: patched; en: engrailed.
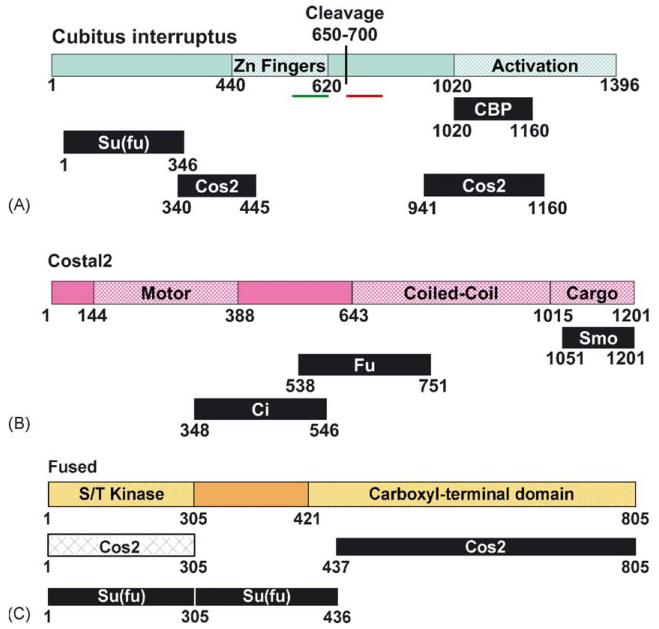
Fig. 2.
Functional domains of Hh signaling components. Indicated are the consensus interaction domains for each of the trimeric HSC components. Additional low affinity interactions that are not indicated may exist, and be involved in maintenance of HSC integrity. (A) Cubitus interruptus (Ci). Schematic of the transcription factor Ci, diagramming the Zinc finger domain [54], putative cleavage site [32] and activation domain. The putative nuclear localization sequence (amino acids 581–616 [96]) is indicated in by a green bar. The putative nuclear export sequence (amino acids 675–860 [97]) is indicated by the red bar. Interaction domains for binding partners CBP [70], Su(fu) and Cos2 [83,84] are also indicated. (B) Costal2 (Cos2). Schematic of the KRP Cos2, diagramming the motor domain, coiled-coil and cargo domains. Also indicated are interaction domains for Hh pathway binding partners Smo [75,76,94,95], Fu and Ci [83,84]. (C) Fused (Fu). Schematic of the serine/threonine protein kinase Fu, diagramming the kinase and carboxyl-terminal functional domains [82]. Also indicated are interaction domains for Hh pathway binding partners Cos2 and Su(fu) [83-85].
Cos2, Fu and Ci exist in a large, multi-protein complex termed the Hedgehog Signaling Complex (HSC) [29]. The HSC has been demonstrated to associate with microtubules (MT) in the absence of Hh stimulation [29]. A non MT associated, Su(fu)-containing tetrameric HSC has also been described [30]. In the absence of Hh, the HSC stimulates the processing of the 155 kDa Ci protein into a ~75 kDa transcriptional repressor (Ci75) [23,31,32]. Hh blocks Ci processing, concomitant with release of the trimeric HSC from MTs. Conversion between the MT-associated trimeric complex and the soluble, tetrameric complex is not completely understood. Also unclear is the exact mechanism by which Smo signals to the HSC to release it from MTs and halt Ci processing. Here, we will review what is known about regulation of individual HSC components, paying particular attention to how Hh-activated Smo communicates with the HSC to block production of Ci75 and stimulate production of Ci activator(s).
2. Hedgehog and patched
Hh proteins undergo an intramolecular processing event whereby the carboxyl terminal domain cleaves itself to release the amino terminal signaling domain (reviewed in [15,33]). Hh signaling molecules are lipid modified by palmitate at the amino-terminus and cholesterol at the carboxyl-terminus of the signaling domain (reviewed in [15]). The mechanism by which Hh is lipid modified following intra-molecular processing, and the functionality provided by the lipid modifications, are not entirely clear. However, cholesterol modification of Hh appears to be important in Hh receiving cells. Ptc contains a sterol sensing domain (SSD) that, while not required for Hh binding, is required for transduction of the Hh signal [34]. Support for functionality of this domain in regulation of Ptc activity comes from studies demonstrating that mutations in the SSD block Ptc-mediated repression of Smo [34,35]. The mechanism by which Ptc inhibits Smo function, and the process by which this inhibition is alleviated by Hh binding to Ptc have not been defined. However, based on its similarity to prokaryotic RND permeases, Ptc has been proposed to transport a small-molecule Smo inhibitor [36-38]. Binding of Hh to Ptc is proposed to block transport of the small molecule, thus allowing Smo to activate the pathway. While this process has yet to be clearly demonstrated, Hh has been demonstrated to affect Ptc sub-cellular localization. Binding by Hh has been documented to decrease the amount of Ptc on the plasma membrane (PM) [39]. This is achieved primarily through internalization of Hh-bound Ptc into endocytic vesicles [35,40]. Binding of Hh to Ptc also functions to limit the range of Hh signaling, as Ptc expressing cells near the A/P boundary can sequester Hh and prevent its diffusion far into the anterior [4].
3. Smoothened
Smo is a member of the Frizzled (Fz) family of G-Protein Coupled Receptors (GPCR) [5,41]. In response to Hh, it is phosphorylated and stabilized to allow for activation of the signaling cascade [39,42]. Both stabilization and phosphorylation appear to be actively repressed by Ptc, as these Smo modifications can occur in response to Ptc dysfunction or loss [39,42,43]. Conversely, over-expression of Ptc reduces Smo concentration and phosphorylation in both embryos and wing discs [39,43,44]. Ptc appears to function catalytically in repression and destabilization of Smo, as Ptc can block Smo signaling at sub-stochiometric levels [45]. As mentioned above, Smo repression has been suggested to occur via Ptc-mediated transport of an as yet unidentified small-molecule inhibitor [45,46]. This hypothesis is supported by recent work describing small molecule inhibitors of Smo that functionally mimic Ptc over-expression [36,46]. These antagonists appear to target the hepta-helical bundle of Smo, the domain demonstrated to be affected by Ptc [46].
Changes in Smo subcellular localization correlate with its conversion to an active signaling molecule. In cells not receiving Hh, Smo localizes to discrete endosomal vesicles, where it is believed to be degraded [39,47]. In response to Hh, Smo moves from internal structures to the plasma membrane (PM), presumably to activate the signaling cascade [39,44,47]. These results suggest that localization of Smo to the PM is a requisite step for functional signaling. Consistent with this argument, Smo targeted to the PM via a lipid anchor triggers activation of Hh target genes in a Hh independent manner [44]. It is not clear how Hh-stimulated PM localization of Smo allows for activation of the signaling cascade. However, it has been proposed that relocation of Smo to the PM may result from Hh blockage of Ptc-mediated transport of a small molecule inhibitor [45]. It is possible that one function of this inhibitor may be to bind Smo and alter its conformation to prevent PM localization. Another possibility is that PM localization may facilitate interaction with an as yet unidentified G-protein binding partner. However, no candidate G-protein has been implicated, genetically or biochemically, to be immediately down-stream of Smo.
Recent studies, which utilized Smo chimeras to examine the mechanism by which Smo signals to the HSC, further diminished the likelihood of G-protein involvement. A Smo chimera consisting of the extracellular and transmembrane domains of the Wingless receptor (Wg) Fz, fused to the carboxyl-terminal domain of Smo (FFS) was capable of fully activating the Hh signaling pathway, albeit in a Wg-dependent fashion [48]. These results demonstrate that the intracellular carboxyl-terminal tail of Smo contains all necessary components to activate Hh signaling. This is supported by results obtained from a Smo construct consisting of the intracellular carboxyl-terminal domain of Smo fused to a myristate membrane targeting sequence (SmoC). While this construct was not sensitive to Hh, it was capable of activating low level signaling events in a Hh independent manner [48]. These results argue against involvement of a traditional G-protein, as GPCRs do not typically associate with G-proteins via their carboxyl-terminal domains [49-51].
A chimera consisting of the Smo extracellular and transmembrane domains fused to the Fz cytoplasmic tail (SSF) suggested another aspect of Smo signaling to the HSC: the formation of Smo multimers may be required to facilitate high-level signaling. Over-expression of SSF in a wild type background blocked high Hh responses, but did not interfere with low Hh responses [48]. Phenotypes of these flies mimicked those of fu mutants (discussed below), where both the gradient of Ci activator across the wing disc and Ci nuclear entry were disrupted. This resulted in decreased expression of genes normally responsive to high-level Hh stimulation, without affecting genes activated in response to low-level Hh stimulation. Over-expression of Fu was not able to suppress the SSF phenotype, suggesting that SSF blocked high level signaling by interfering with a factor upstream of Fu. Accordingly, over-expression of wild-type Smo in the presence of SSF was able to effectively suppress the SSF phenotype [48]. These results suggest that SSF may exert a dominant negative effect by disrupting endogenous Smo dimeric or multimeric complexes.
4. Cubitus interruptus
The transcription factor Ci is the most down-stream component of the Hh signaling pathway, and can function as either a transcriptional activator or repressor in response to varying levels of Hh stimulation [32,52-55]. In the absence of Hh, the repressor form of Ci, Ci75, holds target genes in a repressed state. Ci75 is produced through proteolytic cleavage of full-length Ci in cells receiving little or no Hh stimulation [32]. Cleavage of Ci occurs in a proteasome-dependent manner, and requires phosphorylation by Protein Kinase A (PKA), Glycogen Synthase Kinase 3β (GSK3β) and Casein Kinase 1δ (CK1δ) [56-60]. Also involved in production of Ci75 is the F-box protein Supernumerary limbs (Slimb), which has been proposed to target phosphorylated full-length Ci for ubi-quitination [61]. Mosaic analysis reveals accumulation of full-length Ci, decrease of Ci75, and up-regulation of the TGF-β homologue, decapentaplegic (dpp), in clones of cells lacking Slimb, GSK3β or CK1δ [57,59,61]. Genetic evidence also reveals the ring finger protein Roc1α as a putative component in Ci processing [62]. Roc1α is a member of the SCF E3 complex, which facilitates substrate recognition for the proteolytic machinery (reviewed in [63]). Disruption of Roc1α function results in accumulation of full-length Ci and activation of dpp [62]. Roc1α appears to be specific to Ci processing, as it does not affect proteolysis of Armadillo, a down-stream signaling component of the Wg pathway, which is also degraded in a ligand sensitive, Slimb-dependent manner (reviewed in [64]).
In response to Hh, Ci processing to Ci75 is blocked, allowing for accumulation of full-length Ci155. Hh is required to convert the default Ci155 to a functional transcriptional activator, as evidenced by clones of cells expressing a non-cleavable form of Ci [65]. These cells accumulate full-length Ci155, but do not activate target genes in the absence of Hh stimulation. In response to Hh, Ci155 is converted to either of two activator form(s), Ciact or Ci* [66]. The less robust activator form, Ciact, functions in cells receiving intermediate to high levels of Hh stimulation [66,67]. It is involved in activation of ptc and dpp target genes in the wing imaginal disc. The enhanced activator form of Ci, Ci*, has been proposed to function in wing imaginal discs of late third instar larvae. Its function requires the highest level of Hh stimulation and is dependent on Cos2, Fu and PKA [67,68]. Conversion of Ci to Ci* is not presently understood. Biochemical analysis of Ci* has been difficult, as the activated protein has yet to be visualized by available methods. It can be evidenced only through its activity: up-regulation of anterior engrailed (en) expression in A cells directly adjacent to the A/P compartment boundary of wing imaginal discs [67]. Ci* activator function correlates with decreased protein detection by an antibody recognizing full-length Ci (2A1 mAb, Fig. 3). The loss of epitope recognition of Ci by the 2A1 antibody, and its relationship to Ci*, are not presently understood. However, it is clear that Ci* is a distinct Ci species, as clones of cells at the A/P boundary that do not express Ci do not activate anterior en [65].
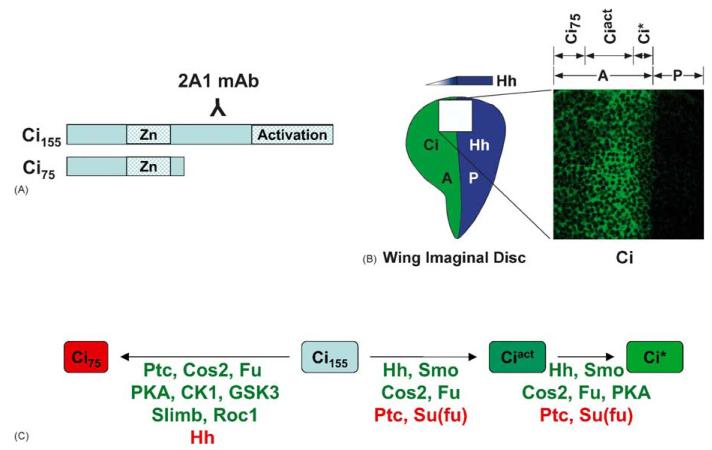
Fig. 3.
Hedgehog sensitive processing of Cubitus interruptus. (A) Schematic of Ci155 and Ci75 depicting the epitope of the 2A1 monoclonal antibody. (B) The Ci gradient in the Drosophila wing imaginal disc. Late third instar wing imaginal discs were immuno-stained with 2A1 monoclonal antibody. Hedgehog diffuses from the Posterior (P) compartment to affect production of Ci activator species in Anterior (A) compartment cells. Cells near the A/P compartment boundary, receiving the highest level of Hh stimulation convert Ci155 to the enhanced transcriptional activator Ci*. It is evidenced by decreased detection of Ci by the 2A1 monoclonal antibody. Cells further into the A compartment accumulate Ciact, as evidenced by more intense Ci staining, which results from decreased Ci proteolysis. Cells far into the A compartment contain higher amounts of Ci75, which is not detected by the 2A1 antibody. (C) Hedgehog sensitive Ci processing is dependent on multiple Hedgehog signaling components. Factors listed in green promote conversion to the indicated Ci species. Factors listed in red prevent conversion.
The only co-activator thus far demonstrated to directly associate with Ci activator(s) is the acetyl transferase CREB binding protein (CBP) [69,70]. CBP binds the carboxyl-terminal activation domain of Ci, presumably to facilitate modifications of target gene chromatin structure. Coincidentally, the CBP binding site overlaps one of the two known Cos2 interaction domains [71], suggesting that Cos2 may regulate Ci function by occluding the Ci activation domain. Cos2 is also involved in regulating the nuclear localization of Ci: Cos2 has been demonstrated to block Ci nuclear entry [71]. Ci that does gain access to the nucleus, does so without other members of the HSC [30], implying that Cos2, Fu and Su(fu) regulation of Ci occurs in the cytoplasm.
5. Costal2
Based on genetic studies, Cos2 is categorized as both a negative and positive regulator of the Hh pathway. cos2 mutant Drosophila are embryonic lethal and exhibit segment-polarity defects [72-74]. However, the effects of cos2 loss of function can be examined in adult structures through mosaic analysis. Clones of cells in far anterior regions of the A compartment of the wing imaginal disc that lack cos2 trigger anterior overgrowth phenotypes, and activation of low level Hh target genes, suggesting a negative regulatory role [23,66,67,71]. However, cos2 clones near the A/P compartment boundary do not express high level Hh-target genes, suggesting a role for Cos2 in activation of Ci [48,66,71]. These studies demonstrate that Cos2 plays a role in establishing both activated and repressed Hh states. It is likely that the requirement for Cos2 in both activation and repression of Hh signaling is based, at least in part, on a proposed interaction and localization with Smo, and the mechanism by which it connects Smo with the HSC. Accordingly, target gene activation can be dramatically affected by changing the ratio of Cos2 to Smo [75]. Increased expression of exogenous Cos2 with a constant concentration of Smo facilitates target gene activation until Cos2 concentration far exceeds that of Smo. At this concentration, reporter activation is dramatically decreased, indicating that a higher relative concentration of Cos2 favors repression while a higher relative concentration of Smo shifts Cos2 towards a positive regulatory role. Accordingly, biochemical studies demonstrate that Hh-mediated accumulation of Smo appears to trigger degradation of Cos2/Fu complexes [75,76]. This may serve to alter the Smo/Cos2 ratio, allowing Smo to push Cos2 towards its activating function.
Cos2 shares homology with the family of Kinesin-related protein (KRP) molecular motors [23]. KRPs bind vesicular membranes and microtubules (MT) in a highly regulated manner, and can facilitate movement along MTs through ATP hydrolysis (reviewed in [77]). Although ATPase activity by Cos2 has yet to be demonstrated, evidence supporting Cos2-mediated transport activity in Hh signaling is growing [44,76,78]. Studies using Drosophila salivary glands as a model system demonstrated that over-expression of Cos2 resulted in redistribution of Smo from the plasma membrane to discrete intracellular vesicles in a MT-dependent manner [44]. Expression of exogenous Hh in the presence of over-expressed Cos2 could trigger movement of Smo from vesicles to the PM [44]. Thus, a primary role for Cos2 may be to facilitate movement of Smo in response to Hh: inactive Smo is transported away from the PM while activated Smo is transported to the PM. Consistent with this hypothesis, Cos2 appears to associate with cellular membranes in a Hh-sensitive, Smo independent manner [78]. Based on these results, and the results described above, it is tempting to speculate that Cos2-mediated changes in Smo localization dictate whether the pathway is active (PM localized), or inactive (vesicular localization). The question then arises, if Cos2 does function as the KRP responsible for changes in Smo and HSC localization, what regulates the movement of Cos2? Cos2 is phosphorylated in response to Hh [29,79], but the kinase responsible for phosphorylation, and what that phosphorylation might be regulating are not clearly defined.
6. Fused
Fu kinase activity is required for proper Hh signal transduction, as mutations within the conserved residues required for kinase activity disrupt signaling [22,67,80-82]. However, while these results are consistent with the kinase domain of Fu being required for Hh signaling, such activity has been difficult to demonstrate biochemically. Thus, the direct substrates(s) of Fu have not been clearly elucidated. Fu is an integral member of both trimeric and tetrameric HSCs [29,30]. It is therefore likely that Fu targets other complex members for phosphorylation. The most likely substrates of Fu kinase activity are Cos2 and Su(fu), as both proteins have been found to interact directly with Fu [30,83-85]. Additionally, there is some indirect evidence indicating that both Cos2 and Su(fu) are phosphorylated by Fu in response to Hh [75,79].
Genetic interactions between fu and Su(fu) reveal a second functional domain within the Fu carboxyl-terminus, which plays two distinct roles in Hh signaling [80,82]. The carboxyl-terminal domain of Fu interacts directly with Cos2 and Su(fu) [83,84], and has been proposed to function as the targeting subunit, required to connect the kinase domain with its substrate(s) [80]. Mutations within the carboxyl-terminal domain are predicted to block Fu’s ability to locate its substrate(s), thereby preventing proper Hh signaling. Consistent with this prediction, Fu proteins truncated within the carboxyl-terminal domain lack the ability to associate with Cos2 [29]. These results suggest that, despite an intact kinase domain, mutants unable to bind Cos2 do not facilitate proper Hh signaling [31,67]. Accordingly, when over-expressed, the carboxyl-terminal domain of Fu acts as a dominant inhibitor of Hh signaling by preventing endogenous Fu from interacting with endogenous Cos2 [85].
The carboxyl-terminus of Fu also plays a role in the conversion of Ci to Ci75. Analysis of Ci in mutant flies that lack Su(fu) and the carboxyl-terminal domain of fu show little to no Ci75, increased levels of full-length Ci, and ectopic accumulation of Ci throughout the entire anterior compartment of the wing imaginal disc [31,86]. Accordingly, over-expression of Fu-tail in wild type imaginal discs prevents the formation of Ciact and Ci*, but does not affect conversion of Ci to Ci75 [85]. It was proposed that although Fu tail could block endogenous Fu association with Cos2, Ci processing was unaffected because Fu kinase activity is not required for processing of Ci to Ci75.
7. Suppressor of fused
Su(fu) does not share homology with any known proteins [26]. Thus, speculation regarding its role in the Hh pathway has been difficult. Drosophila with Su(fu) mutation(s) are phenotypically normal, suggesting that it is not required for functional Hh signaling [26,31,80]. However, since Su(fu) has been demonstrated to interact directly with both Fu and Ci, but not Cos2, it has been suggested that Su(fu) function may be partially redundant with that of Cos2 [30,71]. This is supported by genetic studies demonstrating that loss of cos2 suppresses fu mutations [80].
The most prevalent model regarding Su(fu)’s role as a negative regulator of Hh signaling is that it regulates nuclear translocation of Ci. Clones of cells over-expressing Su(fu) demonstrate decreased nuclear accumulation of full-length Ci activator(s) [87]. Conversely, cells lacking Su(fu) demonstrate increased nuclear Ci, consistent with Su(fu) acting in a partially redundant fashion with Cos2 to block Ci nuclear entry [66,71,87]. Studies involving mammalian Su(fu) support this hypothesis. Over-expression of human SU(FU) in cultured mammalian cells triggers increased cytoplasmic GLI-1 and decreased expression of Gli reporter construct(s) [88-90].
Evidence supporting a role for Su(fu) as a transcriptional co-repressor also exists. In vertebrate systems, Su(fu) has been demonstrated to interact with SAP18, a member of the mSin3a histone deacetylase complex, to trigger decreased expression of Gli reporter construct(s) [91,92]. Accordingly, studies in chicken demonstrate endogenous Su(fu) to be predominantly nuclear, and to enhance the ability of both Gli1 and Gli3 to bind DNA [93]. Thus, Su(fu) may associate with DNA bound Ci (or Gli) transcription factors to recruit HDAC complexes to facilitate target gene repression in the absence of Hh. Fu-mediated phosphorylation of Su(fu) and Cos2 in response to Hh may block these negative regulatory effects on Ci [67]. Differential phosphorylation of Cos2 and Su(fu) might then allow for graded levels of transcriptional activator function in response to changing levels of Hh stimulation.
Any model for the role of Su(fu), or Fu, in Hh signaling must take into account the observation that a kinase inactive fu allele in a Su(fu) background yields a grossly wild type fly [26]. What then are the roles of Fu and Su(fu) if their activities, when both genes are mutated, are not necessary? It is possible that the only roles for Fu kinase activity and Su(fu) are to oppose each other. Thus, loss of both proteins would not have an obvious effect on Hh signaling. Another possibility is that because Hh signaling is critical for proper development, other proteins may play redundant functions, as a fail-safe mechanism, to ensure proper pattern formation following the loss of Fu or Su(fu).
8. Smoothened communicates directly with the Hedgehog Signaling Complex to regulate Ci
Until recently, one of the most puzzling questions regarding Hh signaling involved how Smo signaled to the HSC to regulate Ci. Although Smo shares homology with GPCRs, current evidence argues against the involvement of a traditional G-protein. For example, the Smo mutants SmoC and FFS, which lack the domains one would expect to interact with a G-protein, are still capable of propagating Hh signaling [48]. Additionally, reducing the expression of all known Drosophila heterotrimeric GTP binding proteins, through use of RNA interference (RNAi), had little effect on Hh responses in cultured cells [56,75]. This lack of compelling evidence for G-protein involvement in the Hh pathway led multiple groups to look for direct interactions between Smo and HSC components. A series of recent publications, demonstrating an interaction between the carboxyl terminal tail of Smo and the cargo domain of Cos2, have begun to shed light onto the mechanistic events involved in Smo-mediated signaling to the HSC [75,76,94,95]. While there are some differences in the published studies, there seems to be a consensus on the following points: (1) Smo binds Cos2 directly; (2) the interaction is necessary for functional Hh signaling; (3) Cos2 appears to tether significant amounts of Fu to Smo, while Ci and Su(fu) binding are not as obvious. A requirement for direct Smo–Cos2 binding in signal transduction is most obvious when examining target gene expression following loss of interaction. The Smo carboxyl-terminal tail was demonstrated to contain the Cos2 interaction domain [75,94,95]. Over-expression of this domain appears to have a dominant negative effect, resulting in a dose-dependent loss of reporter gene expression [95]. Similarly, over-expression of Smo proteins lacking the Cos2 binding domain and/or Cos2 constructs lacking the Smo binding domain demonstrate compromised Hh responses [75,76]. These results clearly demonstrate a requirement for Cos2–Smo interaction for proper Hh signal transduction.
The studies differ on some points, and leave some questions unanswered. The primary difference between the studies involves the putative Hh-mediated regulation of Smo–Cos2 association. One biochemical study suggests that the amount of Cos2 binding to Smo is increased, and that the complexes enrich on membranes in response to Hh [76]. While there is supporting evidence for a PM targeting requirement in activation of Smo signaling, there is not additional evidence for Hh-mediated increase in Smo–Cos2 binding [75,94,95]. The modest Hh-induced increase in Smo–Cos2 binding observed in the other studies was attributed to the Hh-mediated stabilization of Smo.
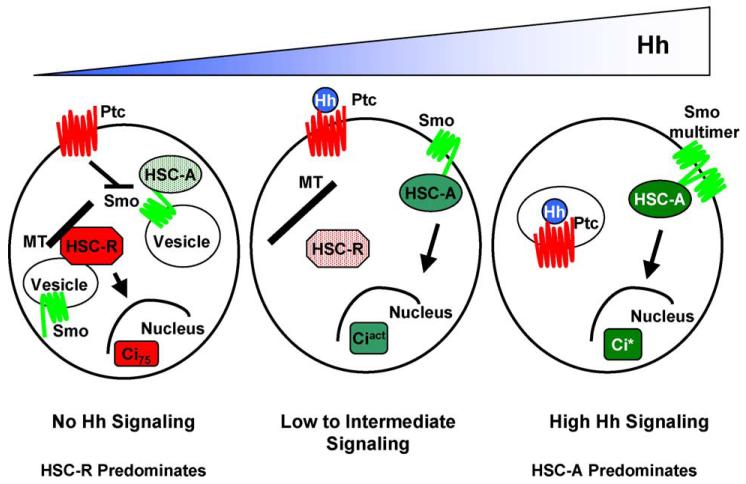
The observation that Smo/HSC and Smo/HSC/Ci associations do not appear to be dramatically affected by Hh stimulation is somewhat surprising. How is production of Ci repressor blocked and Ci activator(s) stimulated if association between the upstream and downstream effectors does not change? Additionally, the amount of membrane associated HSC appears to decrease in response to Hh [76,78]. This observation is seemingly inconsistent with the model whereby the Smo/Cos2 association remains constant or even increases. To explain this apparent paradox we have proposed that there may be two distinct HSCs, one involved in converting Ci into its repressor form, HSC-R, and one involved in converting Ci into its activated forms, HSC-A [78]. In the absence of Hh, HSC-R is on and HSC-A is off, while in response to maximal Hh, HSC-A is turned on and HSC-R is turned off. In between this two switch system, numerous intermediates exist. For example, in response to 25% maximal Hh signaling the majority of HSC-R would still be making Ci75, which would function in the presence of activation of 25% of HSC-A, producing activated Ci. Thus, various ratios of Ci75 and the activated forms of Ci would combine to establish unique cell fates within a specific position in a Hh gradient. A simplified diagram of this two HSC model is shown in Fig. 4, which depicts how three cells in a Hh gradient might use these two distinct HSCs to interpret its position in that gradient. We show HSC-R anchored to vesicular membranes through a central domain of Cos2, in a manner independent of Smo [78], where it converts Ci into a form suitable for proteosome degradation. In response to Hh, Cos2 is phosphorylated and releases from the vesicular membranes, attenuating Ci75 production. In the absence of Hh, HSC-A is also associated with vesicular membranes but in this case the association is indirect, as it is through Smo association. In response to Hh, HSC-A accumulates on the PM where Ci is activated. This activation requires Fu kinase activity to overcome the Su(fu)-mediated repression of Ci activator(s) [67]. An enhanced activation of Ci occurs in cells receiving the highest concentration of Hh. This super-activated form of Ci, Ci*, may require Smo dimerization [48], as is depicted in the cell nearest the Hh source. This two HSC model begins to resolve the paradox of Smo/Cos2 association increasing in the face of decreasing HSC/membrane association, and is consistent with the complex genetics observed between the various components of the Hh pathway [80]. Additionally, this model is consistent with the observation that Ci accumulation does not correlate with strength of target gene activation [67], as we predict that the bulk of Ci is in HSC-R and is not converted into an activated form. Instead the Ci in HSC-A, which is the minor form, would need to be activated in order to get higher levels of Hh signaling.
Fig. 4.
Model depicting how the Hh gradient regulates Hedgehog Signaling Complex activator and repressor functions. In the cell receiving no Hh (left cell), HSC-R is producing Ci75 and HSC-A is inactive. In the middle cell, HSC-A is producing Ciact. However, Ciact may be acting in the presence of a lower amount of Ci75. In the cell receiving the greatest amount of Hh (right cell), HSC-A is maximally activated by Smo multimerization, and HSC-R is completely off.
Acknowledgments
We thank Joan Hooper for helpful discussion of our model and John A. Goetz for review of the manuscript. This work was supported by grant CA82628 (to D.J.R.). M.A. is an Albert J. Ryan Fellow.
References
- [1].Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–87. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- [2].Taylor AM, Nakano Y, Mohler J, Ingham PW. Contrasting distributions of patched and hedgehog proteins in the Drosophila embryo. Mech Dev. 1993;42(1/2):89–96. doi: 10.1016/0925-4773(93)90101-3. [DOI] [PubMed] [Google Scholar]
- [3].Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ. Biochemical evidencethat patchedis the Hedgehog receptor. Nature. 1996;384(6605):176–9. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- [4].Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87(3):553–63. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- [5].Alcedo J, Ayzenzon M, Von Ohlen T, Noll M, Hooper JE. The Drosophila smoothened gene encodes a seven-pass membrane protein, a putative receptor for the hedgehog signal. Cell. 1996;86(2):221–32. doi: 10.1016/s0092-8674(00)80094-x. [DOI] [PubMed] [Google Scholar]
- [6].Hooper JE, Scott MP. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell. 1989;59(4):751–65. doi: 10.1016/0092-8674(89)90021-4. [DOI] [PubMed] [Google Scholar]
- [7].Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, Noll M, Hooper JE, de Sauvage F, Rosenthal A. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384(6605):129–34. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- [8].Murone M, Rosenthal A, de Sauvage FJ. Hedgehog signal transduction: from flies to vertebrates. Exp Cell Res. 1999;253(1):25–33. doi: 10.1006/excr.1999.4676. [DOI] [PubMed] [Google Scholar]
- [9].Cohen MM., Jr The hedgehog signaling network. Am J Med Genet. 2003;123A(1):5–28. doi: 10.1002/ajmg.a.20495. [DOI] [PubMed] [Google Scholar]
- [10].Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411(6835):349–54. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- [11].Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez-del Castillo C, Yajnik V, Antoniu B, McMahon M, Warshaw AL, Hebrok M. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425(6960):851–6. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422(6923):313–7. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- [13].Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman JR, Watkins DN, Beachy PA. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425(6960):846–51. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- [14].Johnson RL, Tabin C. The long and short of hedgehog signaling. Cell. 1995;81(3):313–6. doi: 10.1016/0092-8674(95)90381-x. [DOI] [PubMed] [Google Scholar]
- [15].Goetz JA, Suber LM, Zeng X, Robbins DJ. Sonic Hedgehog as a mediator of long-range signaling. Bioessays. 2002;24(2):157–65. doi: 10.1002/bies.10056. [DOI] [PubMed] [Google Scholar]
- [16].Rodriguez I, Basler K. Control of compartmental affinity boundaries by hedgehog. Nature. 1997;389(6651):614–8. doi: 10.1038/39343. [DOI] [PubMed] [Google Scholar]
- [17].Porter JA, von Kessler DP, Ekker SC, Young KE, Lee JJ, Moses K, Beachy PA. The product of hedgehog autoproteolytic cleavage active in local and long-range signalling. Nature. 1995;374(6520):363–6. doi: 10.1038/374363a0. [DOI] [PubMed] [Google Scholar]
- [18].Tabata T, Eaton S, Kornberg TB. The Drosophila hedgehog gene is expressed specifically in posterior compartment cells and is a target of engrailed regulation. Genes Dev. 1992;6(12B):2635–45. doi: 10.1101/gad.6.12b.2635. [DOI] [PubMed] [Google Scholar]
- [19].Tabata T, Kornberg TB. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell. 1994;76(1):89–102. doi: 10.1016/0092-8674(94)90175-9. [DOI] [PubMed] [Google Scholar]
- [20].Lee JJ, von Kessler DP, Parks S, Beachy PA. Secretion and localized transcription suggest a role in positional signaling for products of the segmentation gene hedgehog. Cell. 1992;71(1):33–50. doi: 10.1016/0092-8674(92)90264-d. [DOI] [PubMed] [Google Scholar]
- [21].Eaton S, Kornberg TB. Repression of ci-D in posterior compartments of Drosophila by engrailed. Genes Dev. 1990;4(6):1068–77. doi: 10.1101/gad.4.6.1068. [DOI] [PubMed] [Google Scholar]
- [22].Forbes AJ, Nakano Y, Taylor AM, Ingham PW. Genetic analysis of hedgehog signalling in the Drosophila embryo. Dev Suppl. 1993:115–24. [PubMed] [Google Scholar]
- [23].Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90(2):235–45. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- [24].Preat T, Therond P, Lamour-Isnard C, Limbourg-Bouchon B, Tricoire H, Erk I, Mariol MC, Busson D. A putative serine/threonine protein kinase encoded by the segment-polarity fused gene of Drosophila. Nature. 1990;347(6288):87–9. doi: 10.1038/347087a0. [DOI] [PubMed] [Google Scholar]
- [25].Schwartz C, Locke J, Nishida C, Kornberg TB. Analysis of cubitus interruptus regulation in Drosophila embryos and imaginal disks. Development. 1995;121(6):1625–35. doi: 10.1242/dev.121.6.1625. [DOI] [PubMed] [Google Scholar]
- [26].Preat T. Characterization of suppressor of fused, a complete suppressor of the fused segment polarity gene of Drosophila melanogaster. Genetics. 1992;132(3):725–36. doi: 10.1093/genetics/132.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Horabin JI, Walthall S, Vied C, Moses M. A positive role for patched in hedgehog signaling revealed by the intracellular trafficking of sex-lethal, the Drosophila sex determination master switch. Development. 2003;130(24):6101–9. doi: 10.1242/dev.00865. [DOI] [PubMed] [Google Scholar]
- [28].Vied C, Horabin JI. The sex determination master switch, sex-lethal, responds to Hedgehog signaling in the Drosophila germline. Development. 2001;128(14):2649–60. doi: 10.1242/dev.128.14.2649. [DOI] [PubMed] [Google Scholar]
- [29].Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90(2):225–34. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- [30].Stegman MA, Vallance JE, Elangovan G, Sosinski J, Cheng Y, Robbins DJ. Identification of a tetrameric hedgehog signaling complex. J Biol Chem. 2000;275(29):21809–12. doi: 10.1074/jbc.C000043200. [DOI] [PubMed] [Google Scholar]
- [31].Alves G, Limbourg-Bouchon B, Tricoire H, Brissard-Zahraoui J, Lamour-Isnard C, Busson D. Modulation of Hedgehog target gene expression by the Fused serine-threonine kinase in wing imaginal discs. Mech Dev. 1998;78(1/2):17–31. doi: 10.1016/s0925-4773(98)00130-0. [DOI] [PubMed] [Google Scholar]
- [32].Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89(7):1043–53. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- [33].Jeong J, McMahon AP. Cholesterol modification of Hedgehog family proteins. J Clin Invest. 2002;110(5):591–6. doi: 10.1172/JCI16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Strutt H, Thomas C, Nakano Y, Stark D, Neave B, Taylor AM, Ingham PW. Mutations in the sterol-sensing domain of Patched suggest a role for vesicular trafficking in Smoothened regulation. Curr Biol. 2001;11(8):608–13. doi: 10.1016/s0960-9822(01)00179-8. [DOI] [PubMed] [Google Scholar]
- [35].Martin V, Carrillo G, Torroja C, Guerrero I. The sterol-sensing domain of Patched protein seems to control Smoothened activity through Patched vesicular trafficking. Curr Biol. 2001;11(8):601–7. doi: 10.1016/s0960-9822(01)00178-6. [DOI] [PubMed] [Google Scholar]
- [36].Frank-Kamenetsky M, Zhang XM, Bottega S, Guicherit O, Wichterle H, Dudek H, Bumcrot D, Wang FY, Jones S, Shulok J, Rubin LL, Porter JA. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J Biol. 2002;1(2):10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16(21):2743–8. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406(679):1005–9. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- [39].Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell. 2000;102(4):521–31. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- [40].Incardona JP, Gruenberg J, Roelink H. Sonic hedgehog induces the segregation of patched and smoothened in endosomes. Curr Biol. 2002;12(12):983–95. doi: 10.1016/s0960-9822(02)00895-3. [DOI] [PubMed] [Google Scholar]
- [41].van den Heuvel M, Ingham PW. Smoothened encodes a receptor-like serpentine protein required for hedgehog signalling. Nature. 1996;382(6591):547–51. doi: 10.1038/382547a0. [DOI] [PubMed] [Google Scholar]
- [42].Alcedo J, Zou Y, Noll M. Posttranscriptional regulation of smoothened is part of a self-correcting mechanism in the Hedgehog signaling system. Mol Cell. 2000;6(2):457–65. doi: 10.1016/s1097-2765(00)00044-7. [DOI] [PubMed] [Google Scholar]
- [43].Ingham PW, Nystedt S, Nakano Y, Brown W, Stark D, van den Heuvel M, Taylor AM. Patched represses the Hedgehog signalling pathway by promoting modification of the Smoothened protein. Curr Biol. 2000;10(20):1315–8. doi: 10.1016/s0960-9822(00)00755-7. [DOI] [PubMed] [Google Scholar]
- [44].Zhu AJ, Zheng L, Suyama K, Scott MP. Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev. 2003;17(10):1240–52. doi: 10.1101/gad.1080803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418(6900):892–7. doi: 10.1038/nature00989. [DOI] [PubMed] [Google Scholar]
- [46].Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA. 2002;99(22):14071–6. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Incardona JP, Lee JH, Robertson CP, Enga K, Kapur RP, Roelink H. Receptor-mediated endocytosis of soluble and membrane-tethered Sonic hedgehog by Patched-1. Proc Natl Acad Sci USA. 2000;97(22):12044–9. doi: 10.1073/pnas.220251997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hooper JE. Smoothened translates Hedgehog levels into distinct responses. Development. 2003;130(17):3951–63. doi: 10.1242/dev.00594. [DOI] [PubMed] [Google Scholar]
- [49].Lu ZL, Saldanha JW, Hulme EC. Seven-transmembrane receptors: crystals clarify. Trends Pharmacol Sci. 2002;23(3):140–6. doi: 10.1016/S0165-6147(00)01973-8. [DOI] [PubMed] [Google Scholar]
- [50].Yeagle PL, Choi G, Albert AD. Studies on the structure of the G-protein-coupled receptor rhodopsin including the putative G-protein binding site in unactivated and activated forms. Biochemistry. 2001;40(39):11932–7. doi: 10.1021/bi015543f. [DOI] [PubMed] [Google Scholar]
- [51].Yeagle PL, Albert AD. A conformational trigger for activation of a G protein by a G protein-coupled receptor. Biochemistry. 2003;42(6):1365–8. doi: 10.1021/bi0270539. [DOI] [PubMed] [Google Scholar]
- [52].Alexandre C, Jacinto A, Ingham PW. Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev. 1996;10(16):2003–13. doi: 10.1101/gad.10.16.2003. [DOI] [PubMed] [Google Scholar]
- [53].Von Ohlen T, Hooper JE. Hedgehog signaling regulates transcription through Gli/Ci binding sites in the wingless enhancer. Mech Dev. 1997;68(1/2):149–56. doi: 10.1016/s0925-4773(97)00150-0. [DOI] [PubMed] [Google Scholar]
- [54].Von Ohlen T, Lessing D, Nusse R, Hooper JE. Hedgehog signaling regulates transcription through cubitus interruptus, a sequence-specific DNA binding protein. Proc Natl Acad Sci USA. 1997;94(6):2404–9. doi: 10.1073/pnas.94.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Methot N, Basler K. An absolute requirement for Cubitus interruptus in Hedgehog signaling. Development. 2001;128(5):733–42. doi: 10.1242/dev.128.5.733. [DOI] [PubMed] [Google Scholar]
- [56].Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299(5615):2039–45. doi: 10.1126/science.1081403. [DOI] [PubMed] [Google Scholar]
- [57].Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature. 2002;416(6880):548–52. doi: 10.1038/nature733. [DOI] [PubMed] [Google Scholar]
- [58].Jiang J, Struhl G. Protein kinase A and hedgehog signaling in Drosophila limb development. Cell. 1995;80(4):563–72. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- [59].Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 2002;108:823–35. doi: 10.1016/s0092-8674(02)00664-5. [DOI] [PubMed] [Google Scholar]
- [60].Price MA, Kalderon D. Proteolysis of Cubitus interruptus in Drosophila requires phosphorylation by Protein Kinase A. Development. 1999;126:4331–9. doi: 10.1242/dev.126.19.4331. [DOI] [PubMed] [Google Scholar]
- [61].Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391(6666):493–6. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- [62].Noureddine MA, Donaldson TD, Thacker SA, Duronio RJ. Drosophila Roc1a encodes a RING-H2 protein with a unique function in processing the Hh signal transducer Ci by the SCF E3 ubiquitin ligase. Dev Cell. 2002;2(6):757–70. doi: 10.1016/s1534-5807(02)00164-8. [DOI] [PubMed] [Google Scholar]
- [63].Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10(10):429–39. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- [64].Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science. 2000;287(5458):1606–9. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- [65].Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96(6):819–31. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- [66].Wang QT, Holmgren RA. Nuclear import of cubitus interruptus is regulated by hedgehog via a mechanism distinct from Ci stabilization and Ci activation. Development. 2000;127(14):3131–9. doi: 10.1242/dev.127.14.3131. [DOI] [PubMed] [Google Scholar]
- [67].Ohlmeyer JT, Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396(6713):749–53. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- [68].Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev. 1999;13(21):2828–37. doi: 10.1101/gad.13.21.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Akimaru H, Chen Y, Dai P, Hou DX, Nonaka M, Smolik SM, Armstrong S, Goodman RH, Ishii S. Drosophila CBP is a co-activator of Cubitus interruptus in hedgehog signalling. Nature. 1997;386(6626):735–8. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- [70].Chen Y, Goodman RH, Smolik SM. Cubitus interruptus requires Drosophila CREB-binding protein to activate wingless expression in the Drosophila embryo. Mol Cell Biol. 2000;20(5):1616–25. doi: 10.1128/mcb.20.5.1616-1625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wang G, Amanai K, Wang B, Jiang J. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of Cubitus interruptus. Genes Dev. 2000;14(22):2893–905. doi: 10.1101/gad.843900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- [73].Simpson P, Grau Y. The segment polarity gene costal-2 in Drosophila. II. The origin of imaginal pattern duplications. Dev Biol. 1987;122(1):201–9. doi: 10.1016/0012-1606(87)90345-9. [DOI] [PubMed] [Google Scholar]
- [74].Grau Y, Simpson P. The segment polarity gene costal-2 in Drosophila. I. The organization of both primary and secondary embryonic fields may be affected. Dev Biol. 1987;122(1):186–200. doi: 10.1016/0012-1606(87)90344-7. [DOI] [PubMed] [Google Scholar]
- [75].Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, Weis-Garcia F, Gong R, Wang B, Beachy PA. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell. 2003;12(5):1261–74. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- [76].Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Therond PP. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol. 2003;5(10):907–13. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- [77].Goldstein LS. With apologies to scheherazade: tails of 1001 kinesin motors. Annu Rev Genet. 1993;27:319–51. doi: 10.1146/annurev.ge.27.120193.001535. [DOI] [PubMed] [Google Scholar]
- [78].Stegman MA, Goetz JA, Ascano M, Jr, Ogden SK, Nybakken KE, Robbins DJ. The kinesin related protein Costal2 associates with membranes in a hedgehog sensitive, smoothened independent manner. J Biol Chem. doi: 10.1074/jbc.M311794200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Nybakken KE, Turck CW, Robbins DJ, Bishop JM. Hedgehog-stimulated phosphorylation of the kinesin-related protein Costal2 is mediated by the serine/threonine kinase fused. J Biol Chem. 2002;277(27):24638–47. doi: 10.1074/jbc.M110730200. [DOI] [PubMed] [Google Scholar]
- [80].Preat T, Therond P, Limbourg-Bouchon B, Pham A, Tricoire H, Busson D, Lamour-Isnard C. Segmental polarity in Drosophila melanogaster: genetic dissection of fused in a Suppressor of fused background reveals interaction with costal-2. Genetics. 1993;135(4):1047–62. doi: 10.1093/genetics/135.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Therond PP, Knight JD, Kornberg TB, Bishop JM. Phosphorylation of the fused protein kinase in response to signaling from hedgehog. Proc Natl Acad Sci USA. 1996;93(9):4224–8. doi: 10.1073/pnas.93.9.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Therond P, Alves G, Limbourg-Bouchon B, Tricoire H, Guillemet E, Brissard-Zahraoui J, Lamour-Isnard C, Busson D. Functional domains of fused, a serine-threonine kinase required for signaling in Drosophila. Genetics. 1996;142(4):1181–98. doi: 10.1093/genetics/142.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Monnier V, Ho KS, Sanial M, Scott MP, Plessis A. Hedgehog signal transduction proteins(2003) contacts of the Fused kinase and Ci transcription factor with the Kinesin-related protein Costal2. BMC Dev Biol. 2002;2(1):4. doi: 10.1186/1471-213X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Monnier V, Dussillol F, Alves G, Lamour-Isnard C, Plessis A. Suppressor of fused links fused and Cubitus interruptus on the hedgehog signalling pathway. Curr Biol. 1998;8(10):583–6. doi: 10.1016/s0960-9822(98)70227-1. [DOI] [PubMed] [Google Scholar]
- [85].Ascano M, Jr, Nybakken KE, Sosinski J, Stegman MA, Robbins DJ. The carboxyl-terminal domain of the protein kinase fused can function as a dominant inhibitor of hedgehog signaling. Mol Cell Biol. 2002;22(5):1555–66. doi: 10.1128/mcb.22.5.1555-1566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Lefers MA, Wang QT, Holmgren RA. Genetic dissection of the Drosophila Cubitus interruptus signaling complex. Dev Biol. 2001;236(2):411–20. doi: 10.1006/dbio.2001.0345. [DOI] [PubMed] [Google Scholar]
- [87].Methot N, Basler K. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development. 2000;127(18):4001–10. doi: 10.1242/dev.127.18.4001. [DOI] [PubMed] [Google Scholar]
- [88].Murone M, Luoh SM, Stone D, Li W, Gurney A, Armanini M, Grey C, Rosenthal A, de Sauvage FJ. Gli regulation by the opposing activities of fused and suppressor of fused. Nat Cell Biol. 2000;2(5):310–2. doi: 10.1038/35010610. [DOI] [PubMed] [Google Scholar]
- [89].Kogerman P, Grimm T, Kogerman L, Krause D, Unden AB, Sandstedt B, Toftgard R, Zaphiropoulos PG. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat Cell Biol. 1999;1(5):312–9. doi: 10.1038/13031. [DOI] [PubMed] [Google Scholar]
- [90].Ding Q, Fukami S, Meng X, Nishizaki Y, Zhang X, Sasaki H, Dlugosz A, Nakafuku M, Hui C. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol. 1999;9(19):1119–22. doi: 10.1016/s0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- [91].Paces-Fessy M, Boucher D, Petit E, Paute-Briand S, Blanchet-Tournier MF. The negative regulator of Gli, suppressor of fused (Sufu), interacts with SAP18, Galectin3 and other nuclear proteins. Biochem J Pt. doi: 10.1042/BJ20030786. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cheng SY, Bishop JM. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc Natl Acad Sci USA. 2002;99(8):5442–7. doi: 10.1073/pnas.082096999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Pearse RV, 2nd, Collier LS, Scott MP, Tabin CJ. Vertebrate homologs of Drosophila suppressor of fused interact with the gli family of transcriptional regulators. Dev Biol. 1999;212(2):323–36. doi: 10.1006/dbio.1999.9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Jia J, Tong C, Jiang J. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes Dev. 2003;17(21):2709–20. doi: 10.1101/gad.1136603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Ogden SK, Ascano M, Jr, Stegman MA, Suber LM, Hooper JE, Robbins DJ. Identification of a functional interaction between the transmembrane protein Smoothened and the kinesin-related protein Costal2. Curr Biol. 2003;13(22):1998–2003. doi: 10.1016/j.cub.2003.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Wang QT, Holmgren RA. The subcellular localization and activity of Drosophila cubitus interruptus are regulated at multiple levels. Development. 1999;126(22):5097–106. doi: 10.1242/dev.126.22.5097. [DOI] [PubMed] [Google Scholar]
- [97].Chen CH, von Kessler DP, Park W, Wang B, Ma Y, Beachy PA. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell. 1999;98(3):305–16. doi: 10.1016/s0092-8674(00)81960-1. [DOI] [PubMed] [Google Scholar]