Abstract
Ribosomes, constituting protein factories in living cells, translate genetic information carried by messenger RNAs into proteins, and are thus involved in virtually all aspects of cellular development and maintenance. The few available structures of the eukaryotic ribosome1–6 reveal its increased complexity compared to its prokaryotic counterpart7–8, manifested mainly in the presence of eukaryotic-specific ribosomal proteins and additional rRNA insertions, called expansion segments (ES)9. These structures also point out some differences among species, in part represented by the size and arrangement of these ESs. Such differences are extreme in kinetoplastids, unicellular eukaryotic parasites often infectious to humans. Here we present a high-resolution cryo-electron microscopy structure of the ribosome from Trypanosoma brucei, the parasite transmitted by the Tse-tse fly and causing African sleeping sickness. Our atomic model reveals the unique features of this ribosome, characterized mainly by the presence of extraordinarily large ESs and ribosomal proteins extensions leading to the formation of four additional intersubunit bridges, and additional rRNA insertions including one large rRNA domain nonexistent in other eukaryotes. The kinetoplastid large ribosomal subunit (LSU) rRNA chain is uniquely cleaved into six pieces10–12; our structure reveals the five cleavage sites and suggests the importance of the cleavage for the maintenance of the T. brucei ribosome in the observed structure. We discuss several possible implications of the large rRNA ESs for the translation regulation process. Our structure could serve as a basis for future experiments aiming at understanding the functional importance of these kinetoplastid-specific ribosomal features in the protein translation regulation, an essential step toward finding effective and safe kinetoplastid-specific drugs.
The uniqueness of the T. brucei ribosomes, and those of other kinetoplastids, has been highlighted early on by the discovery of the LSU rRNA chain processing into six pieces10–12. The rRNA secondary structure of T. brucei (Fig. S1a and b) already pointed out the existence of several interesting features, such as the size of several ESs. A comparison of T. brucei ribosomal protein sequences with those of eukaryotic homologs13 also reveals the presence of T. brucei-specific large protein extensions. However, the arrangement and the structure of these features have remained so far unknown, although a low-resolution cryo-EM study of the T. cruzi ribosome has been reported previously14. It is worth mentioning that gene expression in trypanosomes is primarily regulated post-transcriptionally (see ref. 15), at the protein translation level. Several observations suggest differences both in translation and translational regulation in T. brucei and kinetoplastids in general, compared to other eukaryotes. For example, kinetoplastid mRNAs are capped by a 35 nucleotides conserved spliced-leader (SL) that is trans-spliced onto their 5' ends (see ref. 16) and has an extraordinarily modified cap. Additionally, unique protein factors are involved in the maturation of the T. brucei ribosomes17, while several eukaryotic initiation factors (eIF-4B, eIF-3 subunit P135, eIF-3 subunit 3) and the eukaryotic ribosomal recycling factor (eRRF) are absent18.
Here we present a high-resolution cryo-EM structure of the T. brucei ribosome which allowed us to model the atomic structure using a combination of map segmentation19,20, homology modeling, ab initio rRNA modeling based on co-variation analysis using S2S and Assemble21,22, and Molecular Dynamics Flexible Fitting (MDFF)23 (see Methods).
The cryo-EM map was reconstructed from ~164,000 particles (see Methods). The T. brucei ribosome is observed in the intersubunit-rotated state, with a tRNA at the E site (Fig. 1a and b). A measurement of local resolution (see Methods) shows that the resolution is ~5 Å in most parts of the map (Fig. S2a) except for small regions of high variance where it reaches 9 Å. Consistently, the 3D variance24 of the T. brucei ribosome map (Fig. S2b) shows little structural variability, with the exception of the intersubunit space where tRNA- and factor-binding takes place, and areas around the L1 and P stalks. At the tRNAs binding sites, the 3D variance mostly points out the intermittent presence of tRNAs at the A and P sites, as our ribosome purification is prepared from T. brucei cells extract, including a large fraction of translating ribosomes. Unsupervised data classification (see Methods) was attempted on a subset of the particles, but presumably because of the local nature of the variability it was not possible to identify distinct classes. Most of the T. brucei ribosome particles appear to exist in the same state. Phosphate density bumps along the rRNA strands and indications of protein side chains are visible (Fig. S2c) in most regions of the map, and a comparison of the experimental cryo-EM map with simulated density maps generated from the atomic model, at a resolution of 5.0 Å (Fig. S2d), validate our resolution estimation.
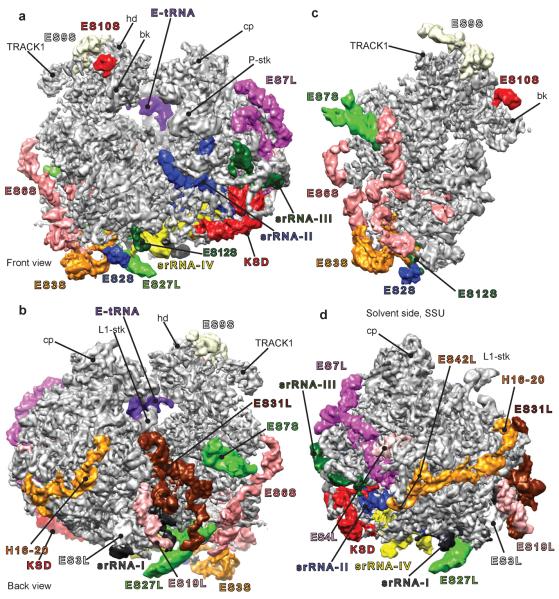
Fig. 1.
High-resolution cryo-EM structure of the T. brucei ribosome. The density map was filtered with a locally varying bandpass, according to the local resolution measurements (see Methods and figure S2A). In all following panels, largest rRNA expansion segments (ESs) are rendered in different colors. (a) The T. brucei ribosome viewed from the front (P-stalk and beak side), (b) viewed from the back (L1-stalk and platform side), (c) solvent side of the small subunit (SSU) and (d) solvent side of the large subunit (LSU). KSD = kinetoplastid-specific domain, hd = head, bk = beak and cp = central protuberance.
The first striking observation is the unusual size of several expansion segments, (Fig. 1). In the small ribosomal subunit (SSU), ESs 3S, 6S, 7S and 9S appear to be several times larger than in other 80S ribosomes of known structures (Fig. 1c). The same holds true for ESs 4L, 7L, 19L, 27L and 31L of the LSU (Fig. 1d). An additional domain on the LSU, which we call Kinetoplastid-Specific Domain (KSD), is situated in the vicinity of ES7L (Fig. 1, in red). These observations are consistent with the rRNA sequence and secondary structure in both ribosomal subunits25 (Fig. S1). We were able to model all proteins of the T. brucei ribosome (figs. 2 and S2d), except for some unstructured tails, and the rRNAs in their entirety, with the exception of a part of ES27L and ES9S where the density is poorly resolved, indicative of their dynamic behavior.
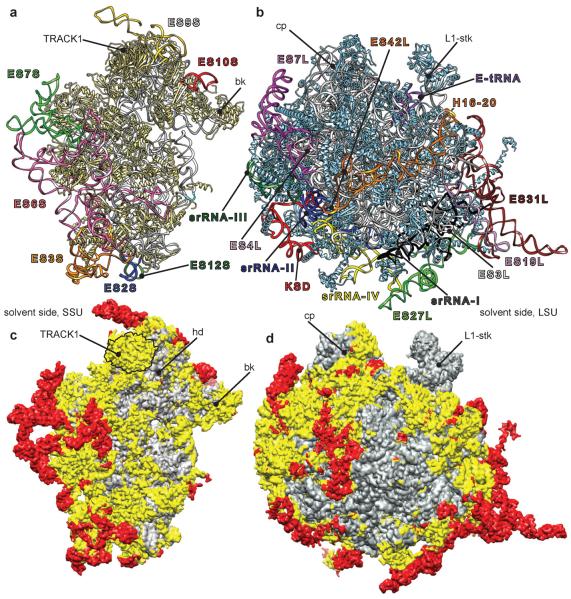
Fig. 2.
Atomic model of the T. brucei ribosome. (a) SSU and (b) LSU: atomic models with the ESs colored and annotated similarly as in Fig. 1. (c) SSU and (d) LSU: atomic models in surface presentations. Gray regions indicate the location of conserved common core elements of all ribosomes, prokaryotic and eukaryotic. Yellow regions highlight eukaryotic-specific conserved elements, including those for trypanosomes. Red regions indicate trypanosome-specific elements, nonexistent in other 80S ribosomes.
Our atomic model of the T. brucei ribosome reveals the structure of its ESs and additional rRNA insertions in both subunits (Fig. 2a and b), representing a substantial additional mass compared to other known eukaryotic ribosomes (Fig. 2c and d). In spite of this addition and the change in rRNA arrangement at several sites (Fig. S3a and c), the structures and binding sites of ribosomal proteins are conserved (Fig. S3b and d), as exemplified by proteins S7e on the SSU and L19e on the LSU (Fig. 3a). The high structural similarity of these proteins contrasts with the difference in the arrangement of the surrounding rRNA context, formed by ES6S, ESs 27L and 31L (Fig. 3a), when comparing T. brucei to yeast.
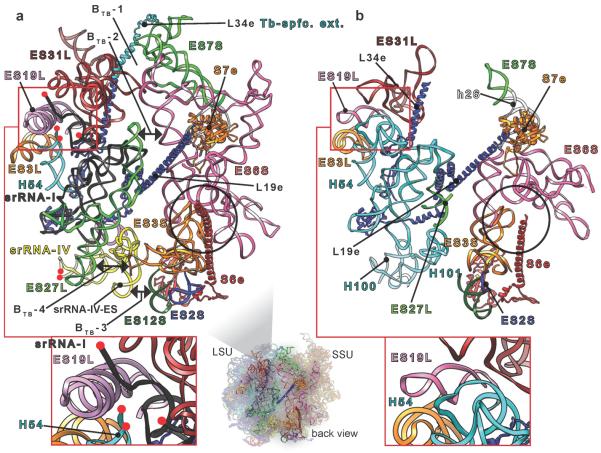
Fig. 3.
Comparison between T. brucei and yeast ribosomes. View of rRNA expansion segments (ES) at the back of the T. brucei (a) and yeast (b) ribosomes. T. brucei-specific intersubunit bridges are indicated by double arrows in (a). srRNA-I cleavage site in T. brucei and its counterpart region in yeast are blown up at the bottom and highlighted by red frames. Black circles highlight S6e interaction with ESs 3S & 6S in both ribosomes. Red spheres in (a) denote srRNA I and IV cleaved ends. Tb-spfc. ext. = T. brucei-specific extension.
The largest ESs are mostly concentrated in the regions of the LSU L1 stalk and the platform of the SSU. These large ESs form four additional intersubunit bridges, not seen in either of the two available 80S structures1–4 (Fig. 3a). These bridges are defined and designated as follows: (i) BTB-1, formed by three contacts (figs. 3a and S4a); BTB-1a, SSU ES7S helix (h) A with LSU ES31L-hC. BTB-1b, SSU ES7S-hA with L34e C-terminal extension accompanying ES31-hC. BTB-1c, SSU ES6S-hF with LSU ES31L-hC. This first bridge appears stronger in the previously published T. cruzi density map (Fig. S4a). (ii) BTB-2, formed by SSU ES6S-hG and LSU ES27L-hA (figs. 3a and S4b). This bridge is not established in T. cruzi despite the presence of its components. (iii) BTB-3, formed by SSU ES12S and a trypanosome-specific ES grafted on srRNA-IV, which we name srRNA-IV-ES (figs. 3a and S4c). This bridge is the strongest among the four, as seen from the T. brucei and T. cruzi density maps. (iv) BTB-4, formed by SSU ES3S-hC and LSU ES27L-hB (figs. 3a and S4d). However, the cryo-EM density of this latter bridge is poorly resolved and can only be observed at a low density threshold in T. brucei, probably due to the inherent flexibility of ES3S and ES27L. This contrasts with T. cruzi, where it is observable at a relatively high density threshold.
As for the enigmatic processing of LSU rRNA in six pieces, our structure shows the arrangement of these pieces and appears to indicate the structural necessity of such processing. While the processing of the LSU rRNA in two pieces, a and b, has been observed in other organisms (see literature cited in ref. 10), only trypanosomes and other kinetoplastids10 exhibit such extensive processing. The first srRNA, ~220 nucleotides11,12, is generated from the main immature rRNA of LSU by the removal of two flanking sequences, (Fig. S5a), splitting the main immature rRNA chain of the LSU into two components, α and β (Fig. 3a, red frame). These flanking sequences are localized at the position of LSU ES20L and ES26L, respectively, found in other eukaryotic ribosomes9, right after H54 on LSU domain III12. It is possible that LSU ESs 20L and 26L in kinetoplastids have evolved to become cleavage sites.
srRNA-I is structurally very similar to its homologous region on domain III in yeast (Fig. S5b), except that it bears an ES, which we name srRNA-I-ES. It is interesting to note that LSU ES19L in T. brucei is two to three times the size that it has in other 80S ribosomes (Fig. 3). srRNA-I cleavage in T. brucei apparently removes a piece of rRNA that would otherwise collide with ES19L. Conceivably, this cleavage could be an expedient evolutionary solution to allow room for the unusually large ES19L in its observed structure. However, the functional importance of ES19L is unknown.
The remaining srRNAs II, III and IV (180, 70 and 140 nucleotides, respectively) were suggested previously, based on sequence conservation and 2D modeling, to interact with one another and mimic the missing last domain of the LSU β rRNA in the T. brucei ribosome, corresponding to domain VI in yeast and other 80S ribosomes12. Our structure confirms this hypothesis and shows, in addition, that srRNAs II–IV interact directly with the KSD (Fig. 4a, left and middle) so as to form one single stable domain similar to domain VI in other 80S ribosomes (Fig. 4a, right). Similarly to srRNA-I, the processing of srRNAs II–IV could be explained, at least in part, as a structural necessity. Indeed, the existence of KSD in the observed structure and conformation is possible only if several pieces of rRNA are cleaved, as they would otherwise collide with KSD. The cleavage sites correspond to LSU ESs 36L and part of 39L in other eukaryotic ribosomes, and the cleavage yields the three srRNAs. The bundle formed by srRNAs II–IV and KSD is anchored in place through two specific protein extensions on L14e and L31e that hug the rRNA bundle (Fig. 4a, middle). There is also a unique interaction between srRNA-II and domain I of LSU α rRNA (Fig. 4a, green arrow). This interaction occurs between a proximal stem-loop on srRNA-II and another stem-loop, part of a T. brucei ES between helices 19 and 20 on the α rRNA domain I, which we call ES42L. Although the effect of this connection is unknown, we speculate it may contribute to maintaining the structure of the srRNAs bundle. 5S and 5.8S rRNAs in T. brucei ribosome are very similar to their counterparts in yeast, except that ES3L on the 5.8S rRNA is larger.
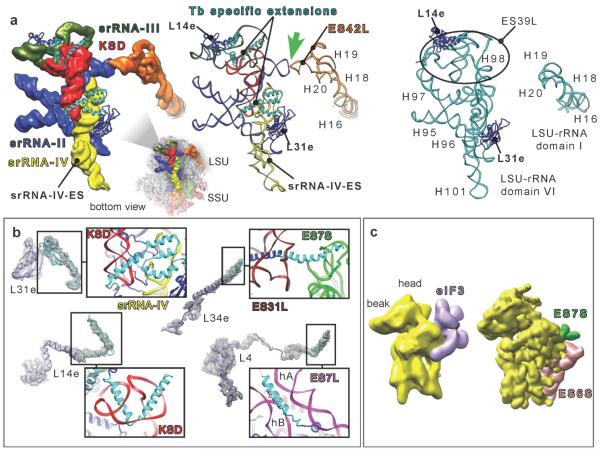
Fig. 4.
T. brucei srRNAs and protein extensions. (a) Left and middle, srRNAs II to IV and KSD cryo-EM density segmentations along with their atomic models compared to yeast domain VI (right). Thick Green arrow highlights the kissing-loop interaction. Black circle surrounds ES39L in yeast ribosome. (b) Ribosomal proteins presenting specific extensions in T. brucei (cyan ribbon) with zooms on the interactions of their extensions with the surrounding rRNA. (c) Left, cryo-EM-derived model of the eIF3-binding site (recreated according to ref. 21). Right, segmented map of the SSU, from T. brucei ribosome, filtered to 20Å. KSD = kinetoplastid-specific domain.
No trypanosome-specific proteins were found, based on the analysis of the density map. However, several T. brucei ribosomal proteins have evolved specific helical extensions that interact with the additional rRNA mass and contribute toward stabilizing the extended ESs in their observed conformations (Fig. 4b). In other cases, ribosomal protein tails have adopted different structures. On the LSU, L4 presents a long N-terminal helical tail, different in the other available structures, which is likely due to the presence of a large, trypanosome-specific rRNA helical insertion in LSU ES7L, hA (Fig. 4b). On the SSU, the trypanosomal RACK1 protein (TRACK1) is found in its conserved binding site (Fig. 2a), which contrasts with T. cruzi ribosome14 where TRACK1 was missing from the density map, for a reason that is unresolved26.
An interesting protein-rRNA interaction is formed by S6e with ESs 3S and 6S (Fig. 3a, black circle). S6e possesses an unstructured C-terminal tail, unresolved in both yeast 80S1,2 and T. thermophila 40S subunit structures5. This tail is involved in translation regulation by phosphorylation of several serine residues recognized by S6e kinases (RSK and S6K) in higher eukaryotes and many lower eukaryotes (see ref. 27). The T. brucei S6e C-terminal tail contains a conserved RxxSS motif, recognizable by S6 kinases, and appears to have a helical structure (Fig. 3a, black circle, and Fig. S6). The S6e C-terminal tail is surrounded by additional parts of SSU ESs 3S and 6S shielding the phosphorylation site, Ser 233 and 234 (Fig. S6), and could render it inaccessible for kinases on the ribosome. This is suggestive of a difference in translation regulation between trypanosomes and higher eukaryotes.
The role of the kinetoplastid-specific ESs in the regulation of the translation process remains unknown. In fact, very little is known about the translation regulation in kinetoplastids. However, the localization and the arrangement of the specific ESs, revealed by our structure, already suggest the importance of several elements. In T. brucei, SSU ESs 6S and 7S are localized at a very special site, near the mRNA channel exit, poised for a potential role in the regulation of translation initiation. The positions of ESs 6S and 7S on the SSU would put them in reach of the mRNA spliced-leader and its extraordinarily modified cap, as well as with the eukaryotic initiation factor 3 (eIF3)28,29. It is interesting to note that SSU ESs 6S and 7S would collide with the proposed binding site of eIF329 (Fig. 4c). eIF3 does exist in trypanosomes18 (Fig. S7); in T. brucei, it contains at least seven or eight subunits. Our structure suggests that in T. brucei, eIF3 may interact with the SSU in a way different from other eukaryotes, and that ESs 6S and 7S may have an important role in this interaction, or, alternatively, that the proposed binding site of eIF329 may require reconsideration.
It was observed that the T. brucei ribosome interacts with specific factors involved in translation regulation, such as L34 and L3717 (not to be confused with the LSU proteins of the same name), required in the maturation and cytoplasmic exportation of the LSU. It is possible that kinetoplastid-specific ESs are involved in the binding of such specific factors regulating the translation process. This hypothesis is supported by the observation that T. brucei ESs contain several pyrimidine-rich internal loops, previously suggested to be important for protein recognition or binding to other RNA elements30.
In conclusion, our cryo-EM study reveals an unusual structure for the T. brucei ribosome and shows the unique arrangement of its ESs, which may have consequences for the regulation of translation in trypanosomes. Our results may explain, in structural terms, the rationale for the processing of LSU-rRNA in six pieces. More generally, our structure highlights the extraordinary diversity of ribosome structure among eukaryotes. We believe it constitutes a basis for future investigations of translation regulation in kinetoplastids and the function of the specific expansion segments, opening the door to the development of new drugs.
Methods
Trypanosome Culture and Ribosomal Isolation
Procyclic forms of T. brucei strain TREU 667 were grown at 27°C in a semi-defined medium (SM) supplemented with 10% heat-inactivated fetal bovine serum and 25 mM HEPES, as previously described31. For ribosomal isolation, cells were grown to a density of 0.5–1.2 x 107 cells / mL in a 4 L volume, and harvested by 10 min centrifugation at 6000xg.
Ribosomes were prepared using a previously described method32 with slight modifications. Briefly, cell pellets were washed three times with SKS buffer and resuspended in SKS buffer containing 5 mM EGTA, 0.5% Triton-X- 100 and protease inhibitor cocktail (0.5mM TLCK, 0.1mM PMSF, 25 μg/mL aprotinin, 10 μg/mL leupeptin). The cells were freeze-thawed five times and further ruptured using 10 strokes of a Dounce homogenizer. The described1 centrifugation steps were used to obtain a ribosome-enriched pellet and the pellet was resuspended in a buffer containing 20 mM Hepes pH 7.2, 10mM MgCl2, 500 mM KCl and 5 mM β-me. The suspension was clarified and ribosomes were purified using the described methods1, however 20 mM Hepes pH 7.2 was substituted for Tris in all buffers to reduce ribosomal aggregation.
Electron Microscopy
3 μL of each sample was applied to holey carbon grids (carbon-coated Quantifoil 2/4 grid, Quantifoil Micro Tools GmbH) containing an additional continuous thin layer of carbon33 and glow-discharged. Grids were blotted for 6 sec. at 4°C in 100% humidity and vitrified by rapidly plunging into liquid ethane at −180°C with a Vitrobot (FEI)34–35. Data acquisition was done under low-dose conditions (25 e−/Å2) on a FEI Tecnai F30 Polara (FEI, Eindhoven) operating at 300 kV. The micrographs were taken at a nominal resolution of 59000x on Kodak SO-163 film using the automated data collection system AutoEMation6 and scanned on a Nikon Super Coolscan 9000 ED with a step size of 6.35 μm, resulting in a pixel size of 1.09 Å on the object scale.
Image Processing
The data were processed using SPIDER37. ~700,000 particles windows were automatically extracted from 1,100 film-recorded micrographs using the lfcpick.spi spider batch file8 with a window size of 500 pixels and a pixel size of 1.09 Å. The windows were inspected manually; only particles with high contrast were selected, and those in contact with other particles were eliminated. This conservative manual verification step yielded ~164,000 particles in total. Reconstruction was done following standard SPIDER protocols for reference-based reconstruction38, except that contrast transfer function (CTF) of the reconstructions was corrected by phase-flipping the particles using the defocus value estimated for each micrograph and a single reconstruction was obtained from the entire dataset using conjugate gradients with regularization (BP CG command in SPIDER). The resolution at FSC=0.5 for the final reconstruction was estimated at 5.57 Å (fig. S2C). However, this resolution is an average overall the ribosome and it underestimates most of the ribosome volume that is resolved at ~5.0 Å (see below).
Maximum-likelihood classification39, on a subset of 40,000 particles taken from the full dataset, was attempted using the XMIPP package40. The full T. brucei reconstruction was used as a seed for four classes. The resulting classes were all very similar, as expected based on the 3D variance calculation (see below) and the resolution of the map, probably as a consequence of most of the ribosomes being in the same state. No further classification was attempted.
We attribute the higher resolution obtained in our study to several factors; (i) The CTF correction procedure by Phase-flipping, which has the advantage of correcting the CTF more precisely at the micrograph level. (ii) Performing the 3D reconstructions including all the phase-flipped particles at once, instead of summing different reconstructions issued from different defocus groups. (iii) The global homogeneity of the sample in our study. (iv) The very carful particles selection.
Local Resolution Estimation and Filtering
The local resolution was estimated on the final map using the “blocres” module, part of Bernard's Software Package (bsoft) (URL: http://lsbr.niams.nih.gov/bsoft/)41,42. The 3D map depicting local resolutions showed that the ribosome volume has a resolution of ~5.0 Å (at FSC=0.5) that concerns mostly the core (fig. S2A), and a resolution between 5 and 6 Å at the outer shell, while regions of higher variability have resolutions ranging between 7 and 9 Å. The T. brucei cryo-EM map was filtered according to the estimated local resolution by the following procedure: The map was segmented in accordance with the local resolution using the Segger module43 implemented in Chimera UCSF44. The segments obtained were filtered according to their local resolution and then merged together in one map.
Map Segmentation
A detailed description of the segmentation protocol can be found in the online SI section. A preliminary segmentation of the T. brucei map was performed using Segger43 implemented in Chimera44, and VolRover45, now (also see protocols described in 46–48). These preliminary segments were used during the atomic modeling process. After the atomic model was obtained, we used it to re-segment the T. brucei map more accurately (see online SI).
3D Variance Estimation
The particles windows were binned six times for the 3D variance estimation step in order to reduce the tremendous amount of time and memory required for this operation, resulting in a pixel size of 6.54 Å. The 3D variance map was computed using the bootstrapping method, developed by several groups49–52, in the following way: Forty thousand bootstrap reconstructions were generated, each of which was reconstructed from N particle projections that were randomly sampled with replacement from the total set of ~164,000 particles. In order to enhance the signal-to-noise ratio of the 3D variance, the bootstrap volumes were filtered to about halfway of the first zero-crossing of the contrast transfer functions (CTFs). The structural variance was estimated as the sample variance of the bootstrap volumes minus the variance of the noise, and the difference was then multiplied by N50. In this estimation, the noise variance is assumed to be uniform across the map50. Ideally, the 3D variance map should reflect only regions of high conformational changes, including presence versus absence of a ligand such as a protein or a tRNA. In our case, the low-pass filtration was necessary to reduce the noise variance. However, the filtering also introduces spatial correlation of the noise, and the resulting variance map is distorted to some extent by the noise variance (as verified via simulations).
Ribosomal RNA Modeling
The T. brucei rRNAs were modeled using a workflow combining ab initio and homology approaches and can be found in the online SI section, schematized in fig. S8. The 18S rRNA (SSU rRNA) was modeled based on the T. thermophila 40S crystal structures5. The LSU rRNA was modeled based on the yeast ribosome crystal structure2. The LSU and SSU rRNAs were modeled by homology using the S2S tool21. T. brucei and other kinetoplastid rRNA sequences were fetched from the SILVA webserver (http://www.arbsilva.de/)53 and the comparative RNA web site and project (CRW site) (http://www.rna.icmb.utexas.edu/)54. T. brucei expansion segments (ESs) were modeled mostly ab initio by performing a covariation analysis on each one of these ESs sequences against a number of orthologous rRNA sequences found in other related kinetoplastids, using the mlocarna algorithm55. This allowed deriving a consensus secondary structure that was used in the Assemble tool22 and 3D models were generated for each ES and fitted into the EM map.
Modeling of Ribosomal Proteins
The modeling workflow can be found in the online SI section, schematized in fig. S10. The ribosomal proteins were modeled by homology to mainly three crystal structures, the yeast ribosome2, the Tetrahymena thermophila 40S subunit in complex with eIF15, and the 60S subunit with eIF66. The T. brucei ribosomal protein sequences, used as inputs for the homology modeling, were fetched from the NCBI protein databases (http://www.ncbi.nlm.nih.gov/). The tool we used is Swiss-Model webserver (http://swissmodel.expasy.org/)56–57. We used the Phyre and the Phyre2 webservers58 for the ab initio modeling of the missing fragments and extensions into the density map. We assessed the structure of each model using the MolProbity webserver (http://molprobity.biochem.duke.edu/)59–60.
Molecular Dynamics Flexible Fitting (MDFF)
A detailed description can be found in the online SI section. The initial system was prepared for MDFF61 using VMD62 and ran in NAMD63 for 1.5 ns of simulation time.
Supplementary Material
Acknowledgements
We thank Giovanni Cardone for assistance in the local resolution computation, Melissa Thomas for her assistance with the preparation of figures. We wish to thank Dr. Noreen Williams particularly for an enlightening discussion about the T. brucei LSU rRNA processing. This work was supported by HHMI and NIH R01 GM29169 (to J.F.), ANR project AMIS ARN ANR-09-BLAN-0160 (E.W. and F.J.), as well as NIH R01-EB004873 and R01-GM074258 (to Q.Z. and C.B.). S.N.B. was supported by a CDC EID fellowship program.
Footnotes
Author Contributions Y.H., A.G., S.N.B., F.J., Q.Z., C.B., S.M.A., E.W. and J.F. interpreted the data and wrote the manuscript. S.N.B. purified The T. brucei ribosomes. Y.H., J. Fu and B.G. performed the cryo-EM experiments. H.Y.L. performed the 3D variance estimation. Y.H., A.J. and Q.Z. performed the density map segmentations. Y.H., A.G., J. Fu, A.J. and H.Y.L. performed the cryo-EM data processing. Y.H and F.J. modeled the rRNA. Y.H. and Q.Z. modeled the ribosomal proteins. J.F. conducted research.
The electron microscopy (EM) map has been deposited in the EMBL-European Bioinformatics Institute EM Data Bank under accession codes EMD-2239. Coordinates of EM-based model have been deposited in the RCSB Protein Data Bank under accession numbers 3ZEQ, 3ZEX, 3ZEY and 3Z7F. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Ben-Shem A, et al. Crystal Structure of the Eukaryotic Ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Shem A, et al. The Structure of the Eukaryotic Ribosome at 3.0 Å Resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 3.Armache JP, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5 Å cryo-EM map of the 80S eukaryotic ribosome. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armache JP, et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19748–19753. doi: 10.1073/pnas.1009999107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2010;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 6.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 7.Klinge S, Voigts-Hoffmann F, Leibundgut M, Ban N. Atomic structures of the eukaryotic ribosome. Trends. Biochem. Sci. 2012;37:189–198. doi: 10.1016/j.tibs.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Wilson DN, Doudna Cate JH. The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol. 2012;4:a011536. doi: 10.1101/cshperspect.a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoyama T, Suzuki T. Ribosomal RNAs are tolerant toward genetic insertions: evolutionary origin of the expansion segments. Nucl. Acids Res. 2008;36:3539–3551. doi: 10.1093/nar/gkn224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White TC, Rudenko G, Borst P. Three small RNAs within the 10 kb trypanosome rRNA transcription unit are analogous to domain VII of other eukaryotic 28S rRNA. Nucl. Acids Res. 1986;14:9471–9489. doi: 10.1093/nar/14.23.9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordingly JS, Turner MJ. 6.5 S RNA; preliminary characterization of unusual small RNAs in Trypanosoma brucei. Molecular & Biochemical Parasitology. 1980;1:91–96. doi: 10.1016/0166-6851(80)90003-1. [DOI] [PubMed] [Google Scholar]
- 12.Campbell DA, Kubo K, Graham Clark C, Boothroyd JC. precise identification of cleavage sites involved in the unusual processing of Trypanosome ribosomal RNA. J. Mol. Biol. 1987;196:113–124. doi: 10.1016/0022-2836(87)90514-6. [DOI] [PubMed] [Google Scholar]
- 13.Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 14.Gao H, Juri Ayub M, Levin ML, Frank J. The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc. Natl. Acad. Sci. 2005;102:10206–10211. doi: 10.1073/pnas.0500926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Molecular & Biochemical Parasitology. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011;6:459–474. doi: 10.2217/fmb.11.20. [DOI] [PubMed] [Google Scholar]
- 17.Ciganda M, Williams N. Characterization of a novel association between two trypanosome-specific proteins and 5S rRNA. PLoS One. 2012;7:e30029. doi: 10.1371/journal.pone.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivens AC, et al. The Genome of the Kinetoplastid Parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Q, Bettadapura R, Bajaj C. Macromolecular structure modeling from 3D EM using VolRover 2.0. Biopolymers. 2012;97:709–731. doi: 10.1002/bip.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pintilie G, Zhang J, Goddard T, Chiu W, Gossard D. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. JSB. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jossinet F, Westhof E. Sequence to Structure (S2S): display, manipulate and interconnect RNA data from sequence to structure. Bioinformatics. 2005;21:3320–3321. doi: 10.1093/bioinformatics/bti504. [DOI] [PubMed] [Google Scholar]
- 22.Jossinet F, Ludwig TE, Westhof E. Assemble: an interactive graphical tool to analyze and build RNA architectures at the 2D and 3D levels. Bioinformatics. 2010;26:2057–2059. doi: 10.1093/bioinformatics/btq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trabuco L, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao HY, Frank J. Classification by bootstrapping in single particle methods. IEEE Int. Symp. Biom. Imaging. 2010:169–172. doi: 10.1109/ISBI.2010.5490386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cannone JJ, et al. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayub MJ, Atwood J, Nuccio A, Tarleton R, Levin MJ. Proteomic analysis of the Trypanosoma cruzi ribosomal proteins. Biochem. Biophys. Res. Commun. 2009;382:30. doi: 10.1016/j.bbrc.2009.02.095. [DOI] [PubMed] [Google Scholar]
- 27.Meyuhas O. Physiological roles of ribosomal protein S6: One of its kind. International Review of Cell and Molecular Biology. 2008;268:1–37. doi: 10.1016/S1937-6448(08)00801-0. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava S, Verschoor A, Frank J. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit-subunit interface. J. Mol. Biol. 1992;226:301–304. doi: 10.1016/0022-2836(92)90946-h. [DOI] [PubMed] [Google Scholar]
- 29.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 30.Lescrinier EMHP, et al. Structure of the pyrimidine-rich internal loop in the poliovirus 30'-UTR: The importance of maintaining pseudo-2-fold symmetry in RNA helices containing two adjacent non-canonical base-pairs. J. Mol. Biol. 2003;331:759–769. doi: 10.1016/s0022-2836(03)00787-3. [DOI] [PubMed] [Google Scholar]
- 31.Kaminsky R, Beaudoin E, Cunningham I. Cultivation of the life cycle stages of Trypanosoma brucei sspp. Acta Trop. 1988;45:33–43. [PubMed] [Google Scholar]
- 32.Gómez EB, Medina G, Ballesta JP, Levin MJ, Téllez-Iñón MT. Acidic ribosomal P proteins are phosphorylated in Trypanosoma cruzi. International Journal for Parasitology. 2001;31:1032–1039. doi: 10.1016/s0020-7519(01)00219-3. [DOI] [PubMed] [Google Scholar]
- 33.Grassucci RA, Taylor DJ, Frank J. Preparation of macromolecular complexes for cryo-electron microscopy. Nat Protoc. 2007;2:3239–3246. doi: 10.1038/nprot.2007.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubochet J, et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 35.Wagenknecht T, Frank J, Boublik M, Nurse K, Ofengand J. Direct localization of the tRNA--anticodon interaction site on the Escherichia coli 30 S ribosomal subunit by electron microscopy and computerized image averaging. J. Mol. Biol. 1988;203:753–760. doi: 10.1016/0022-2836(88)90207-0. [DOI] [PubMed] [Google Scholar]
- 36.Lei J, Frank J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 2005;150:69–80. doi: 10.1016/j.jsb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 38.Shaikh T, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nature Protocols. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheres SHW, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo JM. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 40.Scheres SHW, Nuñez-Ramirez R, Sorzano COS, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using Xmipp. Nat. Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heymann JB. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 2001;133:156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- 42.Heymann JB, Cardone G, Winkler DC, Steven AC. Computational resources for cryo-electron tomography in Bsoft. J. Struct. Biol. 2007;161:232–242. doi: 10.1016/j.jsb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pintilie G, Zhang J, Goddard T, Chiu W, Gossard D. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. JSB. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;13:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Baker ML, Yu Z, Chiu W, Bajaj C. Automated segmentation of molecular subunits in electron cryomicroscopy density maps. J. Struct. Biol. 2006;156:432–441. doi: 10.1016/j.jsb.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 46.Yu Z, Bajaj C. Automatic ultrastructure segmentation of reconstructed cryoEM maps of icosahedral viruses. IEEE Transactions on Image Processing: Special Issue on Molecular and Cellular Bioimaging. 2005;14:1324–1337. doi: 10.1109/tip.2005.852770. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Q, Bettadapura R, Bajaj C. Macromolecular structure modeling from 3D EM using VolRover 2.0. Biopolymers. 2012;97:709–731. doi: 10.1002/bip.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu Z, Bajaj C. Computational approaches for automatic structural analysis of large biomolecular complexes. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2008;5:568–582. doi: 10.1109/TCBB.2007.70226. [DOI] [PubMed] [Google Scholar]
- 49.Penczek PA, Yang C, Frank J, Spahn CM. Estimation of variance in single-particle reconstruction using the bootstrap technique. J. Struct. Biol. 2006;154:168–183. doi: 10.1016/j.jsb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W, Kimmel M, Spahn CM, Penczek PA. Heterogeneity of large macromolecular complexes revealed by 3D cryo-EM variance analysis. Structure. 2008;16:1770–1776. doi: 10.1016/j.str.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liao HY, Frank J. Classification by bootstrapping in single particle methods. IEEE Int. Symp. Biom. Imaging. 2010:169–172. doi: 10.1109/ISBI.2010.5490386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simonetti A, et al. Structure of the 30S translation initiation complex. Nature. 2008;455:416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 53.Pruesse E, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nuc. Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cannone JJ, et al. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Will S, Joshi T, Hofacker IL, Stadler PF, Backofen R. LocARNA-P: accurate boundary prediction and improved detection of structural RNAs. RNA. 2012;18:900–914. doi: 10.1261/rna.029041.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 57.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucl. Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 59.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica. 2010;D66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucl. Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trabuco L, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;1:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 63.Phillips JC, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;16:1781. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.