Abstract
Ginseng (Panax ginseng Meyer) has been shown to have anti-aging effects in animal and clinical studies. However, the molecular mechanisms by which ginseng exerts these effects remain unknown. Here, the anti-aging effect of Korean red ginseng (KRG) in rat testes was examined by system biology analysis. KRG water extract prepared in feed pellets was administered orally into 12 month old rats for 4 months, and gene expression in testes was determined by microarray analysis. Microarray analysis identified 33 genes that significantly changed. Compared to the 2 month old young rats, 13 genes (Rps9, Cyp11a1, RT1-A2, LOC365778, Sv2b, RGD1565959, RGD1304748, etc.) were up-regulated and 20 genes (RT1-Db1, Cldn5, Svs5, Degs1, Vdac3, Hbb, LOC684355, Svs5, Tmem97, Orai1, Insl3, LOC497959, etc.) were down-regulated by KRG in the older rats. Ingenuity Pathway Analysis of untreated aged rats versus aged rats treated with KRG showed that the affected most was Cyp11a1, responsible for C21-steroid hormone metabolism, and the top molecular and cellular functions are organ morphology and reproductive system development and function. When genes in young rat were compared with those in the aged rat, sperm capacitation related genes were down-regulated in the old rat. However, when genes in the old rat were compared with those in the old rat treated with KRG, KRG treatment up-regulated C21-steroid hormone metabolism. Taken together, Cyp11a1 expression is decreased in the aged rat, however, it is up-regulated by KRG suggesting that KRG seems enhance testes function via Cyp11a1.
Keywords: Panax ginseng, Korean red ginseng, Aging, Testis, System biology, Cyp11a1
INTRODUCTION
In East Asian countries, ginseng (Panax ginseng Meyer) has been used medicinally, and the major active ingredients are known as ginsenosides [1]. Ginseng is used as an alternative/supplementary medicine and has anti-cancer, anti-inflammatory, and anti-diabetic activities. Korean red ginseng (KRG) is produced by steaming, and then drying fresh ginseng. The anti-cancer and antioxidative activities of KRG seem to be superior to those of white ginseng [1].
A number of anti-aging effects of ginseng on testes have been documented. Ginseng induces spermatogenesis as well as activates glial cell-derived neurotrophic factor [2] and cAMP-responsive element modulator [3] in rat testes. Moreover, ginseng administration into guinea pigs or rats exposed to 2,3,7,8-tetrachlorodibenzo-pdioxin improves the survival rate and sperm quality [4,5]. Studies with ginsenosides also proved that ginsenoside- Rb1 increases secretion of luteinizing hormone by acting directly on rat anterior pituitary gland cells [6]. Further, ginsenoside Rb2 promoted sperm progression and ginsenoside Rc enhanced both sperm motility and progression in vitro [7]. Ginsenoside Re promotes human sperm capacitation and acrosome reactions through the nitric oxide/cGMP pathway [8]. In addition, a clinical trial also proved that P. ginseng extract increased a number of markers including: spermatozoa number/mL, progressive oscillating motility, plasma total and free testosterone, dihydrotestosterone, follicle-stimulating hormone and luteinizing hormone levels [9]. Despite these findings, the mechanism of action ginseng on testes at the molecular level still remains unknown. Therefore, we investigated the anti-aging activity of KRG on rat testes at the molecular level using system biology analysis and selected the Cyp11a1 (cytochrome P450, family 11, subfamily a, polypeptide 1) gene. The roles of Cyp11a1 include cholesterol binding, cholesterol monooxygenase (side-chaincleaving) activity, electron carrier activity, heme binding, and monooxygenase activity. It is noteworthy that this gene is related to many processes such as C21-steroid hormone biosynthsis, Leydig cell differentiation, cellular response to follicle-stimulating hormone and peptide hormone, estrogen biosynthesis, male gonadal development, maternal processes involved in female pregnancy, mating behavior, progesterone biosynthesis, response to estrogen stimulus, response to genistein, response to gonadotropin stimulus, response to peptide hormone stimulus, response to steroid hormone stimulus, steroid biosynthetic process, steroid metabolic process, and testosterone biosynthetic process. In summary, Cyp11a1 has important role of sex hormone control both males and females. Moreover, based on our experiments, the Cyp11a1 gene plays a central role in rejuvenating rat testes due to aging.
MATERIALS AND METHODS
Preparation of water extract from Korean red ginseng
Water extract from 6-year-old KRG was kindly supplied by Korea Ginseng Corporation (Seoul, Korea).
Experimental animals
Eighteen, 12-month-old (750±20 g) and six, 2-monthold (280±10 g) male Sprague-Dawley rats were purchased from Samtako Bio Korea (Osan, Korea) and acclimatized for at least 1 wk prior to the experiments. They were provided with standard pellet diets and water ad libitum as well as kept at constant temperature (23±2℃) and relative humidity (55±10%) while maintained on a 12-/12-hour light/dark cycle. Rats were kept in the Regional Innovation Center Experimental Animal Facility, Konkuk University in accordance with the Institutional Animal Care and Use Committee Guidelines. The study was approved by the Animal Ethics Committee in accordance with the 14th article of Korean Animal Protection Law.
The rats were divided into three groups of six rats each: young rats, old rats, old rats receiving KRG (old+KRG) at a dose of 200 mg/kg, daily for 4 mo. KRG which was received as a water extract, was mixed evenly in sterilized standard diet and then pelleted for consumption. The appropriate content of KRG was controlled by weighting the rats weekly and intake of the food daily. Young and old control groups received feed pellets without extract. At the end of the experiment, all food was removed 24 h prior to sacrifice. The rats were euthanized under general anesthesia with diethyl ether.
RNA isolation and mRNA level determination
Total RNA of rat testes was isolated by using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and purified using RNeasy mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The amount of isolated RNA was determined by UV spectrophotometry at an absorbance of 260 nm. One μg of isolated RNA was converted to cDNA using the M-MLV Reverse Transcriptase kit (RexGene Biotech, Cheongwon, Korea) according to the manufacturer’s instruction. Nucleotide primers used in this study for reverse-transcriptase polymerase chain reaction (RT-PCR) is shown in Table 1. PCR was performed using a thermal cycler (GeneAmp PCR system 2700; Applied Biosystems, Foster City, CA, USA), with β-actin serving as a control.
Table 1.
List of primers and PCR product size used for RT-PCR
| Genes | Primer sequence (5'-3') | Size (b.p.) |
|---|---|---|
|
| ||
| Cyp11a1 | Forward: ggt tca tcg acg ccgtct ac | 648 |
| Reverse: tac cgg aag tgc gtg gtg tt | ||
| RT1-Db1 | Forward: gag tac acg cgc ttc gac ag | 423 |
| Reverse: gct cca cct ggc agg tat aa | ||
| Sv2b | Forward: aac gcg gtg agc atc tca gc | 1,074 |
| Reverse: cca gcc gat cat cgc aga ct | ||
| β-actin | Forward: ttg taa cca act ggg acg ata tgg | 1,312 |
| Reverse: ccg gac tcg tca tac tcc tg | ||
PCR, polymerase chain reaction; RT, reverse-transcriptase; Cyp11a1, cytochrome P450, family 11, subfamily A, polypeptide 1; Sv2b, synaptic vesicle glycoprotein 2B.
DNA microarray and data analysis
RNA samples from 5 rat testes per group were pooled and subjected to transcriptional profiling with oligonucleotide microarrays. RNA samples were processed for hybridization according to the manufacturer’s protocol. One hundred ng of biotin-labeled cRNA samples were hybridized to Rat-Ref-12-v1 Expression Bead Chips containing gene-specific oligonucleotides (Illumina Inc., San Diego, CA, USA) containing over 22,000 transcripts. Hybridization was detected with 1 μg/mL of Cy3-streptavidine (Amersham Biosciences, Piscataway, NJ, USA) and the chips were scanned with an Illumina BeadArray Reader. Subsequent to image scans, quantile normalization of the data was carried out using Genomestudio software (Illumina Inc., San Diego, CA, USA). The genes which showed a ratio of more than the pre-set threshold were considered significantly changed (≥1.5 fold, detection p-value <0.05) and were categorized as up-regulated or down-regulated genes. In order to check the variation between arrays, box plots and principle component analysis were performed using GeneSpring ver. 8.0.0 (Agilent Technologies, Santa Clara, CA, USA).
Ingenuity Pathway Analysis
Data sets of significantly changed genes were analyzed using Ingenuity Pathway Analysis (IPA) ver. 8.5 (Ingenuity systems, Redwood City, CA, USA). IPA is web-based bioinformatics software to identify canonical pathways and bio-functions from omics data. In function/ pathway data, bar length indicates gene expression change associated with function/pathway. Ingenuity Knowledge Base generated from a manually created data base of the scientific literature revealed both direct and indirect relationships among genes, proteins, interactions, activation, inhibition, reaction and expressions. Canonical pathway analysis identified the pathways containing the genes in the user’s omics data. Briefly, it indicates the pathways implicated within the user’s uploaded list. The p-value calculated by Fisher’s exact test was used to determine the probability between the data set and pathway, randomly. The p-value was converted to –Log2 (p-value) format. High values correspond to the pathway significantly associated with the data set. Each gene symbol was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. Networks of these genes were algorithmically generated based on their connectivity and then assigned a score. Graphical network models were generated using literature based pathway links to connect the genes with other genes in the knowledge base.
Statistical analysis
Data were analyzed by analysis of variance followed by Dunett’s t-test for comparisons between groups. Probability values of less than 0.05 were considered significant (*p<0.05, **p<0.01, ***p<0.001). Data are expressed as mean±SD of three to five independent experiments
RESULTS
Modulation of gene expression by KRG in rat testes
To analyze gene expression, 5 RNA samples from rat testes group were pooled and gene expression was analyzed by microarray containing over 22,000 genes. Change of gene expression between any two groups was compared using a statistical criteria and a change ≥2-fold was considered significantly. Microarray analysis identified 33 genes that significantly changed either by age or treatment with KRG (Table 2). Comparison of gene expression of the young and the aged rat showed that expression of SerpinB6a (serine [or cysteine] peptidase inhibitor), and LOC684355 (unknown function) genes were significantly increased in the old rat as compared to the young rat whereas expression of Col1a2 (collagen fibril organization protein) and Cyp11a1 genes were decreased in the old rat.
Table 2.
List of genes modulated by ginseng in rat testis
| Gene | Y | N | B | N/Y | B/Y | B/N |
|---|---|---|---|---|---|---|
|
| ||||||
| Serpinb6a | 431 | 1176 | 1299 | 2.73 | 3.01 | 1.10 |
| RT1-A2 | 169 | 155 | 270 | 0.92 | 1.60 | 1.74* |
| RT1-A1 | 364 | 241 | 202 | 0.66 | 0.55 | 0.84 |
| RGD1564560_predicted | 596 | 937 | 920 | 1.57 | 1.54 | 0.98 |
| Cldn5 | 507 | 453 | 278 | 0.89 | 0.55 | 0.61 |
| LOC497920 | 300 | 488 | 429 | 1.63 | 1.43 | 0.88 |
| Hbb | 956 | 493 | 339 | 0.52 | 0.35 | 0.69 |
| RGD1564153_predicted | 212 | 271 | 371 | 1.28 | 1.75 | 1.37 |
| LOC684355 | 316 | 622 | 470 | 1.97* | 1.49 | 0.76 |
| Col1a2 | 854 | 413 | 363 | 0.48 | 0.43 | 0.88 |
| Hba-a2 | 671 | 475 | 415 | 0.71 | 0.62 | 0.87 |
| RGD1304748 | 172 | 183 | 282 | 1.06* | 1.64 | 1.54 |
| LOC501211 | 229 | 354 | 437 | 1.55 | 1.91 | 1.23 |
| Orai1 | 1223 | 1321 | 892 | 1.08 | 0.73 | 0.68 |
| LOC497959 | 3068 | 1843 | 1270 | 0.60 | 0.41 | 0.69 |
| Tmem97 | 5970 | 3878 | 2682 | 0.65 | 0.45 | 0.69 |
| Svs5 | 983 | 550 | 336 | 0.56 | 0.34 | 0.61 |
| Insl3 | 3614 | 2630 | 1826 | 0.73 | 0.51 | 0.69 |
| RGD1565959_predicted | 324 | 399 | 657 | 1.23 | 2.03 | 1.65 |
| Sv2b | 265 | 174 | 334 | 0.65 | 1.26 | 1.92* |
| RGD1307595_predicted | 1800 | 2586 | 2434 | 1.44 | 1.35 | 0.94 |
| Chrnd | 418 | 685 | 555 | 1.64 | 1.33 | 0.81 |
| Vdac3 | 2987 | 2788 | 1846 | 0.93 | 0.62 | 0.66 |
| Egln2 | 963 | 916 | 1359 | 0.95 | 1.41 | 1.48 |
| Hsd11b1 | 1408 | 914 | 737 | 0.65 | 0.52 | 0.81 |
| Rpl10a | 1196 | 1867 | 1702 | 1.56 | 1.42 | 0.91 |
| Syf2 | 560 | 533 | 592 | 0.95 | 1.06 | 1.11 |
| RICS_predicted | 213 | 259 | 328 | 1.22 | 1.54 | 1.27 |
| Cyp11a1 | 714 | 296 | 485 | 0.41* | 0.68 | 1.64* |
| RT1-Db1 | 705 | 1046 | 340 | 1.48 | 0.48 | 0.32* |
| Degs1 | 3025 | 2442 | 1722 | 0.81 | 0.57 | 0.71 |
| LOC365778 | 460 | 244 | 394 | 0.53 | 0.86 | 1.62 |
| Rps9 | 474 | 309 | 657 | 0.65 | 1.39 | 2.12* |
Differential gene expression profiles of 33 genes in rat testis after Korean red ginseng (KRG) treatment.
Significantly changed genes were marked by asterisk and colored boxes.
Y, young rat testis; N, normal old rat; B, KRG administered old rat.
Comparison of genes expression between the aged rat treated with KRG and the young rat (no treatment) showed that SerpinB6a, RGD1565959 (unknown function), and LOC501211 (unknown function) were significantly increased >2 fold in the old rat treated with KRG. However, expression of Hbb (beta globin gene), Col1a2, LOC497959 (unknown function), Tmem97 (transmembrane protein of unknown function), Svs5 (seminal vesicle secretory protein 5), Insl5 (Insulin-like peptide 5), and RT1-Db1 (unknown function) were significantly decreased in testes of the old rat treated with KRG as compared to the young, untreated rat.
When genes expression in the aged rat treated with KRG was compared with those in the untreated aged rat group, expression of RT1-A2 (unknown function), RGD1565959 (unknown function), Sv2b (synaptic vesicle glycoprotein 2B), Cyp11a1, LOC365778 (unknown function), and Rps9 (ribosomal protein S9) genes was increased more than 1.6-fold in the old rat treated with KRG. In contrast, only RT1-Db1 expression in the old rat treated with KRG was decreased significantly to 32% of the untreated aged rat group, which was the most significant decrease in all 3 groups. These results suggest that not many genes are modulated more than 2-fold by KRG administration, and the highest gene expression modulation in all 3 groups was the increase of SerpinB6a in the old rat which was increased 2.7-fold over the young rat group.
Consistency of RT-PCR result with microarray data
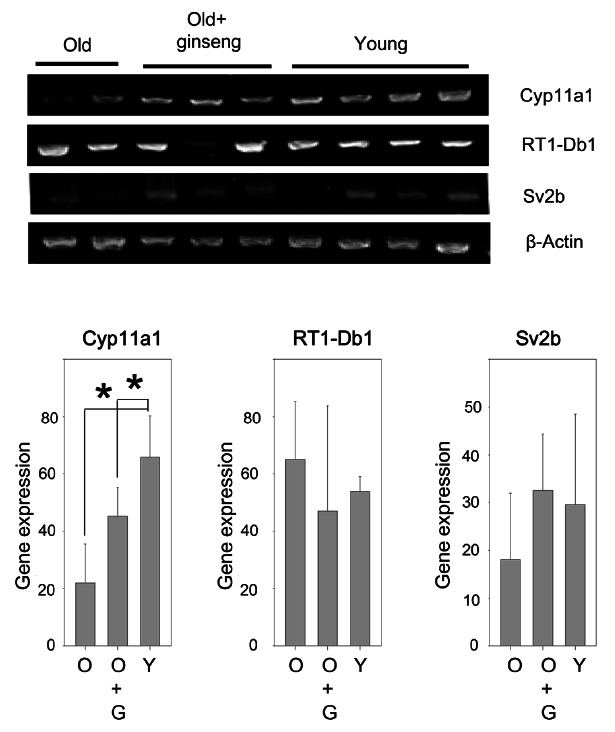
To confirm the microarray data, RT-PCR was performed. Expression of Cyp11a1 gene in the old rat was decreased 33% compared to the young rats. However, KRG treatment increased expression of Cyp11a1 to 2.06- fold of that in the aged rat group (Fig. 1). Moreover, KRG treatment also increased Sv2b gene expression to 1.8-fold of the aged rat group (Fig. 1). These demonstrate a close correlation between microarray data and RT-PCR results.
Fig. 1. Korean red ginseng (KRG) increases Cyp11a1 expression in the old rat testes. Total RNA was isolated from testes of young rats (2 months old), old rats (16 months old) and old rats administered KRG for 4 months, then 5 μg of isolated RNA was converted to cDNA using a reverse transcriptase. Quantification of mRNA levels were carried out by reverse-transcriptase polymerase chain reaction using β-actin as a control. Each lane represents an RNA sample isolated from a single rat testes. O, old rat; O+G, old rat administered KRG; Y, young rat. *p<0.05.
Four classes of gene modulation in rat testis by KRG treatment
The gene expression pattern can be largely classified into 4 groups. 1) Genes decreased in the aged rat but increased by KRG treatment; expression of Cyp11a1, Sv2b, RT1-A2, Syf2, LOC365778, and Rps9 was decreased in the old rat compared to the young rat, but increased by KRG treatment. 2) Genes increased in the old rat but decreased by KRG: expression of RGD1565959, LOC497920, LOC684355, Orai1, RGD1307595, Chrnd, Rpl10a, and RT1-Db1 was increased in the aged rat compared to the young rat, but decreased by KRG administration. 3) Genes decreased in the aged rat and further decreased by KRG treatment: this category included Cldn5 (claudin 5), RT1-A1, Hbb, Col1a2, Hbaa2, LOC497959, Svs5, Tmem97 (transmembrane protein of unknown function), Insl3, Vdac3 (voltage-dependent anion channel 3), Hsd11b1 and Degs1. 4) Genes increased in the aged rat and increased further by KRG: SerpinB6A, RGD1564153, RGD1304748, LOC501211, RGD1565959, and RICS.
Comparative analysis of networks
To check which pathway was affected by aging and by KRG treatment, the expression levels of each gene were analyzed by Ingenuity Pathway Analysis. Interestingly, when genes were compared between 1) old rat vs. young rat, 2) young rat vs. old rat treated with KRG, and 3) old rat vs. old rat+KRG treated, the highest score, 29, was found in the group that compared the old versus young rat. The second high score, 28, was found in the group comparing the old rat versus the old rat treated with KRG group.
Network 1. Down-regulation of sperm capacitation related genes by aging (young vs. old group)
When pathway and network analysis was performed between the young and old rat, it revealed a number of interactions in the old rat group with a score of 29 (Figs. 2 and 3). Interestingly, Hbb (hemoglobin, beta) and NFκB were found at the center of this network and they are comprised functional genes involved in cancer, gastrointestinal disease, reproductive system development and function, developmental disorders, lipid metabolism, and small molecule biochemistry (Fig. 2).
Fig. 2. Pathway analyses of genes expression between the old rat versus the young rat. (A) Functions modulated significantly by aging. (B) Pathways modulated significantly by aging. Ingenuity Pathway Analysis of genes between the old rat and young rat was performed.
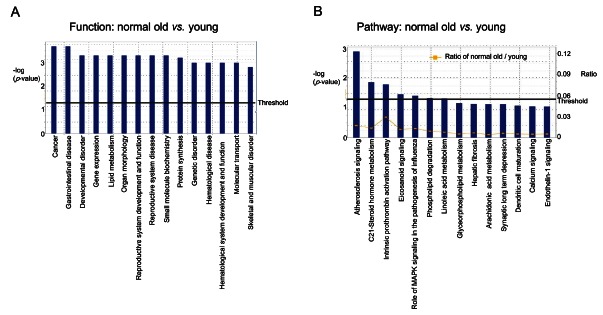
Fig. 3. Down-regulation of sperm capacitation- related genes by aging (young vs. old). Ingenuity Pathway Analysis of genes between the old rat and young rat showed 10 focus molecules (score 29), which function in processes including molecular transport, lipid metabolism, and small molecule biochemistry). The intensity of the red colors indicates the degree of up regulation (Chrnd, Rpl10a, and Serpinb6a) in the old rat compared to the young rat. Uncolored genes were not identified as differentially expressed in our experiment and were integrated into the computationally generated networks on the basis of the evidence stored in the Ingenuity Pathway Analysis knowledge memory. The green color denotes decreased gene expression (Hbb, MBA1, Col1a2, PLA2, PROCA1, Tmem97, Cyp11a1, and Svs5).
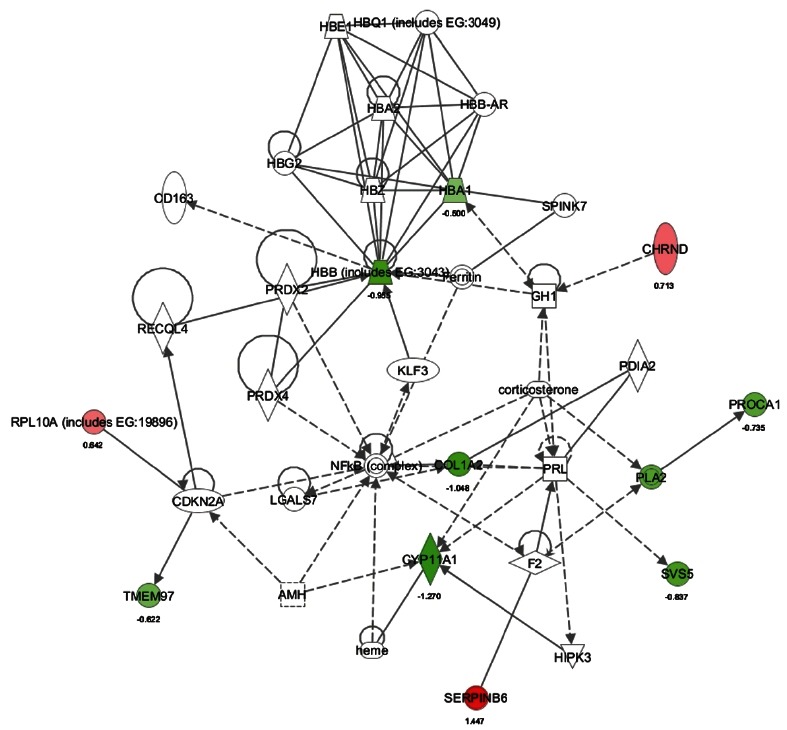
Three genes including Chrnd (cholinergic receptor, nicotinic, delta), Rpl10a (ribosomal protein L10a), and SerpinB6 were up-regulated (Fig. 3). 6 genes including Hba1 (hemoglobin, alpha 1), Hbb, Col1a2 (collagen, type I, alpha 2), Cyp11a1, Tmem97 (transmembrane protein 97), PLA2 (phospholipase A2), PROCA1 (protein interacting with cyclin A1), and Svs5 (seminal vesicle secretory protein 5), were down-regulated (Fig. 3). These down-regulated genes are involved in seminal vesicle secretion (Svs), cholesterol/lipid homeostasis, and sperm capacitation which is the process responsible for the timing of changes in sperm activity and behavior, following ejaculation. In addition, aging induces SerpinB6 which could be involved in gonad development, gametogenesis, and/or fertilization [10].
Network 2. Down-regulation of dendritic cell maturation and immune system in the old-KRG treated rat (young vs. old+KRG group)
When genes expression of old rats treated with KRG was compared with those of the young rats, 2 pathway and networks were revealed (Figs. 4 and 5).
Fig. 4. Pathway analysis of gene expression between the old rat treated with Korean red ginseng (KRG) versus the young rat. (A) Functions modulated significantly between the old rat+KRG versus the young rat. (B) Pathways modulated significantly between the old rat+KRG versus the young rat. Ingenuity Pathway Analysis of genes between the old rat+KRG versus the young rat was performed.
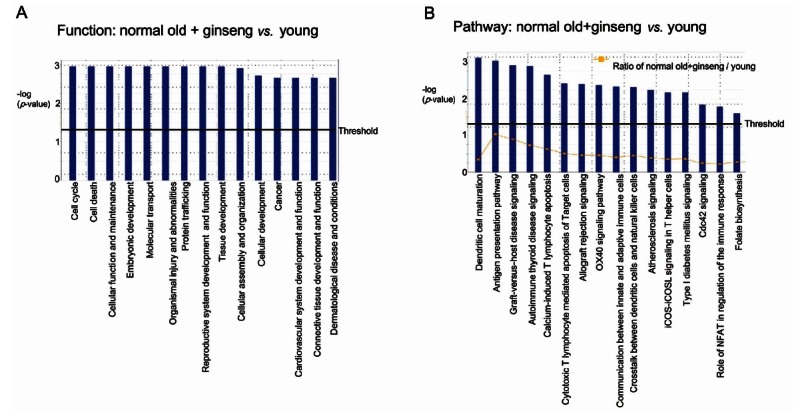
Fig. 5. Down-regulation of dendritic cell maturation and immune system in the old rat treated with Korean red ginseng (KRG, young vs. old rat+KRG groups). Ingenuity Pathway Analysis of genes between the old rat+KRG and the young rat showed 2 networks. (A) The network with a score of 28 has 11 focus molecules, which had functions in developmental disorders, genetic disorders, and the reproductive system. Expression of four genes was increased (Rpl10a, Sv2b, Serpinb6a, and DDX6), and expression of 8 genes was decreased (Cldn5, Hbb, Tmem97, HLADRB1, Degs1, HLAC, Insl3, and HLA-ABC) in the old rat+KRG group. The numbers under the gene name show change of gene expression in log2 basis. Red color indicates increase of gene expression whereas green color decrease of gene expression. (B) The network with a score of 22 has 9 focus molecules, whose functions are involved in cell morphology, lipid metabolism, and molecular transport. Expression of three genes was increased (Syf2, PLEKHA1, and ARHGAP32), and expression of 8 genes was decreased (Col1a2, Hsd11b1, Insl3, Orai1, PROCA1, collagen, PLA2, and Svs5) in the old rat+KRG group.
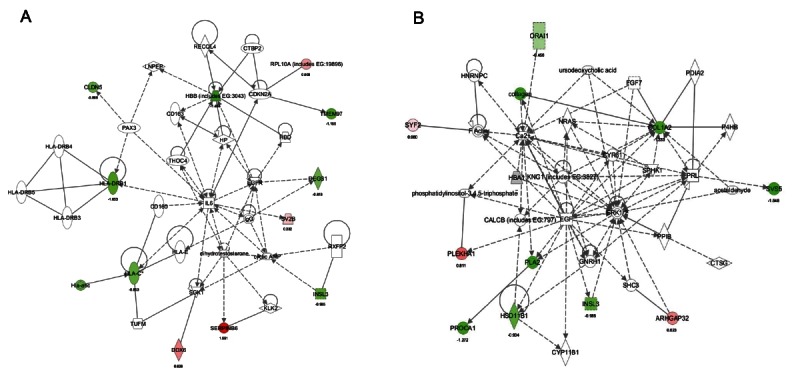
One network (score 28) focused on reproductive system disease, developmental disorders, and genetic disorders (Fig. 4). KRG administration up-regulated 4 genes (SerpinB6, Rpl10a [ribosomal protein L10a], Sv2b [synaptic vesicle glycoprotein 2B], and DDX6 [DEAD (Asp-Glu-Ala-Asp) box polypeptide 6]) than those of the young group as well as down-regulated 8 genes (Cldn5 [claudin 5], Hbb, Tmem97, Hla-drb1 [major histocompatibility complex, class II, DR beta 1], Degs1 [degenerative spermatocyte homolog 1, lipid desaturase], Hla-abc, Hla-c [human leukocyte antigen system], and Insl3) (Fig. 5A).
Another network (score 22) focused on lipid metabolism, cell morphology, and molecular transport (Fig. 4). Compared to young rats, KRG treatment in old rats demonstrated up-regulation of PLEKHA1 (pleckstrin homology domain containing, family A [phosphoinositide binding specific] member 1), ARHGAP32 (Rho GTPase activating protein 32), and Syf2 (Syf2 homolog, RNA splicing factor), in addition to down-regulation of 8 genes comprised of Orai1, collagen, Col1a2 (collagen, type I, alpha 2), Svs5, PLA2, Insl3, PROCA1, and Hsd11b1 (Fig. 5B).
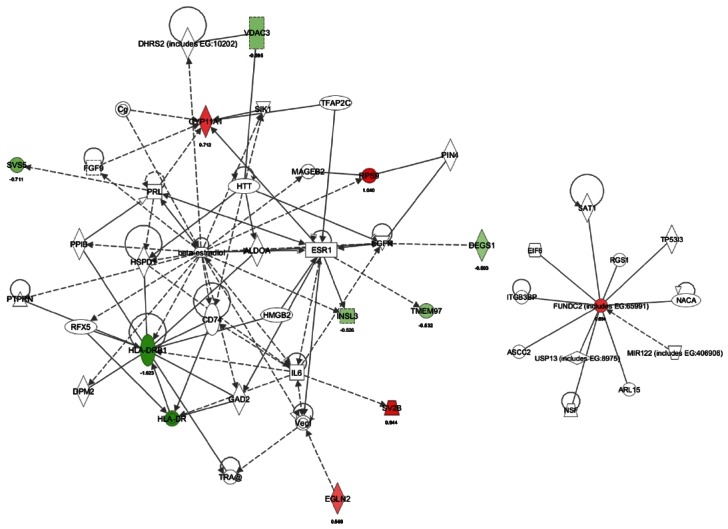
Network 3. Up-regulation of C21-steroid hormone metabolism by KRG treatment (old vs. old+KRG group)
When genes expression of the aged rat was compared with those of the aged rat treated with KRG, KRG treatment up-regulated 4 genes including Rps9 (ribosomal protein S9), Sv2b, Cyp11a1, and Egln2 (egl nine homolog 2). These genes are involved in organ and cellular morphology, reproductive system disease and reproductive system development/function (Figs. 6 and 7). In contrast, the top seven genes down-regulated by KRG were Hla-drb1 (major histocompatibility complex, class II, DR beta 1), Hla-dr, Svs5, Insl3 (insulin-like 3 [Leydig cell]), Tmem97, Degs1 (degenerative spermatocyte homolog 1, lipid desaturase), and Vdac3 (voltage-dependent anion channel 3) (Figs. 6 and 7). Another pathway (score 2) focused on up-regulation of FUNDC2 (Fig. 7), whose function is not currently known.
Fig. 6. Pathway analysis of gene expression between the old rat versus the old rat+Korean red ginseng (KRG). (A) Functions modulated significantly between the old rat+KRG group versus the old rat. (B) Pathways modulated significantly between the old rat+KRG group versus the old rat. Ingenuity Pathway Analysis of genes between the old rat versus the old rat+KRG group was performed.
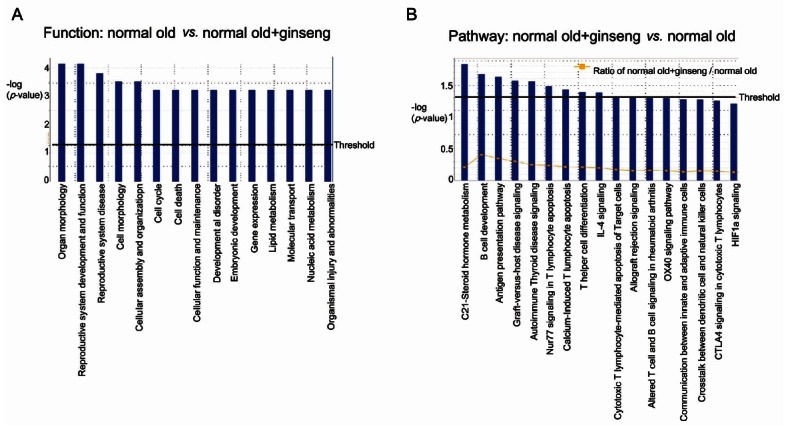
Fig. 7. Up-regulation of C21-steroid hormone metabolism by Korean red ginseng (KRG) treatment (old rat vs. old rat+KRG groups). Ingenuity Pathway Analysis of genes between the old rat+KRG group and the old rat showed 2 networks. (A) The network with a score of 28 has 10 focus molecules, which includes functions in small molecule biochemistry, developmental disorders, and lipid metabolism. Expression of four genes was increased (Cyp11a1, Rps9, Sv2b, and Egln2), and Expression of 7 genes was decreased (Vdac3, Svs5, Degs1, Insl3, Tmem97, HLA-DRB1, and HLA-DR) in the old rat+KRG group. (B) The network with a score of 2 has 1 focus molecule, which has functions in cellular assembly and organization, cellular function and maintenance, and dermatological diseases. Gene expression was increased (FUNDC2) in the old rat+KRG group.
DISCUSSION
Although several lines of evidence strongly suggest that administration of ginseng alleviates and/or reverses aging in testes, the underlying mechanisms of ginseng in testes has not been demonstrated. Here we demonstrated for the first time the whole array of gene expression modulated by ginseng in rat testis.
Steroids are produced throughout life in the interstitial Leydig cells of the testes as well as in the fasciculata, zonae glomerulosa, and reticularis of the adrenal cortex. Mature Leydig cells maintain high levels of the cholesterol side-chain cleavage enzyme, cytochrome P450scc (encoded by Cyp11a1) and 3β-hydroxysteroid dehydrogenase/δ5-δ4 isomerase (encoded by Hsd3b). In response to the pituitary gonadotropin, luteinizing hormone (LH), these rapidly synthesize steroidogenic acute regulatory protein (STARD1) and testosterone. The adult adrenal cortex also expresses high levels of Cyp11a1, Hsd3b, and STARD1 [11]. Therefore, the first steps in steroidogenesis are regulated by these 3 genes, STARD1, Cyp11a1, and Hsd3b in the adrenal, testis, ovary and placenta [11].
Cyp11a1 expression is primarily regulated by gonadotropins in adult gonads. Gonadotropins and cAMP stimulate Cyp11a1 promoter activity in granulosa, luteal, theca, and Leydig cells. In Leydig cells, basal levels of Cyp11a1 are high and increases more slowly than STARD1 in response to cAMP, LH or hCG. Cyp11a1 mRNA is also induced by thyroid hormone and TGFβ in immature rat Leydig cells, but are repressed by glucocorticoids, interferon c, leptin, TGFβ, TNFα, and IL-1 which may be found in serum and be locally produced by Leydig cells or immune cells [11].
In the preliminary study, we found that aged rats testes were shrunk compared to young rats testes but KRG administered rats testes was healthy (data not shown). Based on this observation, we started these experiments. Through microarray analysis, we selected 33 genes significantly changed, and expression of highly significant genes (Cyp11a1, RT1-Db1, Sv2b) was further confirmed by RT-PCR. In this study, Cyp11a1 was found to be decreased in testes of the aged rat, however, KRG administration reversed this decrease (Fig.1). Moreover, KRG administration up-regulates C21-steroid hormone metabolism, of which Cyp11a1 is a pivotal enzyme. Since Cyp11a1 catalyzes the conversion of cholesterol to pregnenolone in mitochondria, the first step of steroid biosynthesis [12], up-regulation of Cyp11a1 by KRG seems to be a critical step pathway. Once cholesterol is converted to pregnenolone, which is subsequently metabolized to progesterone by 3β-hydroxysteroid dehydrogenase/δ5- δ4 isomerases (encoded by HSD3B genes), other steroid hormones can be formed [11]. C21-steroid hormone metabolism significantly changed in both old rats testes versus young rats testes (Fig. 2B) and aged rats treated with KRG versus non-treated aged rats testes (Fig. 6B). In addition, in case of aged rats treated with KRG versus non-treated aged rats testes, β-estradiol was placed in the center of network analysis (Fig. 7). Therefore, KRG has anti aging effect on testes by regulating Cyp11a1 related to C21-steroid hormone metabolism.
Network analysis on young vs. old+KRG group showed that up-regulation of SerpinB6, Rpl10a, Sv2b, DDX6, PLEKHA1, ARHGAP32, and Syf2 as well as down-regulation of Cldn5, Hbb, Tmem97, Hla-drb1, Degs1, Hla-abc, Hla-c, and Insl3. Since these genes function in testes has not been clarified yet, more study would warrant how KRG could effect on these molecules in testes.
When mRNA levels in the old rat were compared with those in the young rat, SerpinB6A was the most elevated gene in testes the old rat. Serpin is an important endogenous regulator of proteolysis [13]. Keratinocytes produce a number of different serpins, including serpinA5 (protein C inhibitor), serpinE1 (plasminogen activator inhibitor-1/ PAI-1), serpinB3 (squamous cell carcinoma antigen-1/ SCCA1), and serpinB6 (protease inhibitor 6/PI-6) [14-16]. Although serpins are present in keratinocytes, the nucleocytoplasmic serpinB6 is also expressed in most epithelial cells, endothelial cells, mast cells, monocytes, neutrophils, and platelets [17-20]. Thus serpin dysfunction could lead to uncontrolled proteolysis. However, the physiological functions and proteases regulated by these serpins in keratinocytes are unknown and further studies are required.
Acknowledgments
This work was supported by the 2009 grant from the Korean Society of Ginseng funded by Korea Ginseng Corporation (DKR and SKK), and by the Basic Science Research Program through the NRF of Korea funded by the Ministry of Education, Science, and Technology (2010-0016256 to SC).
References
- 1.Nam KY. The comparative understanding between red ginsengs and white ginsengs, processed ginsengs (Panax ginseng C. A. Meyer). J Ginseng Res. 2005;29:1–18. doi: 10.5142/JGR.2005.29.1.001. [DOI] [Google Scholar]
- 2.Yang WM, Park SY, Kim HM, Park EH, Park SK, Chang MS. Effects of Panax ginseng on glial cell-derived neurotrophic factor (GDNF) expression and spermatogenesis in rats. Phytother Res. 2011;25:308–311. doi: 10.1002/ptr.3239. [DOI] [PubMed] [Google Scholar]
- 3.Park WS, Shin DY, Kim DR, Yang WM,, Chang MS, Park SK. Korean ginseng induces spermatogenesis in rats through the activation of cAMP-responsive element modulator (CREM). Fertil Steril. 2007;88:1000–1002. doi: 10.1016/j.fertnstert.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Park JS, Hwang SY, Lee WS, Yu KW, Paek KY, Hwang BY, Han K. The therapeutic effect of tissue cultured root of wild Panax ginseng C. A. Meyer on spermatogenetic disorder. Arch Pharm Res. 2006;29:800–807. doi: 10.1007/BF02974082. [DOI] [PubMed] [Google Scholar]
- 5.Hwang SY, Kim WJ, Wee JJ, Choi JS, Kim SK. Panax ginseng improves survival and sperm quality in guinea pigs exposed to 2,3,7,8-tetrachlorodibenzo- p-dioxin. BJU Int. 2004;94:663–668. doi: 10.1111/j.1464-410X.2004.05019.x. [DOI] [PubMed] [Google Scholar]
- 6.Tsai SC, Chiao YC, Lu CC, Wang PS. Stimulation of the secretion of luteinizing hormone by ginsenoside-Rb1 in male rats. Chin J Physiol. 2003;46:1–7. [PubMed] [Google Scholar]
- 7.Chen JC, Chen LD, Tsauer W, Tsai CC, Chen BC, Chen YJ. Effects of ginsenoside Rb2 and Rc on inferior human sperm motility in vitro. Am J Chin Med. 2001;29:155–160. doi: 10.1142/S0192415X01000174. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Zhou Q, Li X, Zhao W, Wang Y, Liu H, Li N. Ginsenoside Re promotes human sperm capacitation through nitric oxide-dependent pathway. Mol Reprod Dev. 2007;74:497–501. doi: 10.1002/mrd.20583. [DOI] [PubMed] [Google Scholar]
- 9.Salvati G, Genovesi G, Marcellini L, Paolini P, De Nuccio I, Pepe M, Re M. Effects of Panax ginseng C.A. Meyer saponins on male fertility. Panminerva Med. 1996;38:249–254. [PubMed] [Google Scholar]
- 10.Charron Y, Madani R, Nef S, Combepine C, Govin J, Khochbin S, Vassalli JD. Expression of serpinb6 serpins in germ and somatic cells of mouse gonads. Mol Reprod Dev. 2006;73:9–19. doi: 10.1002/mrd.20385. [DOI] [PubMed] [Google Scholar]
- 11.Lavoie HA, King SR. Transcriptional regulation of steroidogenic genes: STARD1, CYP11A1 and HSD3B. Exp Biol Med (Maywood) 2009;234:880–907. doi: 10.3181/0903-MR-97. [DOI] [PubMed] [Google Scholar]
- 12.Guo IC, Shih MC, Lan HC, Hsu NC, Hu MC, Chung BC. Transcriptional regulation of human CYP11A1 in gonads and adrenals. J Biomed Sci. 2007;14:509–515. doi: 10.1007/s11373-007-9177-z. [DOI] [PubMed] [Google Scholar]
- 13.Scott FL, Sun J, Whisstock JC, Kato K, Bird PI. SerpinB6 is an inhibitor of kallikrein-8 in keratinocytes. J Biochem. 2007;142:435–442. doi: 10.1093/jb/mvm156. [DOI] [PubMed] [Google Scholar]
- 14.Scott FL, Paddle-Ledinek JE, Cerruti L, Coughlin PB, Salem HH, Bird PI. Proteinase inhibitor 6 (PI-6) expression in human skin: induction of PI-6 and a PI-6/proteinase complex during keratinocyte differentiation. Exp Cell Res. 1998;245:263–271. doi: 10.1006/excr.1998.4241. [DOI] [PubMed] [Google Scholar]
- 15.Abts HF, Welss T, Mirmohammadsadegh A, Kohrer K, Michel G, Ruzicka T. Cloning and characterization of hurpin (protease inhibitor 13): a new skin-specific, UV-repressible serine proteinase inhibitor of the ovalbumin serpin family. J Mol Biol. 1999;293:29–39. doi: 10.1006/jmbi.1999.3159. [DOI] [PubMed] [Google Scholar]
- 16.Jensen PJ, Wu Q, Janowitz P, Ando Y, Schechter NM. Plasminogen activator inhibitor type 2: an intracellular keratinocyte differentiation product that is incorporated into the cornified envelope. Exp Cell Res. 1995;217:65–71. doi: 10.1006/excr.1995.1064. [DOI] [PubMed] [Google Scholar]
- 17.Bird CH, Blink EJ, Hirst CE, Buzza MS, Steele PM, Sun J, Jans DA, Bird PI. Nucleocytoplasmic distribution of the ovalbumin serpin PI-9 requires a nonconventional nuclear import pathway and the export factor Crm1. Mol Cell Biol. 2001;21:5396–5407. doi: 10.1128/MCB.21.16.5396-5407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coughlin P, Sun J, Cerruti L, Salem HH, Bird P. Cloning and molecular characterization of a human intracellular serine proteinase inhibitor. Proc Natl Acad Sci U S A. 1993;90:9417–9421. doi: 10.1073/pnas.90.20.9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott FL, Coughlin PB, Bird C, Cerruti L, Hayman JA, Bird P. Proteinase inhibitor 6 cannot be secreted, which suggests it is a new type of cellular serpin. J Biol Chem. 1996;271:1605–1612. doi: 10.1074/jbc.271.3.1605. [DOI] [PubMed] [Google Scholar]
- 20.Strik MC, Wolbink A, Wouters D, Bladergroen BA, Verlaan AR, van Houdt IS, Hijlkema S, Hack CE, Kummer JA. Intracellular serpin SERPINB6 (PI6) is abundantly expressed by human mast cells and forms complexes with beta-tryptase monomers. Blood. 2004;103:2710–2717. doi: 10.1182/blood-2003-08-2981. [DOI] [PubMed] [Google Scholar]


