Abstract
In order to develop a novel system for the discrimination of five ginseng cultivars (Panax ginseng Meyer), single nucleotide polymorphism (SNP) genotyping assays with real-time polymerase chain reaction were conducted. Nucleotide substitution in gDNA library clones of P. ginseng cv. Yunpoong was targeted for the SNP genotyping assay. From these SNP sites, a set of modified SNP specific fluorescence probes (PGP74, PGP110, and PGP130) and novel primer sets have been developed to distinguish among five ginseng cultivars. The combination of the SNP type of the five cultivars, Chungpoong, Yunpoong, Gopoong, Kumpoong, and Sunpoong, was identified as ‘ATA’, ‘GCC’, ‘GTA’, ‘GCA’, and ‘ACC’, respectively. This study represents the first report of the identification of ginseng cultivars by fluorescence probes. An SNP genotyping assay using fluorescence probes could prove useful for the identification of ginseng cultivars and ginseng seed management systems and guarantee the purity of ginseng seed.
Keywords: Panax ginseng, Nucleotide substitution, Fluorescence probes, Single nucleotide polymorphism genotyping
INTRODUCTION
Ginseng (Panax ginseng Meyer), which belongs to the genus Panax in the family Araliaceae, is a herbaceous perennial plant native to Korea and China; it has been used for over 2,000 years as a medicine in the Oriental countries. The major biologically active components of ginseng include ginsenosides, polysaccharides, peptides, polyacetylenic alcohols, phenolic compounds, and fatty acids [1]. P. ginseng evidences a variety of beneficial biological actions, including anti-stress [2], anti-carcinogenic [3,4], anti-hyperlipidemi [5], anti-aging, anti-amnestic [6], anti-diabetic [7], cardiovascular protection, and neuroprotection activities [8]. For these reasons, P. ginseng has become one of the most sought-after herbal remedies in Korea. The ginseng (P. ginseng) has three variants in Korea: the Jakyung, Chungkyung, and Hwangsook varieties.
Until now, a total of nine cultivars have been bred via pure line selection [9] and were registered in Korea Seed & Variety Service (Anyang, Korea). Among these nine cultivars, the Yunpoong [10], Gopoong [11], Sunpoong, Sunwon, Sunwoon, and Sunhyang cultivars were selected from Jakyung, whereas the Chunpoong, Chungsun and Kumpoong cultivars were selected from the Chungkyung and Hwangsook variants, respectively. Recently, ginseng cultivars are frequently mixed-cultivated on local farms, and the seeds of ginseng cultivars are sold in the market by home seed producers. These illegal practices have caused serious social problems, including breeders’ intellectual property rights infringement, deteriorations in ginseng product quality and monetary damage to farmers caused by the use of mixed seeds.
Therefore, an effective management system should be established to maintain the high quality of ginseng cultivars. DNA marker techniques have been successfully applied for the classification of families and the estimation of genetic divergence among and between tested families. DNA-based markers are less profoundly affected by age, the physiological condition of samples, and environmental factors [12]. Thus, several researchers have developed molecular markers for the authentication of ginseng cultivars, including inter-simple sequence repeat markers [13], randomly amplified polymorphic DNA markers [14], restriction fragment length polymorphism markers [15], simple sequence repeat markers [16-18], single nucleotide polymorphism (SNP) markers [19,20], sequence characterized amplified region markers [21], and expressed sequence tag polymerase chain reaction (PCR) markers [22].
However, these markers failed to develop a robust molecular marker for the authentication of ginseng cultivars, owing to low levels of polymorphism and reproducibility. Additionally, several markers have proven effective only on a limited range of cultivars. In an effort to overcome these limitations, we developed sequence-tagged site (STS) markers containing a cleaved amplified polymorphic sequence (CAPS) system for the authentication of ginseng cultivars at the DNA level [23].
However, STS markers containing CAPS systems require complicated steps and electrophoresis and restriction fragment length polymorphism analysis after PCR amplifications are quite time-intensive.
These disadvantages can be complemented using a SNP genotyping assay with real-time PCR. Real-time PCR has advantages over competitive PCR, in that it is faster, more sensitive, and more robust against contamination due to minimal sample manipulation in closed-tube assays [24]. SNP genotyping assays are used routinely for the quantification of genetically modified organisms in food [25]. Moreover, a recently conducted genotyping assay by real-time PCR has also been employed for the identification of species such as beef [26], peanut [27], and fish [28-30].
The aim of the present study was to develop a novel method using fluorescence probes with a real-time PCR machine for the rapid identification of major ginseng cultivars, in order to protect intellectual property rights, to establish ginseng seed management systems and to guarantee the purity of ginseng seed.
MATERIALS AND METHODS
Plant materials
Fresh leaves of 3-years-old plants from five ginseng cultivars were cut and quickly frozen in a deep freezer prior to use. The five ginseng cultivars were used as follows; Chunpoong, Yunpoong, Gopoong, Kumpoong, and Sunpoong Sunpoong. These plant materials were preserved and cultivated at an experimental field of the National Institute of Horticultural and Herbal Science (NIHHS) of the Rural Development Administration, Chungbuk Province, Korea, and voucher samples were deposited at the Korean medicinal herbarium at NIHHS (Table 1).
Table 1.
Ginseng cultivars used in this study
| No. | Cultivars | Voucher no. | Collection area |
|---|---|---|---|
|
| |||
| 1 | Panax ginseng cv. Chunpoong | MPS002375 | Eumsung, Chungbuk, Korea |
| 2 | P. ginseng cv. Yunpoong | MPS002380 | Eumsung, Chungbuk, Korea |
| 3 | P. ginseng cv. Gopoong | MPS002385 | Eumsung, Chungbuk, Korea |
| 4 | P. ginseng cv. Kumpoong | MPS002390 | Eumsung, Chungbuk, Korea |
| 5 | P. ginseng cv. Sunpoong | MPS002395 | Eumsung, Chungbuk, Korea |
DNA extraction
The samples were frozen in liquid nitrogen and immediately ground to a fine powder in a 1.5 mL micro-centrifuge tube with a micro-pestle. Total genomic DNAs were extracted from fresh leaves of the five cultivars of ginseng (3-year-old) using Dneasy Plant Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol and DNA was eluted in 100 μL elution solution.
Polymerase chain reaction analysis by sequence-tagged site markers
STS-primer sets (UFGp74, MFGp110A, and MF-Gp130A) used in this study were developed from the gDNA library clones of P. ginseng cv. Yunpoong [23]. By using the methylation filtering technique, a genomic library was constructed, in which clone inserts were derived from the hypomethylated regions of ginseng genome. We collected more than 3000 white colonies from the methylation filtered library. Colony PCR was carried out on randomly selected 1099 from the methylation filtered library, to select clones between 0.8 and 1.5 kb as their estimated inserts size, and those clones were then subjected for sequence analysis. STS primer sets UFGp74, MFGp110A and MFGp130A were designed using Primer3 based on sequence information from gDNA library clones of P. ginseng cv. Yunpoong [31]. PCR was conducted using the STS primer sets with genomic DNA from the five ginseng cultivars as a template.
The STS primers synthesized by Bioneer (Daejeon, Korea) and the sequences of the STS primers are listed in Table 2. PCR amplification was performed using the following mixture: 50 ng of genomic DNA, 20 pmole of each primer, 200 uM dNTPs, 0.5 U DNA polymerase (5 U/L uL), 1X PCR buffer (Solgent, Daejeon, Korea); giving a 25 uL reaction mixture according to the manufacturer’s protocol. Amplification reactions were carried out on a Thermal-cycler machine (TProfessional 96; Biometra, Göttingen, Germany); the procedure used was an initial 5 min at 94℃ followed by 40 cycles of 1 min at 94℃, 1 min at 60℃, 1 min at 72℃, and a final 7 min at 72℃. Amplification products were analyzed by electrophoresis on 1.5 % agarose gel in TBE buffer (45 mM Tris-HCl, pH 8.0, 45 mM Boric acid, 1 mM EDTA).
Table 2.
STS primer sets discriminating ginseng cultivars
| gDNA library | STS primer | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Clone information1) | Primer name | Primer sequence2) | PCR information3) | |||
|
|
|
|||||
| ID | Insert size (bp) | Gene bank no. | Forward (5’→3’) Reverse (5’→3’) | PCR product size (bp) | Anneal temp (℃) | |
|
| ||||||
| UFG0074 | 717 | HN339415 | UFGp74 | GCGAATTTCACAGAATTAGACG | 639 | 65 |
| ACAGGTCTCGAAACAATTGAAG | ||||||
| MFG0110 | 1129 | HN339416 | MFGp110A | AGTCCCAACGGAATTTCATC | 930 | 65 |
| GTTTTCCGCTTATGTTGCAG | ||||||
| MFG0130 | 1124 | HN339417 | MFGp130A | GGAAAGCGTTCAGCTCTTACG | 322 | 65 |
| TAAATGCTGTCAAGCCCAGAG | ||||||
1) gDNA library clones of Panax ginseng cv. Yunpoong.
2) Primers were converted from gDNA library clones of P. ginseng cv. Yunpoong and sequence-tagged site (STS) primers were designed by Lee [23].
3) The polymerase chain reaction (PCR) product size by using the recommended annealing temperature (anneal temp) and the genomic DNA of P. ginseng cv. Yunpoong.
Cloning and sequencing
The PCR products from five samples per cultivar were amplified and purified with a PCR purification kit (Qiagen) in accordance with the manufacturer’s instructions. Purified PCR products were cloned into the pGEM-T Easy vector System (Promega, Madison, WI, USA) and transformed into competent Escherichia coli KL1-Blue cells. The plasmids were purified with a plasmid mini kit (Qiagen). Sequencing was performed by Sanger’s method using ABI3730 automatic sequencer and the sequences were edited with the BioEdit program [32].
Probes and primer design
The primers and fluorogenic probes were designed on the basis of the sequences of the PCR-amplified DNA fragments by STS markers (UFGp74, MFGp110A, and MFGp130A). The probes were labelled with the reporter molecules 5′-VIC™ (emission wavelength, 552 nm) or FAM™ (emission wavelength, 518 nm) and the 3′endswere labelled with an minor groove binder (MGB) molecule and a nonfluorescent quencher. The primers and fluorogenic probes were synthesized by Applied Biosystems (Foster city, CA, USA).
Real-time polymerase chain reaction
Real-time PCR was performed by amplification using the ABI Step One Plus system (Applied Biosystems). Each reaction was carried out by duplicate in a Micro Amp Optical 96-well reaction plate. The 10 μL reaction mixtures contained 5 μL TaqMan® Universal Master Mix (which includes the heat-activated Ampli-Taq Gold Enzyme), 300 nM of each specific oligonucleotide primer, MGB probe 200 nM, and 1 μL of different dilution of the DNA mixture, corresponding to 10 to 100 ng of total DNA. The reaction conditions were as follows: 60℃ for 30 s, 95℃ for 10 min followed by 40 cycles of 95℃ for 15 s and 60℃ for 1 min.
Genotyping
For the genotyping assay of ginseng cultivars, six TaqMan® MGB probes and three pair of oligonucleotide primers were employed. The one specific for the wild-type allele was labeled with a VICTM dye (green fluorophore), and the other one specific for the mutant allele was labeled with a FAMTM dye (blue fluorophore). The generation of green fluorescence during amplification indicates homozygous wild-type, blue fluorescence indicates homozygous mutants, and both green and blue fluorescence indicates heterozygotes, respectively. For scatter plot analysis, the automatically generated threshold cycle values from each sample were plotted at coordinates that correspond to the signal of either FAMTM or VICTM .
RESULTS
Polymerase chain reaction analysis by sequence-tagged site markers
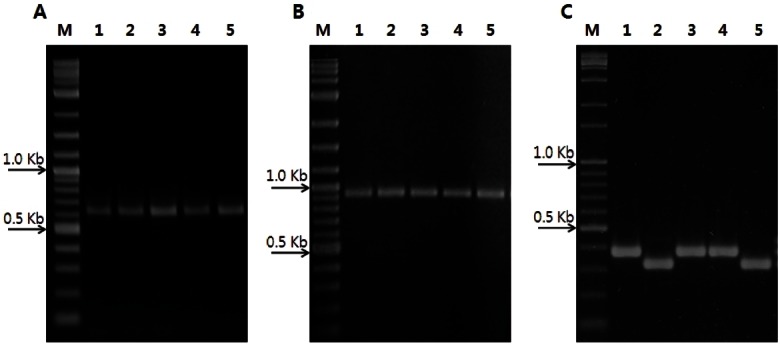
To identify useful variations for the development of a cultivar identification system, UFGp74, MFGp110A, and MFGp130A were tested to detect polymorphisms among ginseng cultivars (Korea). Polymorphic fragments were detected by MFGp130A ranging between 300 and 400 base pairs. MFGp130A generated two alleles among the ginseng cultivars. Chunpoong, Gopoong, and Kumpoong shared the same allele of 375 bp band and Yunpoong and Sunpoong shared another identical allele of the 322 bp band. UFGp74 and MFGp110A amplified uni-bands of 639 bp and 930 bp, respectively (Fig. 1).
Fig. 1. Amplification products of five ginseng cultivars (Korea) by sequence-tagged site-polymerase chain reaction. The monomorphic bands were amplified with (A) UFGp74 and (B) MFGp110A, and in-del polymorphisms were detected with (C) MFGp130 among five ginseng cultivars, respectively. Lane 1, Chunpoong; lane 2, Yunpoong; lane 3, Gopoong; lane 4, Kumpoong; lane 5, Sunpoong; M, size marker (2log DNA ladder; New England Biolabs).
Identification of single nucleotide polymorphisms based on sequences
According to the sequencing results, the amplified DNA with MFGp130A (gene bank accession number: HN339417) observed in one site base substitutions at 212 bp position and an insertion/deletion variation of 53 bp nucleotides at between 1042 bp and 1094 bp position (Fig. 2C). DNA with MFGp110A (gene bank accession number: HN339416) was observed in base substitutions of 6 sites. Chunpoong and Gopoong contain C at the 154 bp nucleotide position, but Yunpoong, Kumpoong, and Sunpoong were replaced with T in the same region. Yunpoong, Kumpoong, and Sunpoong contain C at nucleotide positions of 158 bp, 254 bp, and 630 bp, but Chunpoong and Gopoong were replaced with T at the same region. Chunpoong and Gopoong harbor A at the nucleotide position of 602 bp, but Yunpoong, Kumpoong, and Sunpoong were replaced with G at the same region. Yunpoong, Kumpoong, and Sunpoong harbor G at the nucleotide position of 859 bp, but Chunpoong and Gopoong were replaced with T at the same region (Fig. 2B). At the nucleotide positions 174 bp and 202 bp in the amplified DNA with UFGp74 (gene bank accession number: HN339415), Yunpoong, Gopoong, and Kumpoong harbor nucleotide G, but Chunpoong and Sunpoong contained C (Fig. 2A).
Fig. 2. Nucleotide sequence alignment of polymorphic fragments amplified from Chunpoong, Yunpoong, Gopoong, Kumpoong and Sunpoong by UFGp74 (A), MFGp110A (B) and MFGp 130A (C). The area enclosed by the box indicated single nucleotide polymorphism or ‘in-del’ variation. The position of the target sequence for the forward and reverse primers, and TaqMan minor groove binder (MGB) probes are indicated as arrow and thick underline, respectively.

Probes and primers design
In the sequence alignment, nine SNP sites were detected. Based on the SNP site detected in the amplified region by UFGp74, MFGp110A, and MFGp130A, the primer and probe set were designed for identification of the ginseng cultivars (Korea). The wild-type probes were labeled on the 5’-end with the fluorescent reporter dye (VICTM), whereas the mutant-type probes were labeled with a reporter fluorescent dye (FAMTM). On the 3’-end, all of the probes were labeled with a nonfluorescent quencher and a MGB. To obtain a specific and reliable PCR amplification reaction, each fluorescent probe was placed to target SNP sequence between upstream and downstream primer (Fig. 2). The primers that flank each target were located close to the probe by means of forcing the Primer Express program. The primers and probes used in the present study are shown in Table 3.
Table 3.
Real-time polymerase chain reaction probe and primer combinations used in this study
| Primer | Substitution | Probe | Sequence 5’ → 3’ (fluorophore/quencher)1) |
|---|---|---|---|
|
| |||
| PGP74 | G → A | Probe G | VIC – CGTTTAACGAATCCTC – MGB |
| Probe A | FAM – CGTTTAACAAATCCTC – MGB | ||
| Primer F | GCTCAAGAATCGTCAGATTCTGACT | ||
| Primer R | CGGAATCCGTTAACTTGAAATTCACTT | ||
| PGP110 | C → T | Probe C | VIC – CCTCCCCTAGCTACTCT – MGB |
| Probe T | FAM – CCTCCCCTAGTTACTCT – MGB | ||
| Primer F | GGCCCTTTCATTCTACAACTCTCAA | ||
| Primer R | GTCGCCTTGTACCGAGTCAA | ||
| PGP130 | C → A | Probe C | VIC – ACCTCTGACATCTTG – MGB |
| Probe A | FAM – CCTCTGAAATCTTG – MGB | ||
| Primer F | GCACCCCCAAAAGGGCTAAA | ||
| Primer R | TCGATAGGTGGCTCCATACGAT | ||
1)Bold and underlined nucleotides represent the location of the single nucleotide polymorphism. The wild type probes were labeled on the 5’ end with the fluorescent VIC™ dye, whereas the mutant type probes were labeled with the fluorescent FAM™ dye.
Real-time polymerase chain reaction
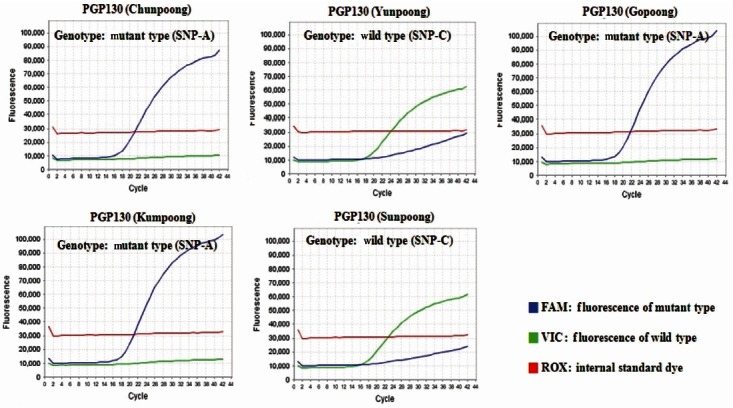
The molecular authentication of major ginseng cultivars (Chunpoong, Yunpoong, Gopoong, Kumpoong, and Sunpoong) was conducted via an SNP genotyping assay with real-time PCR using cultivar-specific probe and primer sets. According to the results of real-time PCR using PGP74, Chunpoong and Sunpoong generated a FAMTM fluorescent (SNP-A type) signal, whereas Yunpoong, Gopoong, and Kumpoong generated a VICTM fluorescent (SNP-G type) signal (Fig. 3). In the case of real-time PCR using PGP110, Chunpoong and Gopoong generated a FAMTM fluorescent (SNP-T type) signal, whereas Yunpoong, Kumpoong, and Sunpoong generated a VICTM fluorescent (SNP-C type) signal (Fig. 4). Finally, the results of realtime PCR using PGP130, Chunpoong, Gopoong, and Kumpoong generated a FAMTM fluorescent (SNP-A type) signal, whereas Yunpoong and Sunpoong generated a VICTM fluorescent (SNP-C type) signal (Fig. 5).
Fig. 3. Single nucleotide polymorphism (SNP) detection of five ginseng cultivars (Korea) by using PGP74. Each diagram shows two real-time polymerase chain reaction fluorescence curve with either SNP-A type or SNP-G type.
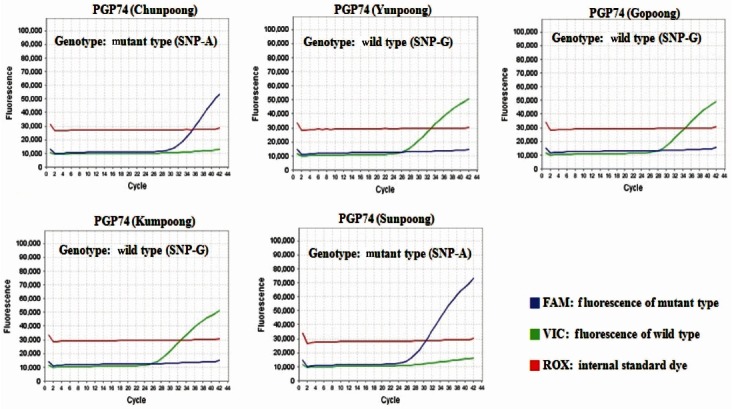
Fig. 4. Single nucleotide polymorphism (SNP) detection of five ginseng cultivars (Korea) by using PGP110. Each diagram shows two real-time polymerase chain reaction fluorescence curve with either SNP-T type or SNP-C type.
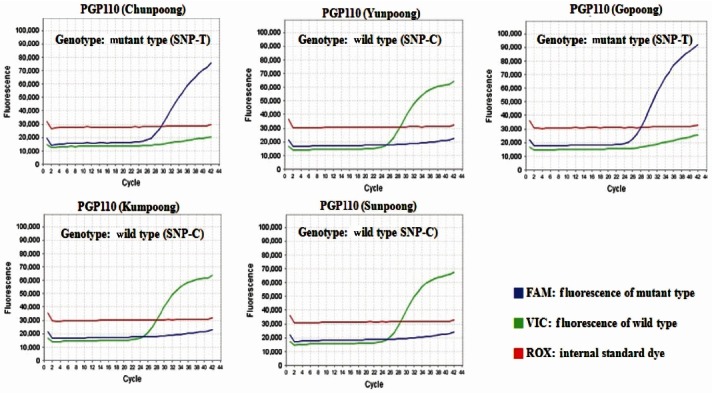
Fig. 5. Single nucleotide polymorphism (SNP) detection of five ginseng cultivars (Korea) by using PGP130. Each diagram shows two real-time polymerase chain reaction fluorescence curve with either SNP-A type or SNP-C type.
Genotyping
The genotypes of ginseng cultivars were determined at the end of the PCR when the accumulated fluorescence was determined as predominantly a VICTM signal, a FAMTM signal, or both VICTM and FAMTM signals. The results were displayed on a scatter plot in which each axis corresponds to a reporter signal. Fig. 6 shows the results of genotyping of the structural variants of PGP74, PGP110, and PGP130, respectively. Two genotypes (SNP-A or SNP-G) were detected in PGP74. Chunpoong and Sunpoong shared the same SNP-A, and Yunpoong, Gopoong, and Kumpoong shared another identical SNP-G. In the case of PGP110, two genotypes (SNP-T or SNP-C) were detected in the five ginseng cultivars (Korea). Chunpoong and Gopoong shared the same SNP-T, whereas Yunpoong, Kumpoong, and Sunpoong shared another identical SNP-C. Finally, two genotypes (SNP-A or SNP-C) of PGP130 in the five cultivars of ginseng have been identified. Chunpoong, Gopoong, and Kumpoong shared the same SNP-A, whereas Yunpoong and Sunpoong shared another SNP allele, allele-C. The allele combination of the SNP type of the five cultivars, Chungpoong, Yunpoong, Gopoong, Kumpoong, and Sunpoong, was identified as ‘ATA’, ‘GCC’, ‘GTA’, ‘GCA’, and ‘ACC’, respectively (Table 4).
Fig. 6. Single nucleotide polymorphism genotyping diagrams of five ginseng cultivars (Korea) by real-time polymerase chain reaction with the three probe pairs (PGP74, PGP110, and PGP130).
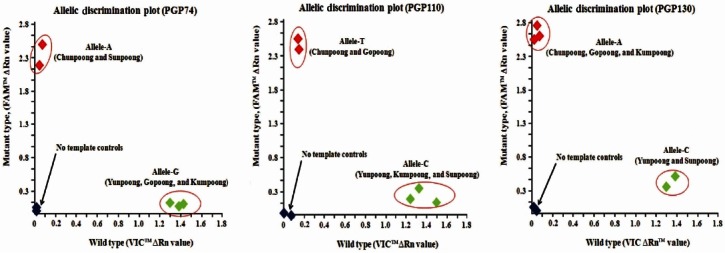
Table 4.
SNP combinations of ginseng cultivars by SNP genotyping
| Probe | Cultivars/allele type | ||||
|---|---|---|---|---|---|
|
| |||||
| Chunpoong | Yunpoong | Gopoong | Kumpoong | Sunpoong | |
|
| |||||
| PGP74 | A | G | G | G | A |
| PGP110 | T | C | T | C | C |
| PGP130 | A | C | A | A | C |
| SNP combinationcombination | ATA | GCC | GTA | GCA | ACC |
SNP, single nucleotide polymorphism.
DISCUSSION
Ginseng (P. ginseng) is a Korea’s national heritage and Korea’s most representative agricultural crop. Its excellent quality has been recognized worldwide, and it has been exported to neighboring Oriental countries, including China. Despite the importance of ginseng, there are no any system and regulation to manage the protection of ginseng cultivars in Korea. In recent years, several efforts have focused principally on the development of various molecular markers to prevent the contamination of cultivars and to authenticate ginseng cultivars (Korea). Among them, the most recently developed STS system containing CAPS markers has proven the most useful in terms of its reproducibility and reliability [23].
STS containing CAPS markers are powerful tools for identification of the ginseng cultivars (Korea). However,
these methods require either restriction enzyme digestion or an agarose gel electrophoresis step. By way of contrast, the SNP genotyping assay using real-time PCR does not require additional post-PCR manipulations such as restriction enzyme digestion or agarose gel electrophoresis.
The authentication of ginseng cultivars (Korea). via SNP genotyping assays is based on allele-specific amplification via real-time PCR. The allelic discrimination of each probe depends on duplex allele-specific probes, labeled with a probe-specific fluorescent dye and a generic quencher that reduces fluorescence in the intact probes. During the amplification of the sequence surrounding the SNP, probes complementary to the DNA target are cleaved by the 5’exonuclease activity of Taq polymerase [33].
The SNP genotyping assay used for identification of the ginseng cultivars (Korea). has several advantages. First, large numbers of samples can be processed within a short time. The conventional PCR method using gel electrophoresis takes approximately 5 h. But, the TaqMan-MGB probe system can analyze 96 samples in less than 2 h. Thus, the TaqMan-MGB probe system has a high-throughput capacity, because it require no post-PCR manipulations. Second, this system can be used with a minimum amount of waste; because this system does not require electrophoresis, toxic substances, such as ethidium bromide and silver staining reagents and the discharge of plastic tubes is avoided. Third, conjugated MGB can increase the melting temperature of probes, thereby increasing probe specificity. Additionally, it allows for the use of shorter probes (usually 13 to 18 nucleotides), which can facilitate probe design, particularly in the AT-rich region. Finally, TaqMan-MGB probes can be employed without restriction enzymes recognizing specific regions. That is, almost all detected DNA sequence variation positions can be employed for probe design [34]. Thus, it is considered a powerful tool for use in SNP genotyping assays.
In this study, the real-time PCR using TaqMan-MGB probes was conducted for the application of SNP genotyping assays to discrimination among the ginseng cultivars (Korea). Consequently, fluorescence probes (PGP74, PGP110, and PGP130) and novel primer sets can be used to distinguish among ginseng cultivars (Chunpoong, Yunpoong, Gopoong, Kumpoong, and Sunpoong). The combination of the SNP type of the five cultivars, Chungpoong, Yunpoong, Gopoong, Kumpoong, and Sunpoong, was identified as ‘ATA’, ‘GCC’, ‘GTA’, ‘GCA’, and ‘ACC’, respectively (Table 4). However, to distinguish the nine cultivars used in the study by using only three pairs of probes would be too limited, as the SNP variations are not sufficient enough. Accordingly, it will be required to build additional probes to distinguish five cultivars out of nine. By discovering additional SNP site, we will try to add the number of probes.
The development of a rapid authentication system of the ginseng cultivars (Korea) using TaqMan-MGB probes has proven helpful in protecting the unique national resources, in establishing a distribution system for Korean herbal markets, in the quality control of the ginseng seed production process, and in the protection of intellectual property.
References
- 1.Park CK, Jeon BS, Yang JW. The chemical components of Korean ginseng. Food Ind Nutr. 2003;8:10–23. [Google Scholar]
- 2.Wang LC, Lee TF. Effect of ginseng saponins on cold tolerance in young and elderly rats. Planta Med. 2000;66:144–147. doi: 10.1055/s-2000-11122. [DOI] [PubMed] [Google Scholar]
- 3.Shin HR, Kim JY, Yun TK, Morgan G, Vainio H. The cancer-preventive potential of Panax ginseng: a review of human and experimental evidence. Cancer Causes Control. 2000;11:565–576. doi: 10.1023/A:1008980200583. [DOI] [PubMed] [Google Scholar]
- 4.Yun TK, Lee YS, Lee YH, Kim SI, Yun HY. Anticarcinogenic effect of Panax ginseng C.A. Meyer and identification of active compounds. J Korean Med Sci. 2001;16 Suppl:S6–S18. doi: 10.3346/jkms.2001.16.S.S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SH, Park KS. Effects of Panax ginseng extract on lipid metabolism in humans. Pharmacol Res. 2003;48:511–513. doi: 10.1016/S1043-6618(03)00189-0. [DOI] [PubMed] [Google Scholar]
- 6.Cheng Y, Shen LH, Zhang JT. Anti-amnestic and anti-aging effects of ginsenoside Rg1 and Rb1 and its mechanism of action. Acta Pharmacol Sin. 2005;26:143–149. doi: 10.1111/j.1745-7254.2005.00034.x. [DOI] [PubMed] [Google Scholar]
- 7.Xie JT, Mehendale SR, Li X, Quigg R, Wang X, Wang CZ, Wu JA, Aung HH, A Rue P, Bell GI , et al. Anti-diabetic effect of ginsenoside Re in ob/ob mice. Biochim Biophys Acta. 2005;1740:319–325. doi: 10.1016/j.bbadis.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Joo SS, Won TJ, Lee DI. Reciprocal activity of ginsenosides in the production of proinflammatory repertoire, and their potential roles in neuroprotection in vivo. Planta Med. 2005;71:476–481. doi: 10.1055/s-2005-864145. [DOI] [PubMed] [Google Scholar]
- 9.Kwon WS, Chung CM, Kim YT, Lee MG, Choi KT. Breeding process and characteristics of KG101, a superior line of Panax ginseng C.A. Meyer. J Ginseng Res. 1998;22:11–17. [Google Scholar]
- 10.Kwon WS, Lee MG, Choi KT. Breeding process and characteristics of Yunpoong, a new variety of Panax ginseng C. A. Meyer. J Ginseng Res. 2000;24:1–7. [Google Scholar]
- 11.Kwon WS, Lee JH, Park CS, Yang DC. Breeding process and characteristics of Gopoong, a new variety of Panax ginseng C. A. Meyer. J Ginseng Res. 2003;27:86–91. doi: 10.5142/JGR.2003.27.2.086. [DOI] [Google Scholar]
- 12.Chan K. Some aspects of toxic contaminants in herbal medicines. Chemosphere. 2003;52:1361–1371. doi: 10.1016/S0045-6535(03)00471-5. [DOI] [PubMed] [Google Scholar]
- 13.Bang KH, Lee SW, Hyun DY, Cho JH, Cha SW, Seong NS, Huh MK. Molecular authentication and genetic polymorphism of Korea ginseng (Panax ginseng C.A. Meyer) by inter-simple sequence repeats (ISSRs) markers. J Life Sci. 2004;14:425–428. doi: 10.5352/JLS.2004.14.3.425. [DOI] [Google Scholar]
- 14.In DS, Kim YC, Bang KH, Chung JW, Kim OT, Hyun DY, Cha SW, Kim TS, Seong NS. Genetic relationships of Panax species by RAPD and ISSR analyses. Korean J Med Crop Sci. 2005;13:249–253. [Google Scholar]
- 15.Kim OT, Bang KH, In DS, Lee JW, Kim YC, Shin YS, Hyun DY, Lee SS, Cha SW, Seong NS. Molecular authentication of ginseng cultivars by comparison of internal transcribed spacer and 5.8S rDNA sequences. Plant Biotechnol Rep. 2007;1:163–167. doi: 10.1007/s11816-007-0019-2. [DOI] [Google Scholar]
- 16.Ma KH, Dixit A, Kim YC, Lee DY, Kim TS, Cho EG, Park YJ. Development and characterization of new microsatellite markers for ginseng (Panax ginseng C.A. Meyer). Conserv Genet. 2007;8:1507–1509. doi: 10.1007/s10592-007-9284-4. [DOI] [Google Scholar]
- 17.Dan NV, Ramchiary N, Choi SR, Uhm TS, Yang TJ, Ahn IO, Lim YP. Development and characterization of new microsatellite markers in Panax ginseng (C.A. Meyer) from BAC end sequences. Conserv Genet. 2010;11:1223–1225. doi: 10.1007/s10592-009-9924-y. [DOI] [Google Scholar]
- 18.Bang KH, Chung JW, Kim YC, Lee JW, Jo IH, Seo AY, Kim OT, Hyun DY, Kim DH, Cha SW. Development of SSR markers for identification of Korean ginseng (Panax ginseng C.A. Meyer) cultivars. Korean J Med Crop Sci. 2011;19:185–190. [Google Scholar]
- 19.Wang H, Sun H, Kwon WS, Jin H, Yang DC. Molecular identification of the Korean ginseng cultivar “Chunpoong” using the mitochondrial nad7 intron 4 region. Mitochondrial DNA. 2009;20:41–45. doi: 10.1080/19401730902856738. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Sun H, Kwon WS, Jin H, Yang DC. A PCR-based SNP marker for specific authentication of Korean ginseng (Panax ginseng) cultivar “Chunpoong”. Mol Biol Rep. 2010;37:1053–1057. doi: 10.1007/s11033-009-9827-5. [DOI] [PubMed] [Google Scholar]
- 21.Lee JW, Kim YC, Jo IH, Seo AY, Lee JH, Kim OT, Hyun DY, Cha SW, Bang KH, Cho JH. Development of an ISSR-derived SACR marker in Korean ginseng cultivars (Panax ginseng C.A. Meyer). J Ginseng Res. 2011;35:52–59. doi: 10.5142/jgr.2011.35.1.052. [DOI] [Google Scholar]
- 22.Sun H, Wang HT, Kwon WS, In JG, Lee BS, Yang DC. Development of molecular markers for the determination of the new cultivar ‘Chunpoong’ in Panax ginseng C. A. Meyer associated with a major latex-like protein gene. Biol Pharm Bull. 2010;33:183–187. doi: 10.1248/bpb.33.183. [DOI] [PubMed] [Google Scholar]
- 23.Lee JW. Development of DNA markers for identification of Korean ginseng cultivars (Panax ginseng C.A. Meyer). University of Dongguk; Seoul: 2010. [Google Scholar]
- 24.Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, et al. 3’-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 2000;28:655–661. doi: 10.1093/nar/28.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alary R, Serin A, Maury D, Jouira HB, Sirven JP, Gautier MF, Joudrier P. Comparison of simplex and duplex real-time PCR for the quantification of GMO in maize and soybean. Food Control. 2002;13:235–244. doi: 10.1016/S0956-7135(02)00015-4. [DOI] [Google Scholar]
- 26.Brodmann PD, Moor D. Sensitive and semi-quantitative TaqMan™ real-time polymerase chain reaction systems for the detection of beef (Bos taurus) and the detection of the family Mammalia in food and feed. Meat Sci. 2003;65:599–607. doi: 10.1016/S0309-1740(02)00253-X. [DOI] [PubMed] [Google Scholar]
- 27.Hird H, Lloyd J, Goodier R, Brown J, Reece P. Detection of peanut using real-time polymerase chain reaction. Eur Food Res Technol. 2003;217:265–268. doi: 10.1007/s00217-003-0726-z. [DOI] [Google Scholar]
- 28.Taylor MI, Fox C, Rico I, Rico C. Species-specific TaqMan probes for simultaneous identification of (Gadus morhua L.) haddock (Melanogrammus aeglefinus L.) and whiting (Merlangius merlangus L.). Mol Ecol Notes. 2002;2:599–601. doi: 10.1046/j.1471-8286.2002.00269.x. [DOI] [Google Scholar]
- 29.Itoi S, Nakaya M, Kaneko G, Kondo H, Sezaki K, Watabe S. Rapid identification of eels Anguilla japonica and Anguilla anguilla by polymerase chain reaction with single nucleotide polymorphism-based specific probes. Fish Sci. 2005;71:1356–1364. doi: 10.1111/j.1444-2906.2005.01102.x. [DOI] [Google Scholar]
- 30.Lopez I, Pardo MA. Application of relative quantification TaqMan real-time polymerase chain reaction technology for the identification and quantification of Thunnus alalunga and Thunnus albacares. J Agric Food Chem. 2005;53:4554–4560. doi: 10.1021/jf0500841. [DOI] [PubMed] [Google Scholar]
- 31.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 32.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 33.De la Vega FM, Lazaruk KD, Rhodes MD, Wenz MH. Assessment of two flexible and compatible SNP genotyping platforms: TaqMan SNP Genotyping Assays and the SNPlex Genotyping System. Mutat Res. 2005;573:111–135. doi: 10.1016/j.mrfmmm.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Lee JW, Bang KH, Choi JJ, Chung JW, Lee JH, Jo IH, Seo AY, Kim YC, Kim OT, Cha SW. Development of peptide nucleic acid (PNA) microarray for identification of Panax species based on the nuclear ribosomal internal transcribed spacer (ITS) and 5.8S rDNA regions. Genes Genomics. 2010;32:463–468. doi: 10.1007/s13258-010-0040-7. [DOI] [Google Scholar]


