Abstract
We investigated whether Korean red ginseng (KRG) and highly active antiretroviral therapy (HAART) affect the frequency of gross deletion in 5’LTR/gag in 20 hemophiliacs. This study is a prospective study in 20 hemophiliacs who were infected with Korean subclade B of HIV-1 from two cash-paid plasma donors in 1990. Over a 13-year period, we obtained 436 amplicons of 5’LTR/gag genes by nested polymerase chain reaction using 147 peripheral blood mononuclear cells. Of the 436 amplicons, 92 (21.1%) showed gross deletion in 5’LTR/gag. Despite of a 2.3-fold higher monthly dose of KRG intake, the frequency of gross deletion in 5’LTR/gag (16.4%) was significantly decreased during HAART compared with 28.1% prior to HAART (p<0.01). Gross deletion in 5’LTR/gag was 10% more detected on KRG-therapy than prior to KRG-therapy (p<0.05). In addition, we also obtained 28 amplicons containing premature stop codon or isoleucine at initiation codon of 254 amplicons sequenced on KRG intake (7.5%) or HAART (13.6%) compared with 0% before KRG intake. These findings indicate that high frequency of gross deletion in 5’LTR/gag and genetic defects prior to HAART are significantly associated with KRG intake and the detection of gross deletion in 5’LTR/gag is decreased by HAART.
Keywords: Panax ginseng, Korean red ginseng, HIV-1 5’LTR/gag, Genetic defects, Highly active antiretroviral therapy, Acquired immuno-deficiency syndrome
INTRODUCTION
Panax ginseng has been used as an herbal medicine for more than 2000 years in the Orient [1]. About 200 constituents from P. ginseng have been isolated and characterized [2,3]. Its major components are ginseng saponins and polysaccharides. The major pharmacological effect of ginseng is adaptogenic effect. In other words, ginseng nonspecifically increases resistance to various hazardous stimuli of physical, chemical, and biological stress through immunomodulation and hyphothalmic-pituitaryadrenal axis [4,5]. Recent literatures show the potential of adjuvant or as immunotherapeutics of ginseng [6-14].
Although there have been many reports on the association between genetic defects and clinical courses in HIV- 1 infected patients [15-22], gross deletion in the nef gene (Δnef) has been reported in very limited proportion even in elite controllers [23]. We previously reported that Korean red ginseng (KRG) increases the frequency of gΔnef and gross deletion in 5’LTR/gag regions as well as slow progression in HIV-1 infected individuals [24-28]. Thus, we could obtain several long-term survivors for more than 20 years in the absence of antiretroviral therapy among our ginseng cohort [24,27]. In Korea, the longest survivor among HIV-1 infected patients was diagnosed in 1987 and in the absence of highly active antiretroviral therapy (HAART) has maintained his CD4 T cell count since 1994. They all reveal genetic defects in the Δnef or 5’LTR/gag although the proportion of defected amplicons was less than 20% and 50%, respectively [27]. In addition, we reported that HAART significantly decreases the frequency of gΔnef caused by KRG intake in 20 hemophiliacs who were infected with Korean subclade B of HIV-1 [26]. In this study, we investigated whether KRG and HAART affect the frequency of gross deletion in 5’LTR/gag in 20 hemophiliacs.
MATERIALS AND METHODS
Study subjects
This study consisted of 20 hemophiliac patients (HPs) infected with Korean subclade of HIV-1 subtype B (KSB) [26,29]. All patients were infected with HIV-1 in between late 1989 and April 1991. The time on the first use of domestic clotting factor was described in Table 1 in the reference 29. Age at HIV-1 diagnosis was from 4 to 39 years. Informed written consent was obtained from all subjects before the initiation of the study.
Table 1.
Summary of 20 hemophiliac individuals infected with Korean subclade B of HIV-1 subtype B
| Patient | Date on diagnosis of HIV-1 infection | Prior to HAART | Plasma RNA copy (/mL) prior to HAART | Amount of KRG (g) | |||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Follow-up mo | Annual decrease in CD4+ T | Monthly KRG (g) | Before HAART | During HAART | |||
|
| |||||||
| HP 1 | 26-11-90 | 51 | 28 | 22 | Not tested | 1,140 | No HAART |
| HP 2 | 21-02-91 | 177 | 27 | 10 | 35,900 | 1,800 | 0 |
| HP 3 | 01-04-91 | 135 | 49 | 15 | Not tested | 2,000 | No HAART |
| HP 4 | 24-08-91 | 111 | 70 | 11 | 8,180 | 1,210 | 4,320 |
| HP 5 | 13-01-92 | 28 | 214 | 21 | Not tested | 600 | No HAART |
| HP 6 | 20-01-92 | 82 | 72 | 0 | Not tested | 0 | >5,760 |
| HP 7 | 22-01-92 | 64 | 140 | 19 | 20,000 | 1200 | 11,640 |
| HP 8 | 15-02-92 | 204 | 1 | 74 | 14,600 | >15,390 | 720 |
| HP 9 | 25-02-92 | 171 | 32 | 3 | 23,700 | 540 | 180 |
| HP 10 | 26-03-92 | 144 | 28 | 5 | Not tested | 780 | 5040 |
| HP 11 | 29-02-92 | 136 | 72 | 1 | 209,000 | 180 | 3300 |
| HP 12 | 25-02-92 | 119 | 49 | 0 | Not tested | 0 | 1500 |
| HP 13 | 29-02-92 | 163 | 28 | 24 | 4,000 | 3,900 | 0 |
| HP 14 | 19-02-92 | 204 | 23 | 109 | 4,146 | >21,078 | 1,980 |
| HP 15 | 16-09-92 | 123 | 0 | 63 | 111 | 7,776 | No HAART |
| HP 16 | 05-12-92 | 158 | 22 | 9 | Not tested | 1,470 | >1,000 |
| HP 17 | 26-02-93 | 102 | 62 | 0 | 3,703 | 0 | 3,120 |
| HP 18 | 02-03-93 | 53 | 38 | 145 | Not tested | 7,680 | 17,400 |
| HP 19 | 26-07-93 | 58 | 84 | 0 | >10,062 | 0 | 9,060 |
| HP 20 | 04-08-94 | 51 | 26 | 113 | 12,000 | 5,760 | 7,680 |
| Total | Mean±SD | 117±54 | 53±50 | 32±44 | 30,485±60,149 | 3,625±5,624 | 4,544±4,862 |
HAART, highly active antiretroviral therapy; KRG, Korean red ginseng; HP, hemophiliac patient.
Antiretroviral therapy
As of December 2009, only HP-15 had not received HAART. In addition to two HPs that died (Table 1), 7 HPs (numbers 2, 8, 10, 11, 13, 14, and 16) had not been treated with HAART for the first 10 years after their diagnosis of HIV-1. Thereafter, HPs (numbers 2, 8, 10, 11, 13, 14, and 16) have been treated from April 2006, June 2007, January 2005, November 2003, February 2006, April 2009, and February 2006, respectively (Table 1).
Therapy with Korean red ginseng
Outpatient-based clinical trial of KRG treatment in HIV-1-infected patients was initiated at the Korean National Institute of Health in late 1991. The daily dose of KRG was 5.4 g (six 300 mg capsules three times per day) for men [28,30,31]. KRG has been supplied since November 1991 although the supply of KRG was not consistent before year 2000.
CD4 T cell count and plasma RNA copy number
At about 6-month intervals, blood was drawn from each patient and peripheral blood mononuclear cells (PBMCs) in each sample were incubated with phycoerythrin- and fluorescein isothiocyanate-conjugated antibodies against CD4 and CD8 antigens, respectively (Simultest reagent; Becton Dickinson, San Jose, CA, USA). Levels of CD4 and CD8 T cells were measured by FACScan (Becton-Dickinson) flow cytometry. Plasma HIV-1 RNA copy (/mL) were measured using the Amplicor HIV-1 Monitor kit (Roche Diagnostics, Branchburg, NJ, USA).
Amplification and sequencing of 5’LTR/gag
One-hundred forty seven PBMCs from 20 HPs were obtained between 1991 and 2010. Proviral 5’LTR/gag sequences (1,125 bp) were amplified from the PBMC using nested polymerase chain reaction, as previously described [25,27].
Statistical analysis
The data are expressed as the means±one standard deviations (continuous variables) or as counts and percentages (categorical variables). Continuous variables were analyzed by using correlation analyses method; categorical variables were compared with the χ2 statistics. A p-value of <0.05 were considered statistically significant.
Sequences
The GenBank accession numbers are EF370244-260, EF370362-366, EF370369-372, EU047688-692, and JN613584-804.
RESULTS
Significant correlation between Korean red ginseng intake and decrease in CD4 T cells
CD4 T cell counts were measured during a follow-up period of 117±54 mo prior to HAART. All patients took KRG for various durations. The amount of KRG used prior to the initiation of HAART was 3,625±5,624 g (32 g/mo) for 117±54 mo and the annual decrease in the CD4 T cell count was 53±50/μL (Table 1). When we focused on 13 patients who were followed up for more than 100 mo prior to HAART, there was a significant inverse correlation between amount of monthly KRG intake and annual decrease in CD4 T cells (r=-0.612) (p<0.05).
Number of amplicons
We obtained 436 amplicons for 5’LTR/gag genes from 147 PBMCs obtained over 10 yr (range, 3 to 19 yr). Of those, 92 genes (21.1%) from 53 PBMCs (36.1%) showed gross deletion in 5’LTR/gag.
Frequency of gross deletion in 5’LTR/gag is associated with Korean red ginseng intake
We obtained 30 amplicons at baseline (before KRG intake). Of the 30 amplicons, 3 showed gross deletion in 5’LTR/gag (10%) (Fig. 1). During KRG intake prior to HAART, we obtained 192 amplicons. Of those, 54 showed gross deletion in 5’LTR/gag (28.1%, p<0.05) (Fig. 1).
Fig. 1. Changes of the proportion of gross deletion in the 5’ LTR/gag gene (gΔ). Proportion of gΔ was significantly increased on Korean red ginseng (KRG) intake only (28.1%, 54/192) compared with that prior to KRG intake (10%, 3/30) (p<0.05). The detection of gΔ was significantly inhibited during highly active antiretroviral therapy (HAART) and KRG intake compared with during KRG only (16.4%, 35/214) (p<0.01).
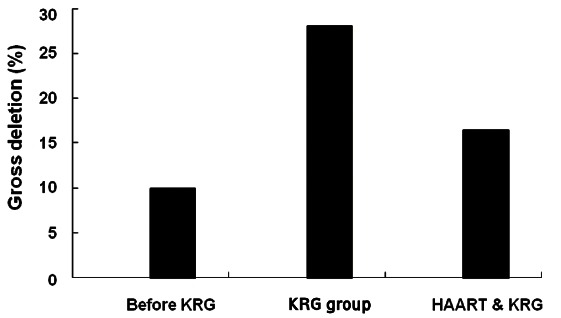
Frequency of gross deletion in 5’LTR/gag was significantly inhibited during HAART
Among the 20 patients, 16 patients received HAART for 58.5±43.0 mo (range, 4 to 124 mo; median, 66.5 mo). During the HAART period, 12 patients also received KRG. The average amount of KRG and monthly KRG were 4,544±4,862 g (range, 0 to 17,400 g) and 75 g, respectively (Table 1). In regard to HAART, 192 and 214 amplicons were obtained on KRG only and during KRG plus HAART, respectively. Of those, 54 (28.1%) and 35 (16.4%) showed gross deletion in 5’LTR/gag for the corresponding periods (Fig. 1). Thus, proportion of gross deletion in 5’LTR/gag was significantly inhibited during HAART (p<0.01).
Nature of gross deletion and a large amplicon
We determined sequences in 82 of 92 gross deletions in 5’LTR/gag from 20 HPs and found that the size of the deletions was all different in all patients although in HP-4, the same size deletions (689 bp) were repeatedly detected (Table 2). Interestingly, there were a case of inversion (HP-5), duplication (EF370252 in HP-8), recombination (HP-14), and insertion (HP-20) together with gross deletion in an amplicon, respectively. Among the 82 amplicons containing gross deletion in 5’LTR/gag, only 4 amplicons revealed gross deletion at two sites and 78 amplicons revealed it at one site. All the 82 amplicons revealed gross deletion in 5’LTR region (21 to 470 bp). Ten of the 82 amplicons did not reveal gross deletion in the gag region (Table 2). Therefore, the proportion of gross deletion in 5’LTR/gag was higher in 5’LTR (82/82) than that in the gag region (72/82).
Table 2.
Characteristics of gross deletions in the 5’LTR/gag gene
| Patient code | Day of sampling | No. of bands by PCR1) | No. of PCR reaction with gΔ2) | Beginning nt. relative to HIV-1 NL4-3 | Size of deletion in 5’ LTR (bp)3) | Size of deletion in gΔ (bp) | A representative GenBank accession no. |
|---|---|---|---|---|---|---|---|
|
| |||||||
| HP-2 | 21-10-01 | 1 | 1 | 330 | 459 | 463 | JN613586 |
| 13-09-07 | 1 | 1 | 701 | 84 | 0 | ||
| 12-06-08 | 1 | 1 | 607 | 262 | 144 | JN613587 | |
| HP-3 | 29-08-02 | 2 | 1 | 440 | 349 | 407 | JN613584 |
| HP-4 | 11-10-02 | 1 | 4 | 484 | 306 | 383 | JN613588 |
| 27-02-03 | 1 | 3 | 484 | 306 | 383 | JN613589 | |
| 04-01-08 | 1 | 2 | 484 | 306 | 383 | JN613591 | |
| 30-06-08 | 1 | 2 | 484 | 306 | 383 | JN613593 | |
| 22-12-08 | 1 | 1 | 657 | 132 | 261 | JN613595 | |
| HP-5 | 28-12-94 | 2 | 3 | 579 | 212 | 437 | 6082s |
| 27-02-95 | 1 | 1 | 627 | 162 | 373 | 6083s | |
| HP-6 | 17-11-02 | 1 | 1 | 443 | Inversion | JN613661 | |
| HP-7 | 20-10-96 | 1 | 1 | 432 | 358 | 531 | JN613596 |
| 2 | 1 | 483 | 288 | 0 | JN613597 | ||
| 27-02-97 | 2 | 1 | 350 | 435 | 0 | JN613598 | |
| 07-01-03 | 2 | 1 | 603 | 186 | 126 | 3568s1 | |
| HP-8 | 19-04-95 | 1 | 1 | 657 | 133 | 322 | EF370245 |
| 15-02-01 | 1 | 1 | 632 | 158 | 228 | EF370250 | |
| 12-12-02 | 1 | 3 | 632 | 158 | 478 | EF370252 | |
| 03-05-04 | 1 | 1 | 110 | 159 | 0 | EF370256 | |
| 06-02-06 | 1 | 1 | 163 | 8 Plus 17 | 0 | EF370259 | |
| 27-09-07 | 1 | 3 | 623 | JN613599 | |||
| 26-04-10 | 1 | 1 | 727 | 62 | 457 | 9198s | |
| HP-9 | 13-12-02 | 2 | 1 | Different | ND | ||
| 2 | 1 | 625 | 164 | 587 | JN613600 | ||
| 2 | 1 | 385 | 406 | 245 | JN613601 | ||
| 2 | 1 | 328 | 462 | 110 | |||
| 20-11-03 | 1 | 1 | 412 | 375 | 36 | 1651s | |
| 07-09-06 | 2 | 1 | 713 | 77 | 480 | JN613602 | |
| 3 | 1 | 722 | 68 | 590 | JN613603 | ||
| 342 | 447 | 334 | JN613604 | ||||
| 19-12-07 | 2 | 1 | ND | JN613605 | |||
| 2 | 2 | 484 | 306 | 5 | JN613606 | ||
| HP-10 | 30-08-02 | 1 | 1 | 637 | 153 | 467 | JN613608 |
| 2 | 1 | ND | ND | ||||
| 2 | 1 | 453 | 337 | 271 | JN613607 | ||
| 13-09-07 | 1 | 1 | 639 | 151 | 19 | JN613609 | |
| HP-11 | 30-10-02 | 2 | 1 | 320 | 470 | 153 | EF370364 |
| 1 | 1 | 304 | 197 | 0 | EF370366 | ||
| 09-11-03 | 1 | 1 | 420 | 370 | 177 | 6021 | |
| 2 | 2 | 613 | 178 | 555 | JN613610 | ||
| HP-12 | 25-10-02 | 1 | 1 | 390 | 343 | 0 | JN613612 |
| 1 | 1 | 602 | 21 | 0 | JN613711 | ||
| 09-04-07 | 1 | 1 | 677 | 113 | 321 | JN613613 | |
| 1 | 1 | ND | JN613614 | ||||
| 01-04-09 | 1 | 1 | 635 | 118 | 0 | 6339 | |
| 24-08-09 | 1 | 1 | 736 | 43 | 0 | ||
| 14-05-10 | 1 | 1 | 667 | 123 | 310 | 9197s | |
| HP-14 | 11-12-96 | 2 | 3 | ND | |||
| 08-01-03 | 2 | 2 | 635 | Rec | 3564s | ||
| 23-05-07 | 1 | 1 | ND | ||||
| 15-10-09 | 1 | 1 | 371 | 420 | 347 | ||
| 27-05-10 | 1 | 1 | 667 | 123 | 552 | JN613621 | |
| HP-13 | 10-03-99 | 1 | 1 | 591 | 199 | 605 | JN613615 |
| 11-28-02 | 2 | 1 | 424 | 366 | 108 | JN613616 | |
| 03-04-03 | 2 | 1 | 627 | 163 | 372 | JN613617 | |
| 2 | 1 | 644 | 146 | 533 | JN613618 | ||
| 19-04-07 | 1 | 3 | 354 | 436 | 119 | JN613619 | |
| 89+ | |||||||
| HP-15 | 08-03-03 | 2 | 1 | ND | |||
| HP-16 | 12-09-02 | 1 | 1 | 579 | 211 | 362 | EU047689 |
| HP-17 | 03-11-02 | 2 | 1 | JN613624 | |||
| 20-02-03 | 2 | 2 | 426 | 364 | 21 | JN613622 | |
| 10-08-04 | 1 | 1 | 671 | 119 | 65 | JN613759 | |
| 16-01-07 | 1 | 1 | About 200 | ND | ND | ||
| HP-18 | 12-04-95 | 2 | 2 | ND | |||
| 27-04-06 | 1 | 1 | 659 | 131 | 505 | JN613625 | |
| HP-19 | 07-11-07 | 1 | 1 | 655 | 135 | 593 | JN613627 |
| HP-20 | 27-02-03 | 2 | 1 | 567 | 223 | 493 | JN613628 |
| 08-07-03 | 1 | 2 | 535 | 255 | 587 | JN613629 | |
| 05-06-08 | 1 | 1 | 727 e | 63 | 210 | JN613630 | |
| 16-03-10 | 1 | 1 | 669 | 121 | 334 | JN613631 | |
PCR, polymerase chain reaction; gΔ, gag gene; HP, hemophiliac patient; ND, not determined; Rec, recombination.
1)1 and 2 denote single short band only and wild type together with short band, respectively.
2)No. of PCR product with gΔ out of 4 PCR reactions.
3)Base pair.
High frequency of nonfunctional genes by G-to-A hypermutations
We determined sequences in 27 amplicons prior to KRG. There was no premature stop codon among the 27 amplicons. We determined sequences for 147 amplicons on KRG only and for 179 amplicons on KRG plus HAART. Maximum number of premature stop codon was 3 in an amplicon. Compared with that prior to KRG therapy (0/27), the amplicons during KRG plus HAART (at 36 codons) revealed significantly higher frequency of stop codons (p<0.05). There was also a significant difference between KRG only (at 15 codons) and KRG plus HAART (p<0.05). However, when we strictly exclude the amplicons containing gross deletion in the gag gene, the number of amplicons prior to KRG, on KRG only, and on KRG plus HAART were 18, 107, and 147, respectively (Table 3).
Table 3.
Frequency of genetic defects in Gag protein
| Patient | Prior to KRG intake | On KRG only | On KRG plus HAART | p-value1) |
|---|---|---|---|---|
|
| ||||
| No. of PCR amplicons | 18 | 107 | 147 | |
| No. of premature stop codon | 0 | 15 | 36 | ˂ 0.05 |
| No. of I at initiation codon | 0 | 6 | 9 | |
| No. of total codons (%) | 02) | 521 (19.6)2) | 45 (30.6) | ˂ 0.01 |
| No. of PCR amplicon with genetic defects3) | 0 | 8 (7.5) | 20 (13.6) | |
| No. of PCR amplicon with G-to-A hypermutations only without genetic defects | 0 | 3 | 6 | |
| No. of total amplicons with G-to-A hypermutations (%) | 0 | 11 (10.3) | 26 (17.7) | |
KRG, Korean red ginseng; HAART, highly active antiretroviral therapy; PCR, polymerase chain reaction.
1)p -value between prior to KRG intake and on KRG plus HAART.
2)p˂0.05 between prior to KRG intake and on KRG only.
3)Genetic defects denote premature stop codon and isoleucine at the initiation codon.
In addition, isoleucine at the initiation codon of Gag protein was detected in six and nine amplicons in KRG only and KRG plus HAART, respectively (Table 3). It was not detected in 18 amplicons before KRG intake. Among the six and nine isoleucines, 5 and 8 were detected together with premature stop codon, respectively. These genetic defects resulted from G-to-A hypermutations (by Hypermut 2.0; Los Alamos National Laboratory, Los Alamos, NM, USA). However, 3 on KRG only and 6 amplicons on KRG plus HAART did not showed genetic defects although they belong to G-to-A hypermutations (Table 3). Differing from the frequency of gross deletion, frequency of G-to-A hypermutations seems higher on KRG plus HAART than KRG only.
Proportion of overall genetic defects
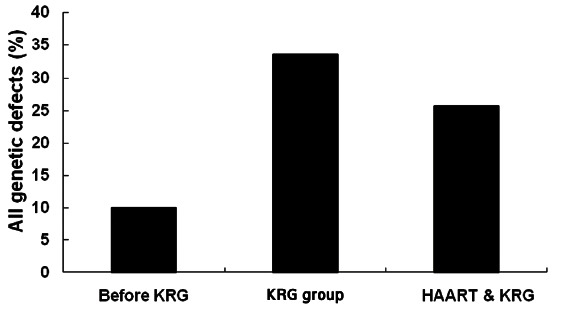
Adding the gross deletions to premature stop codon or isoleucine at initiation codon based on amplicon, the proportions of genetic defects were 10% (3/30) before KRG intake, 33.5% (63/188) on KRG only before HAART and 25.7% (55/214) on KRG plus HAART, respectively. The latter two groups showed significant high proportion of genetic defects compared with data before KRG intake (p< 0.001 for both) (Fig. 2).
Fig. 2. Changes of the proportion of all genetic defects including gross deletion in the 5’LTR/gag gene (gΔ). Proportion of genetic defects significantly increased on Korean red ginseng (KRG) intake only (63/188) and KRG plus highly active antiretroviral therapy (HAART, 55/214) compared with that prior to KRG intake (3/30) (respectively, p<0.001) although there was no significant difference between KRG intake only and KRG plus HAART. However, G-to-A hypermutations were higher on KRG plus HAART than KRG only.
Novel insertion, inversion, or duplication in 5’LTR
Novel insertions of 7 bp and 8 bp in 5’LTR were detected in an amplicon in HP-8 (GenBank numbers, EF370257 and EF370258, respectively). Deletion of 9 bp from position 108 and inversion of 609 bp and deletion of 5 bp was observed in seq 3476 in HP-6. A gross deletion and duplication of 105 bp in HP-8 was also observed (GenBank number, EF370252). In HP-13, unusual recombination of terminal 75bp was observed (seq3564s). In HP-20, seq4809 revealed deletion of 273 bp from 727 to 998 and it was replaced with insertion of env gene (367 bp from 8530 to 8896) (Table 2).
DISCUSSION
In this study, we found that frequency of gross deletion in 5’LTR/gag is also associated with KRG intake and that the frequency of gross deletion in 5’LTR/gag is significantly decreased on KRG plus HAART compared with that prior to HAART. These findings are the same as shown in the frequency of gΔnef in the previous study [26]. This observation may reflect a limit dilution because deleted genes might comprise a minor portion compared with to intact genes in vivo, thus decreasing the chance of detection of gross deletion in 5’LTR/gag gene. However, we found a few different findings compared with gΔnef in the same study population. First, there was no difference in the frequency of 5’LTR/gag on KRG only between 20 HPs (28.1%) and 3 HPs (-21, 22, and 23) (24.6%; 15/61) who were infected with non-KSB in other countries [12]. In contrast, our previous report showed that the frequency of gΔnef in 20 HPs (20.6%) was 10-fold higher than that (2/100) in 3 HPs (-21, 22, and 23) infected with non-KSB. Taken together, our data suggest that 5’LTR/gag region might be more sensitive to KRG intake than in the Δnef. Second, we obtained higher frequency of nonfunctional gene (premature stop codon and I instead of M as initiation codon in Gag protein) (10.3%, 11/107) on KRG intake only. In addition to nonfunctional gag gene, there were also frequent G-to-A hypermutations in the vif genes (unpublished data). These frequencies in 5’LTR/gag and vif genes were significantly higher compared to 0.96% (5/522) [26] in the Δnef in the same patients and 0% (0/277) in other patients (p<0.001) [28].
There are many reports on gΔnef reported in many studies. However, there is no report on gross deletion in 5’LTR/gag gene together with gΔnef. In detail, Huang et al. [32] reported genetic defects in the 5’LTR and gag genes of eight long-term survivors. In their work, full viral sequences were obtained. G-to-A hypermutations were seen in only one patient in gag gene and 5’LTR. However, there was no gross deletion in the 5’LTR and Δnef. Recently, Calugi et al. [33] identified gross deletions in the env gene (next to the Δnef). Blankson and Siliciano [34] and Blankson [35] did not detect any gross deletions in full-length sequences of HIV-1 even in elite suppressors, with viral loads of plasma RNA less than 50 copies/mL.
Despite many limitations, this study showed that the frequency of gross deletion in 5’LTR/gag is associated with KRG intake. Although further study is needed to determine the source of high frequency of genetic defects in HIV-1 in hemophiliacs, these data provide a new perspective on HIV-1 pathogenesis.
Acknowledgments
This work was supported by grants from the Korea Ginseng Corporation via Korean Society of Ginseng in 2008-2009. The authors thank professor Seung-Chul Yun of the Division of Biostatistics for statistical consultation.
References
- 1.Li CP, Li RC. An introductory note to ginseng. Am J Chin Med (Gard City N Y) 1973;1:249–261. doi: 10.1142/S0192415X73000279. [DOI] [PubMed] [Google Scholar]
- 2.Attele AS, Wu JA, Yuan CS. Ginseng pharmacology: multiple constituents and multiple actions. Biochem Pharmacol. 1999;58:1685–1693. doi: 10.1016/S0006-2952(99)00212-9. [DOI] [PubMed] [Google Scholar]
- 3.Kim DH. Metabolism of ginsenosides to bioactive compounds by intestinal microflora and its industrial application. J Ginseng Res. 2009;33:165–176. doi: 10.5142/JGR.2009.33.3.165. [DOI] [Google Scholar]
- 4.Singh VK, Agarwal SS, Gupta BM. Immunomodulatory activity of Panax ginseng extract. Planta Med. 1984;50:462–465. doi: 10.1055/s-2007-969773. [DOI] [PubMed] [Google Scholar]
- 5.Fulder SJ. Ginseng and the hypothalamic-pituitary control of stress. Am J Chin Med. 1981;9:112–118. doi: 10.1142/S0192415X81000159. [DOI] [PubMed] [Google Scholar]
- 6.Song X, Hu S. Adjuvant activities of saponins from traditional Chinese medicinal herbs. Vaccine. 2009;27:4883–4890. doi: 10.1016/j.vaccine.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 7.Jia L, Zhao Y, Liang XJ. Current evaluation of the millennium phytomedicine-ginseng (II): collected chemical entities, modern pharmacology, and clinical applications emanated from traditional Chinese medicine. Curr Med Chem. 2009;16:2924–2942. doi: 10.2174/092986709788803204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao H, Wang F, Lien EJ, Trousdale MD. Immunostimulating polysaccharides from Panax notoginseng. Pharm Res. 1996;13:1196–1200. doi: 10.1023/A:1016060119425. [DOI] [PubMed] [Google Scholar]
- 9.Rivera E, Hu S, Concha C. Ginseng and aluminium hydroxide act synergistically as vaccine adjuvants. Vaccine. 2003;21:1149–1157. doi: 10.1016/S0264-410X(02)00518-2. [DOI] [PubMed] [Google Scholar]
- 10.Hu S, Concha C, Lin F, Persson Waller K. Adjuvant effect of ginseng extracts on the immune responses to immunisation against Staphylococcus aureus in dairy cattle. Vet Immunol Immunopathol. 2003;91:29–37. doi: 10.1016/S0165-2427(02)00264-7. [DOI] [PubMed] [Google Scholar]
- 11.Sun HX, Ye YP, Pan HJ, Pan YJ. Adjuvant effect of Panax notoginseng saponins on the immune responses to ovalbumin in mice. Vaccine. 2004;22:3882–3889. doi: 10.1016/j.vaccine.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 12.Sun H, Ye Y, Pan Y. Immunological-adjuvant saponins from the roots of Panax notoginseng. Chem Biodivers. 2005;2:510–515. doi: 10.1002/cbdv.200590032. [DOI] [PubMed] [Google Scholar]
- 13.Qin F, Ye YP, Sun HX. Haemolytic activity and adjuvant effect of notoginsenoside K from the roots of Panax notoginseng. Chem Biodivers. 2006;3:1144–1152. doi: 10.1002/cbdv.200690116. [DOI] [PubMed] [Google Scholar]
- 14.Sun J, Hu S, Song X. Adjuvant effects of protopanaxadiol and protopanaxatriol saponins from ginseng roots on the immune responses to ovalbumin in mice. Vaccine. 2007;25:1114–1120. doi: 10.1016/j.vaccine.2006.09.054. [DOI] [PubMed] [Google Scholar]
- 15.Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 16.Deacon NJ, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker DJ, McPhee DA, Greenway AL, Ellett A, Chatfield C, et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 17.Mariani R, Kirchhoff F, Greenough TC, Sullivan JL, Desrosiers RC, Skowronski J. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J Virol. 1996;70:7752–7764. doi: 10.1128/jvi.70.11.7752-7764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes DI, Ashton L, Solomon A, Carr A, Cooper D, Kaldor J, Deacon N. Characterization of three nef-defective human immunodeficiency virus type 1 strains associated with long-term nonprogression. Australian Long-Term Nonprogressor Study Group. J Virol. 2000;74:10581–10588. doi: 10.1128/JVI.74.22.10581-10588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brambilla A, Turchetto L, Gatti A, Bovolenta C, Veglia F, Santagostino E, Gringeri A, Clementi M, Poli G, Bagnarelli P, et al. Defective nef alleles in a cohort of hemophiliacs with progressing and nonprogressing HIV-1 infection. Virology. 1999;259:349–368. doi: 10.1006/viro.1999.9783. [DOI] [PubMed] [Google Scholar]
- 20.Rodes B, Toro C, Paxinos E, Poveda E, Martinez-Padial M, Benito JM, Jimenez V, Wrin T, Bassani S, Soriano V. Differences in disease progression in a cohort of long-term non-progressors after more than 16 years of HIV-1 infection. AIDS. 2004;18:1109–1116. doi: 10.1097/00002030-200405210-00004. [DOI] [PubMed] [Google Scholar]
- 21.Roger M. Influence of host genes on HIV-1 disease progression. FASEB J. 1998;12:625–632. doi: 10.1096/fasebj.12.9.625. [DOI] [PubMed] [Google Scholar]
- 22.Salvi R, Garbuglia AR, Di Caro A, Pulciani S, Montella F, Benedetto A. Grossly defective nef gene sequences in a human immunodeficiency virus type 1-seropositive long-term nonprogressor. J Virol. 1998;72:3646–3657. doi: 10.1128/jvi.72.5.3646-3657.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miura T, Brockman MA, Brumme CJ, Brumme ZL, Carlson JM, Pereyra F, Trocha A, Addo MM, Block BL, Rothchild AC, et al. Genetic characterization of human immunodeficiency virus type 1 in elite controllers: lack of gross genetic defects or common amino acid changes. J Virol. 2008;82:8422–8430. doi: 10.1128/JVI.00535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho YK, Lim JY, Jung YS, Oh SK, Lee HJ, Sung H. High frequency of grossly deleted nef genes in HIV-1 infected long-term slow progressors treated with Korean red ginseng. Curr HIV Res. 2006;4:447–457. doi: 10.2174/157016206778560072. [DOI] [PubMed] [Google Scholar]
- 25.Cho YK, Jung YS. High frequency of gross deletions in the 5' LTR and gag regions in HIV type 1-infected long-term survivors treated with Korean red ginseng. AIDS Res Hum Retroviruses. 2008;24:181–193. doi: 10.1089/aid.2007.0143. [DOI] [PubMed] [Google Scholar]
- 26.Cho YK, Jung YS, Sung H. Frequent gross deletion in the HIV type 1 nef gene in hemophiliacs treated with Korean red ginseng: inhibition of detection by highly active antiretroviral therapy. AIDS Res Hum Retroviruses. 2009;25:419–424. doi: 10.1089/aid.2008.0178. [DOI] [PubMed] [Google Scholar]
- 27.Cho YK, Jung YS, Sung H, Sim MK, Kim YK. High frequency of gross deletions in 5' LTR/ gag and nef genes in patients infected with CRF02_AG of HIV type 1 who survived for over 20 years: an association with Korean red ginseng. AIDS Res Hum Retroviruses. 2009;25:535–541. doi: 10.1089/aid.2008.0301. [DOI] [PubMed] [Google Scholar]
- 28.Cho YK, Jung YS. Dosage and duration effects of Korean red ginseng intake on frequency of gross deletions in the nef gene. J Ginseng Res. 2010;34:219–225. doi: 10.5142/jgr.2010.34.3.219. [DOI] [Google Scholar]
- 29.Cho YK, Foley BT, Sung H, Kim YB, Kim JH. Molecular epidemiologic study of a human immunodeficiency virus 1 outbreak in haemophiliacs B infected through clotting factor 9 after 1990. Vox Sang. 2007;92:113–120. doi: 10.1111/j.1423-0410.2006.00866.x. [DOI] [PubMed] [Google Scholar]
- 30.Cho YK, Sung H, Kim TK, Lim J, Jung YS, Kang SM. Korean red ginseng significantly slows CD4 T cell depletion over 10 years in HIV-1 infected patients: association with HLA. J Ginseng Res. 2004;28:173–182. doi: 10.5142/JGR.2004.28.4.173. [DOI] [Google Scholar]
- 31.Sung H, Kang SM, Lee MS, Kim TG, Cho YK. Korean red ginseng slows depletion of CD4 T cells in human immunodeficiency virus type 1-infected patients. Clin Diagn Lab Immunol. 2005;12:497–501. doi: 10.1128/CDLI.12.4.497-501.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y, Zhang L, Ho DD. Characterization of gag and pol sequences from long-term survivors of human immunodeficiency virus type 1 infection. Virology. 1998;240:36–49. doi: 10.1006/viro.1997.8913. [DOI] [PubMed] [Google Scholar]
- 33.Calugi G, Montella F, Favalli C, Benedetto A. Entire genome of a strain of human immunodeficiency virus type 1 with a deletion of nef that was recovered 20 years after primary infection: large pool of proviruses with deletions of env. J Virol. 2006;80:11892–11896. doi: 10.1128/JVI.00932-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blankson JN, Siliciano RF. Elite suppression of HIV-1 replication. Immunity. 2008;29:845–847. doi: 10.1016/j.immuni.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Blankson JN. Effector mechanisms in HIV-1 infected elite controllers: highly active immune responses? Antiviral Res. 2010;85:295–302. doi: 10.1016/j.antiviral.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


