Abstract
In order to investigate the diversity of endophytes, fungal endophytes in Panax ginseng Meyer cultivated in Korea were isolated and identified using internal transcribed spacer (ITS) sequences of ribosomal DNA. Three cultivars of 3-year-old ginseng roots (Chunpoong, Yunpoong, and Gumpoong) were used to isolate fungal endophytes. Surface sterilized ginseng roots were placed on potato dextrose agar plates supplemented with ampicilin and streptomycin to inhibit bacterial growth. Overall, 38 fungal endophytes were isolated from 12 ginseng roots. According to the sequence analysis of the ITS1-5.8S-ITS2, 38 fungal isolates were classified into 4 different fungal species, which were Phoma radicina, Fusarium oxysporum, Setophoma terrestris and Ascomycota sp. 2-RNK. The most dominant fungal endophyte was P. radicina in 3 cultivars. The percentage of dominant endophytes of P. radicina was 65.8%. The percentage of colonization frequency of P. radicina was 80%, 52.9%, and 75% in Chunpoong, Yunpoong, and Gumpoong, respectively. The second most dominant fungal endophyte was F. oxysporum. The diversity of the fungal endophytes was low and no ginseng cultivar specificity among endophytes was detected in this study. The identified endophytes can be potential fungi for the production of bioactive compounds and control against ginseng pathogens.
Keywords: Panax ginseng, Fungal endophytes, Internal transcribed spacer (ITS) sequence
INTRODUCTION
Ginseng (Panax ginseng Meyer) is the most valuable traditional herb. Ginseng has well-known, diverse actions and effects on the human body, such as nonspecific resistance to biochemical and physical stresses, and the improvement of vitality, longevity and mental capacity [1-6]. Generally 4 to 6 years of growth are required to produce high quality ginseng roots and the cultivation of ginseng should be under shade conditions. Consecutive cultivation in the same soil causes severe reduction in production mainly due to pathogenic infection [7]. Heavy use of chemical pesticides has been applied to ginseng fields to control pathogens, which results in the contamination of ginseng roots and the surrounding soil. The importance of biological control methods is now widely recognized to produce organic ginseng roots and reduce environmental contamination.
De Barry [8] first used the term ‘endophyte’ to describe microbes that reside inside the living tissues of healthy plants. Endophytes were subsequently described as fungi and bacteria that spend the most or part of their life cycle internally and asymptomatically in the healthy living tissues of plants [9,10]. It is believed that fungal endophytes originated from pathogenic fungi which either lost their virulence or exhibit extended latent periods [11,12]. Fungal endophytes colonize either locally or systemically in inter- or intra-cellular locations [13]. Many studies have shown that fungal endophytes are ubiquitous in most plants and colonize without apparent harm to their hosts [14]. However, fungal endophytes could be pathogenic to other species [13,15]. Various relationship between fungal endophytes and plants has been observed, which ranges from mutualistic or symbiotic to antagonistic or pathogenic [13,16]. There are at least one million estimated species of fungal endophytes in plants [17] as well as in lichens [14]. Most fungal endophytes belong to the Ascomycota clade and some of them belong to the Basidiomycota [18]. Rodriguez et al. [19] identified four classes of fungal endophytes based on host range, colonization pattern, transmission, and ecological function. The function of fungal endophytes might be dependent upon the plant organ-fungus interaction in each plant [20,21]. Beneficial endophytes have several positive effects on plants, including growth, nutrient uptake, and tolerance to abiotic and biotic stresses [22].
Recent studies have shed light on the isolation of fungal endophytes from medicinal plants [23,24]. Medicinal plants produce chemicals that can be used as raw material for pharmaceutical, cosmetic, and fragrance industries [25]. Some fungal endophytes produce secondary metabolites that are potential sources for natural products with unique structures and bioactivities [26]. Endophytes in medicinal plants may be involved in the metabolic pathways of medicinal plants and produce analogous or novel bioactive metabolites (e.g., taxol) [27]. Fungal endophytes produce metabolites with potential use for medicinal, agricultural and industrial purposes, such as antibiotics, antimycotics, immuno suppressants, and anticancer compounds [28-31]. In addition, fungi have advantages over plants, such as short cultivation time and large biomass production. However, there are very few reports on fungal endophytes in ginseng [32-34]. Therefore, an extensive investigation of fungal endophytes in ginseng can increase the opportunities of identifying potential fungi that can be used to produce bioactive compounds and control against pathogens. This study has been undertaken to study the diversity of fungal endophytes in three cultivars of Panax ginseng Meyer cultivated in Korea.
MATERIALS AND METHODS
Collection of ginseng roots
Three-year-old roots of ginseng, P. ginseng Meyer, were harvested from Gangwon Province, Korea during sunny days in August 2010. Four roots of each cultivar (Chunpoong, Yunpoong, and Gumpoong; 12 roots in total) were harvested, and these were stored at 4℃ before being processed.
Isolation of fungal endophytes
Fungal endophytes from ginseng roots were isolated according to Xing et al. [33] with modification. Root samples were thoroughly washed with running tap water and cut into 1 cm long segments with a clean razor blade. The root segments were surface sterilized with 75% ethanol for 1 min, 4% NaHCO3 for 3 min, and 75% ethanol for 30 s. The root segments were rinsed three times with sterile distilled water and blotted with sterile tissue paper. Three aliquots of 0.1 mL of the water used for the last washing step were inoculated on potato dextrose agar (PDA) plates with 200 μg/mL ampicillin and 200 μg/ mL streptomycin to ensure the elimination of ephiphytic microorganisms. The sterilized root segments were transferred to PDA plates amended with ampicillin and streptomycin. PDA plates were incubated at room temperature and checked every day to detect mycelial growth out of the roots. The fungal mycelial tips of the emerging mycelia from the edges of the root segments were transferred to new PDA plates supplemented with ampicillin and streptomycin. The transfer of emerging mycelia from the root segments was continued for up to 4 wk.
Identification of fungal isolates
Mycelia were scraped using a sterile scalpel from 1- or 2-week-old PDA fungal cultures. The harvested fungal mycelia were frozen in liquid nitrogen and stored at -80℃ until use. Mycelia were ground to a fine powder using a mortar and pestle in liquid nitrogen with sea sand. DNA was extracted using DNeasy Plant Mini kit (Qiagen, CA, USA) according to manufacturer’s recommendation. Amplification of the internal transcribed spacer (ITS) region was carried out using the universal eukaryotic primers of ITS1 (5′ TCCGTAGGTGAA CCTGCGG 3′) and ITS4 (5′ TCCTCCGCTTATTGATATGC 3′) [32]. PCR was performed in 50 μL reaction containing 0.5 μg of DNA, 25 ρmol of each primer with the following reaction conditions: 2 min initial denaturing step at 95℃, followed by 30 cycles of 1 min denaturation at 95℃, 1 min primer annealing at 55℃, 1 min extension at 72℃, and a final 5 min extension at 72℃. PCR products were analyzed by electrophoresis in 0.8% agarose gel and the PCR products (approximately 550 bp) were excised from the gel. DNA was purified using Wizard SV gel and PCR Clean-up System (Promega, WI, USA). After sequence analysis, ITS sequences were searched using the NCBI BLAST program (http://www.ncbi.nlm.nih.gov).
Morphological identification was also carried out to confirm the results of molecular identification based on macroscopic and microscopic appearances. The percentage of colonization frequency (%CF) of fungal endophytes was calculated based on previous studies [35,36] as follows: %CF=(NCOL/Nt)×100, where NCOL=number of segments colonized by each fungus; Nt=total number of segments.
Phylogenetic analysis
A phylogenetic tree was constructed from ITS1-5.8SITS2 sequences by the Maximum Parsimony method and Mega5 software (http://www.megasoftware.net) [37,38]. The Maximum Parsimony tree was generated using the Close-Neighbor-Interchange algorithm [39]. Sclerotinia sclerotiorum was used as an outgroup fungal taxon.
RESULTS AND DISCUSSION
Total of 38 fungal endophytes were isolated from 12 ginseng roots (184 segments) of 3 cultivars. These were classified into 4 taxonomic species of Ascomycota (Table 1 and Fig. 1). Chunpoong (56 root segments), Yunpoong (74 segments), and Gumpoong (54 segments) were colonized by 5, 17, and 16 fungal isolates, respectively. Only 5 fungal isolates were identified in Chunpoong, which may be due to the contamination or outgrowth of endophytic bacteria. Bacterial growth usually inhibited the outcome of fungal endophytes due to the fast colonization of root segments, even on PDA plates containing antibiotics.
Table 1.
Fungal endophytes isolated from 3-year-old ginseng roots of 3 cultivars in Gangwon province in Korea
| Endophytic fungi cultivar | Phoma radicina FJ427058 | Fusarium oxysporum HQ328030 | Setophoma terrestris JN615482 | Ascomycota sp. 2-RNK EU780424 | Isolate no. in total | CF (%) | Average of CF (%) |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Chunpoong 1 | 1 | 1 | 6.7 | ||||
| Chunpoong 2 | 0 | 0.0 | |||||
| Chunpoong 3 | 2 | 2 | 15.4 | ||||
| Chunpoong 4 | 1 | 1 | 2 | 15.4 | |||
| DE (%) | 80 | 20 | 0 | 0 | 100 | 9.4 | |
| Yunpoong 1 | 5 | 1 | 6 | 46.2 | |||
| Yunpoong 2 | 3 | 2 | 5 | 20.8 | |||
| Yunpoong 3 | 3 | 1 | 4 | 19.0 | |||
| Yunpoong 4 | 1 | 1 | 2 | 12.5 | |||
| DE (%) | 52.9 | 23.6 | 17.6 | 5.9 | 100 | 24.6 | |
| Gumpoong 1 | 2 | 2 | 28.6 | ||||
| Gumpoong 2 | 5 | 1 | 6 | 46.2 | |||
| Gumpoong 3 | 4 | 1 | 5 | 22.7 | |||
| Gumpoong 4 | 1 | 2 | 3 | 25 | |||
| DE (%) | 75 | 18.7 | 0.0 | 6.3 | 100 | 30.6 | |
| Isolate no. in total | 25 | 8 | 3 | 2 | 38 | ||
| DE (%) in total | 65.8 | 21.0 | 7.9 | 5.3 | 100 | 21.5 | |
Four roots of each cultivar were used to isolate fungal endophytes (12 roots in total).
CF, colonization frequency; DE, dominant endophyte.
Fig. 1. Endophytic fungi isolated from ginseng roots cultivated in Korea. Fungal isolates were cultivated in potato dextrose agar media for 5 d at 22℃. (A) Phoma radicina, (B) Fusarium oxysporum, (C) Setophoma terrestris, and (D) Ascomycota sp. 2-RNK.
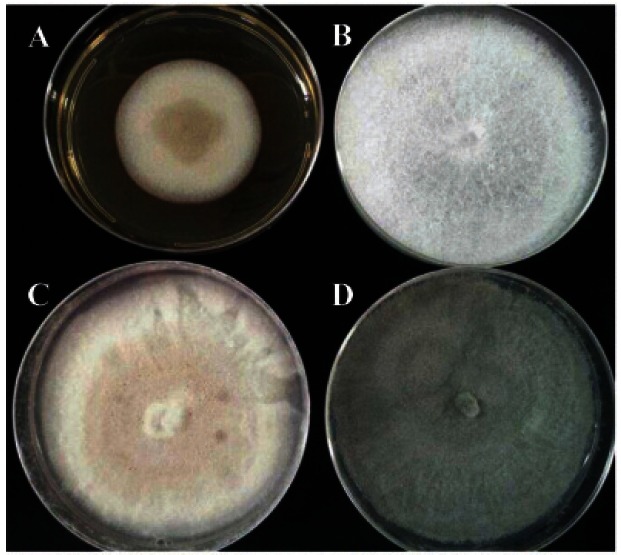
The analyses of ITS1-5.8S-ITS2 regions showed 100% identities of Phoma radicina (FJ427058), Fusarium oxysporum (HQ328030), Setophoma terrestris (JN615482: synonym Phoma trrestris, Pyrenochaeta terrestris) and Ascomycota sp. 2-RNK (EU780424) (Table 1). The highest CF varied among cultivars from 9.4% to 30.6% and the average CF was 21.5% (Table 1), which may also be due to bacterial contamination or growth of bacterial endophytes. Phoma radicina was the most frequent fungal endophyte in three ginseng cultivars: 80%, 52.9%, and 75% of CF in Chunpoong, Yunpoong, and Gumpoong, respectively. In total, the percentage of dominant endophytes (DE) of P. radicina was 65.8%, which is the highest percentage among the detected fungal isolates. The second most dominant species was F. oxysporum: 20%, 23.5%, and 18.7% of DE in Chunpoong, Yunpoong, and Gumpoong, respectively. The remaining fungal endophytes isolated were S. terrestris and Ascomycota sp. 2-RNK and %CF were 7.9% and 5.3% in average, respectively.
The number of fungal endophytes found in this study is lower than expected compared with other studies [24,33,40,41]. However, Dang et al. [34] only reported one fungal isolate (Trichoderma ovalisporum) in P. ginseng. This result indicates that distribution of endophytes depends on plant species and geological locations. As mentioned earlier, there are only a limited number of studies reported on the isolation of fungal endophytes in ginseng species. Xing et al. [33] reported 134 fungal isolates with 27 taxa in American ginseng (P. quinquefolium), in which 11 species were identified in detail. Four species (Alternaria, Collectotrichum, Phoma, and Xylariale) were the most common in 3 different tissues (root, stem and leaf) of American ginseng. These genera are known as common endophytes [23,42-44]. Xing et al. [33] reported tissue specificity for the reported endophytes; Cladosporium sp. were the dominant isolates in roots, but were not detected in stem or leaf tissues. In addition, Cladosporium sp. was not detected in 4-year-old American ginseng roots, but Glomerella cingulata was the dominant species instead. However, we did not find Cladosporium sp. in ginseng roots. They also found that the diversity of fungal endophytes in American ginseng roots decreased with age. This may be because of autotoxic or host defense compounds of American ginseng in the rhizosphere [33,45,46]. It has been known that host defense compounds control endophytic communities [47].
The genus, Phoma sp., which was the most dominant isolate in ginseng, was detected only in leaf tissues of 1-, 2- and 3-year-old American ginseng, but not in 4-year old ginseng. Tissue specificity of endophytes has also been reported in previous studies [48-50]. F. oxysporum was the second dominant isolate in ginseng, while it was only detected in 3-year old roots of American ginseng. The genus Phoma is ubiquitously present in the environment and considered an important fungal plant pathogen [51]. In other plant species, Fusarium and Phoma were the most frequent genera among 142 fungal species isolated in roots of 24 plant species growing at 12 sites in Spain [52]. The genera of Fusarium and Phoma have been known as common endophytes in other studies [13,53,54]. Aveskamp et al. [51] concluded that endophytic communities were dependent on the soil type and plant species, but not on location. The physio-chemical nature of a soil may influence on the colonization of fungal species to plants. While F. oxysporum is commonly present in legume species as a pathogen [55], this fungus can colonize other plants without causing symptoms [54,56]. In general, nonpathogenic endophytes can turn into pathogenic strains after introduction into potentially new host plants [56]. We isolated the unclassified Ascomycota sp. 2-RNK that was also isolated from the roots and fruits of neem tree (Azadirachta indica A. Juss.) [57]. S. terrestris was isolated in ginseng root. Gorenz et al. [58] isolated Pyrenochaeta terrestris as a causal agent of pink root in onions. Fig. 2 shows the maximum parsimony tree grouping fungal endophytes into two clades: 1) P. radicina and S. terrestris, and 2) F. oxysporum and Ascomycota sp. 2-RNK.
Fig. 2. Phylogenetic analysis of fungal endophytes from Panax ginseng Meyer. The phylogenetic tree was constructed from internal transcribed spacer (ITS)1-5.8S-ITS2 sequences using the maximum parsimony method on Mega5 software. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches. GenBank accession numbers are as follows: Phoma radicina (FJ427058), Setophoma terrestris (JN615482), Fusarium oxysporum (HQ328030) and Ascomycota sp. 2-RNK (EU780424). Sclerotinia sclerotiorum (AB607227) was used as outgroup fungal taxa.

We isolated a limited number of fungal endophytes in ginseng root, but our findings can increase the potential use and awareness of fungal endophytes for beneficial applications. There are very few reports on the diversity of fungal endophytes in ginseng. Therefore, our finding can increase the possibilities of identifying potential fungi that can be used to protect ginseng plants and produce bioactive compounds.
Acknowledgments
We thank Dr. Lee (Andong University, Andong, Korea) for assistance in morphologically identifying fungal endophytes. This research was supported by the Yeungnam University Research Grant in 209-A-356-011.
References
- 1.Ernst E. Panax ginseng: an overview of the clinical evidence. J Ginseng Res. 2010;34:259–263. doi: 10.5142/jgr.2010.34.4.259. [DOI] [Google Scholar]
- 2.Lee ST, Chu K, Kim JM, Park HJ, Kim MH. Cognitive improvement by ginseng in Alzheimer’s disease. J Ginseng Res. 2007;31:51–53. doi: 10.5142/JGR.2007.31.1.051. [DOI] [Google Scholar]
- 3.Rausch WD, Weiming L, Gille G, Radad K. Perspectives for ginsenosides in models of Parkinson’s disease. J Ginseng Res. 2007;31:127–136. doi: 10.5142/JGR.2007.31.3.127. [DOI] [Google Scholar]
- 4.Vuksan V, Sievenpipper J, Jovanovski E, Jenkins AL. Current clinical evidence for Korean red ginseng in management of diabetes and vascular disease: a Toronto’s ginseng clinical testing program. J Ginseng Res. 2010;34:264–273. doi: 10.5142/jgr.2010.34.4.264. [DOI] [Google Scholar]
- 5.Nam KY. Clinical applications and efficacy of Korean ginseng (Panax ginseng C.A. Meyer). J Ginseng Res. 2002;26:111–131. doi: 10.5142/JGR.2002.26.3.111. [DOI] [Google Scholar]
- 6.Yuan CS, Wang CZ, Wicks SM, Qi LW. Chemical and pharmacological studies of saponins with a focus on American ginseng. J Ginseng Res. 2010;34:160–167. doi: 10.5142/jgr.2010.34.3.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho DH, Park KJ, Yu YH, Ohh SH, Lee HS. Root-rot development of 2-year old ginseng (Panax ginseng C.A. Meyer) caused by Cylindrocarpon destructans (Zinssm.) Scholten in the continuous cultivation filed. Korean J Ginseng Sci. 1995;19:175–180. [Google Scholar]
- 8.De Bary A. Hofmeister’s handbook of physiological botany. Vol. 2. Leipzig; Amsterdam: 1866. [Google Scholar]
- 9.Sturz AV, Nowak J. Endophytic communities of rhizobacteria and the strategies required to create yield enhancing associations with crops. Appl Soil Ecol. 2000;15:183–190. doi: 10.1016/S0929-1393(00)00094-9. [DOI] [Google Scholar]
- 10.Wilson D. Endophytes: the evolution of a term, and clarification of its use and definition. Oikos. 1995;73:274–276. doi: 10.2307/3545919. [DOI] [Google Scholar]
- 11.Saikkonen K, Faeth SH, Helander M, Sullivan TJ. Fungal endophytes: a continuum of interactions with host plants. Annu Rev Ecol Syst. 1998;29:319–343. doi: 10.1146/annurev.ecolsys.29.1.319. [DOI] [Google Scholar]
- 12.Schardl CL, Clay K. Evolution of mutualistic endophytes from plant pathogens. In: Carrol G. The mycota. V. Plant relationships, part B. Springer; Berlin: 2007. pp. 221–238. [Google Scholar]
- 13.Schulz B, Boyle C. The endophytic continuum. Mycol Res. 2005;109(Pt 6):661–686. doi: 10.1017/S095375620500273X. [DOI] [PubMed] [Google Scholar]
- 14.Li TY, Zeng HL, Ping Y, Lin H, Fan XL, Guo ZG, Zhang CF. Construction of a stable expression vector for Leifsonia xyli subsp. cynodontis and its application in studying the effect of the bacterium as an endophytic bacterium in rice. FEMS Microbiol Lett. 2007;267:176–183. doi: 10.1111/j.1574-6968.2006.00551.x. [DOI] [PubMed] [Google Scholar]
- 15.Shiva P, Pental ND, Bhalla-Sarin N. Regeneration of pigeonpea (Cajanus cajan) from cotyledonary node via multiple shoot formation. Plant Cell Rep. 1994;13:623–627. doi: 10.1007/BF00232933. [DOI] [PubMed] [Google Scholar]
- 16.Arnold AE, Lutzoni F. Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology. 2007;88:541–549. doi: 10.1890/05-1459. [DOI] [PubMed] [Google Scholar]
- 17.Dreyfuss MM, Chapela IH. Potential of fungi in the discovery of novel, low molecular weight pharmaceuticals. In: Gullo VP. The discovery of natural products with therapeutic potential. Butterworth-Heinemann; Boston: 1994. pp. 49–80. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez R, Redman R. More than 400 million years of evolution and some plants still can’t make it on their own: plant stress tolerance via fungal symbiosis. J Exp Bot. 2008;59:1109–1114. doi: 10.1093/jxb/erm342. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez RJ, White JF Jr, Arnold AE, Redman RS. Fungal endophytes: diversity and functional roles. New Phytol. 2009;182:314–330. doi: 10.1111/j.1469-8137.2009.02773.x. [DOI] [PubMed] [Google Scholar]
- 20.Boyle C, Gotz M, Dammann-Tugend U, Schulz B. Endophyte- host interactions. III. Local vs. systemic colonization. Symbiosis. 2001;31:259–281. [Google Scholar]
- 21.Faeth SH. Are endophytic fungi defensive plant mutualists? Oikos. 2002;98:25–36. doi: 10.1034/j.1600-0706.2002.980103.x. [DOI] [Google Scholar]
- 22.Brundrett MC. Understanding the roles of multifunctional mycorrhizal and endophytic fungi. In: Schulz BJ, Boyle CJ, Sieber TN, eds. Microbial root endophytes. Springer; Berlin: 2006. pp. 107–132. [Google Scholar]
- 23.Kumar DS, Hyde KD. Biodiversity and tissue-recurrence of endophytic fungi in Tripterygium wilfordii. Fungal Divers. 2004;17:69–90. [Google Scholar]
- 24.Huang WY, Cai YZ, Hyde KD, Corke H, Sun M. Biodiversity of endophytic fungi associated with 29 traditional Chinese medicinal plants. Fungal Divers. 2008;33:61–75. [Google Scholar]
- 25.Karthikeyan B, Jaleel CA, Lakshmanan GM, Deiveekasundaram M. Studies on rhizosphere microbial diversity of some commercially important medicinal plants. Colloids Surf B Biointerfaces. 2008;62:143–145. doi: 10.1016/j.colsurfb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Weber RW, Kappe R, Paululat T, Mosker E, Anke H. Anti-Candida metabolites from endophytic fungi. Phytochemistry. 2007;68:886–892. doi: 10.1016/j.phytochem.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 27.Stierle A, Strobel G, Stierle D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science. 1993;260:214–216. doi: 10.1126/science.8097061. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q, Kang X, Zhao W. Antiangiogenic effect of low-dose cyclophosphamide combined with ginsenoside Rg3 on Lewis lung carcinoma. Biochem Biophys Res Commun. 2006;342:824–828. doi: 10.1016/j.bbrc.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 29.Tejesvi MV, Kini KR, Prakash HS, Subbiah V, Shetty HS. Genetic diversity and antifungal activity of species of Pestalotiopsis isolated as endophytes from medicinal plants. Fungal Divers. 2007;24:37–54. [Google Scholar]
- 30.Mitchell AM, Strobel GA, Hess WM, Vargas PN, Ezra D. Muscodor crispans, a novel endophyte from Ananas ananassoides in the Bolivian Amazon. Fungal Diviers. 2008;31:37–43. [Google Scholar]
- 31.Aly AH, Debbab A, Kjer J, Proksch P. Fungal endophytes from higher plants: a profilic source of phytochemicals and other bioactive natural products. Fungal Divers. 2010;41:1–16. doi: 10.1007/s13225-010-0034-4. [DOI] [Google Scholar]
- 32.Xu LL, Han T, Wu JZ, Zhang QY, Zhang H, Huang BK, Rahman K, Qin LP. Comparative research of chemical constituents, antifungal and antitumor properties of ether extracts of Panax ginseng and its endophytic fungus. Phytomedicine. 2009;16:609–616. doi: 10.1016/j.phymed.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 33.Xing X, Guo S, Fu J. Biodiversity and distribution of endophytic fungi associated with Panax quinquefolium L. cultivated in a forest reserve. Symbiosis. 2010;51:161–166. doi: 10.1007/s13199-010-0062-6. [DOI] [Google Scholar]
- 34.Dang L, Li G, Yang Z, Luo S, Zheng X, Zhang K. Chemical constituents from the endophytic fungus Trichoderma ovalisporum isolated from Panax notoginseng. Ann Microbiol. 2010;60:317–320. doi: 10.1007/s13213-010-0043-2. [DOI] [Google Scholar]
- 35.White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal genes for phylogenetics. In: Innis M, Gelfand DH, Sninsky JJ, White TJ. PCR protocols: a guide to methods and applications. Academic Press; San Diego: 1990. pp. 315–322. [Google Scholar]
- 36.Hata K, Futai K. Endophytic fungi associated with healthy pine needles and needles infested by the pine needle gall midge, Thecodiplosis japonensis. Can J Bot. 1995;73:384–390. [Google Scholar]
- 37.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 39.Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- 40.Gotz M, Nirenberg H, Krause S, Wolters H, Draeger S, Buchner A, Lottmann J, Berg G, Smalla K. Fungal endophytes in potato roots studied by traditional isolation and cultivation-independent DNA-based methods. FEMS Microbiol Ecol. 2006;58:404–413. doi: 10.1111/j.1574-6941.2006.00169.x. [DOI] [PubMed] [Google Scholar]
- 41.Suryanarayanan TS, Wittlinger SK, Faeth SH. Endophytic fungi associated with cacti in Arizona. Mycol Res. 2005;109(Pt 5):635–639. doi: 10.1017/S0953756205002753. [DOI] [PubMed] [Google Scholar]
- 42.Taylor JE, Hyde KD, Jones EB. Endophytic fungi associated with the temperate palm, Trachycarpus fortunei, within and outside its natural geographic range. New Phytol. 1999;142:335–346. doi: 10.1046/j.1469-8137.1999.00391.x. [DOI] [Google Scholar]
- 43.Kumaresan V, Suryanarayanan TS. Occurrence and distribution of endophytic fungi in a mangrove community. Mycol Res. 2001;105:1388–1391. doi: 10.1017/S0953756201004841. [DOI] [Google Scholar]
- 44.Ananda K, Sridhar KR. Diversity of endophytic fungi in the roots of mangrove species on the west coast of India. Can J Microbiol. 2002;48:871–878. doi: 10.1139/w02-080. [DOI] [PubMed] [Google Scholar]
- 45.Zhao YJ, Wang YP, Shao D, Yang JS, Liu D. Autotoxicity of Panax quinquefolium L. Allelophathy J. 2005;15:67–74. [Google Scholar]
- 46.He CN, Gao WW, Yang JX, Bi W, Zhang XS, Zhao YJ. Identification of autotoxic compounds from fibrous roots of Panax quinquefolium L. Plant Soil. 2009;318:63–72. doi: 10.1007/s11104-008-9817-8. [DOI] [Google Scholar]
- 47.Saunders M, Kohn LM. Evidence for alteration of fungal endophyte community assembly by host defense compounds. New Phytol. 2009;182:229–238. doi: 10.1111/j.1469-8137.2008.02746.x. [DOI] [PubMed] [Google Scholar]
- 48.Fisher PJ, Petrini O. A comparative study of fungal endophytes in xylem and bark of Alnus species in England and Switzerland. Mycol Res. 1990;94:313–319. doi: 10.1016/S0953-7562(09)80356-0. [DOI] [Google Scholar]
- 49.Frohlich J, Hyde KD, Petrini O. Endophytic fungi associated with palms. Mycol Res. 2000;104:1202–1212. doi: 10.1017/S095375620000263X. [DOI] [Google Scholar]
- 50.Ganley RJ, Newcombe G. Fungal endophytes in seeds and needles of Pinus monticola. Mycol Res. 2006;110(Pt 3):318–327. doi: 10.1016/j.mycres.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 51.Aveskamp MM, De Gruyter J, Crous PW. Biology and recent developments in the systematics of Phoma, a complex genus of major quarantine significance. Fungal Divers. 2008;31:1–18. [Google Scholar]
- 52.Macia-Vicente JG, Jansson HB, Abdullah SK, Descals E, Salinas J, Lopez-Llorca LV. Fungal root endophytes from natural vegetation in Mediterranean environments with special reference to Fusarium spp. FEMS Microbiol Ecol. 2008;64:90–105. doi: 10.1111/j.1574-6941.2007.00443.x. [DOI] [PubMed] [Google Scholar]
- 53.Frohlich J, Hyde KD, Petrini O. Endophytic fungi associated with palms. Mycol Res. 2000;104:1202–1212. doi: 10.1017/S095375620000263X. [DOI] [Google Scholar]
- 54.Sieber TN. Fungal root endophytes. In: Waisel Y, Eshel A, Kafkafi U, eds. Plant roots: the hidden half. Marcel Dekker; New York: 2002. pp. 887–917. [Google Scholar]
- 55.Venuto BC, Smith RR, Grau CR. Virulence, legume host specificity, and genetic relatedness of isolates of Fusarium oxysporum from red clover. Plant Dis. 1995;79:406–410. doi: 10.1094/PD-79-0406. [DOI] [Google Scholar]
- 56.Summerell BA, Leslie JF. Genetic diversity and population structure of plant pathogenic species in the genus Fusarium. In: Gillings M, Holmes AJ, eds. Plant microbiology. Bios Science Publishers; Oxford: 2004. pp. 207–223. [Google Scholar]
- 57.Verma VC, Gond SK, Kumar A, Kharwar RN, Strobel G. The endophytic mycoflora of bark, leaf, and stem tissues of Azadirachta indica A. Juss (neem) from Varanasi (India). Microb Ecol. 2007;54:119–125. doi: 10.1007/s00248-006-9179-9. [DOI] [PubMed] [Google Scholar]
- 58.Gorenz AM, Walker JC, Larson RH. Morphology and taxonomy of the onion pink-root fungus. Phytopathology. 1948;38:831–840. [Google Scholar]


