Abstract
Ginsenoside (ginseng saponin), the principal component of ginseng, is responsible for the pharmacological and biological activities of ginseng. We isolated lactic acid bacteria from Kimchi using esculin agar, to produce β-glucosidase. We focused on the bio-transformation of ginsenoside. Phylogenetic analysis was performed by comparing the 16S rRNA sequences. We identified the strain as Lactobacillus (strain 6105). In order to determine the optimal conditions for enzyme activity, the crude enzyme was incubated with 1 mM ginsenoside Rb1 to catalyse the reaction. A carbon substrate, such as cellobiose, lactose, and sucrose, resulted in the highest yields of β-glucosidase activity. Biotransformations of ginsenoside Rb1 were analyzed using TLC and HPLC. Our results confirmed that the microbial enzyme of strain 6105 significantly transformed ginsenoside as follows: Rb1→gypenoside XVII, Rd→F2 into compound K. Our results indicate that this is the best possible way to obtain specific ginsenosides using microbial enzymes from 6105 culture.
Keywords: Panax ginseng, Lactobacillus pentosus, Transformation, Ginsenoside
INTRODUCTION
Panax ginseng Meyer is one of the most valuable Oriental medicinal plants. Ginseng saponin (ginsenoside) is the major pharmacological component in ginseng. More than 100 ginsenosides have been isolated from ginseng plant [1,2]. Among these, six major ginsenosides, such as Rb1, Rb2, Rc, Rd, Re, and Rg1, constitute more than 90% of the total ginsenosides in white and red ginseng [3]. In recent decades, many studies have been reported the successful transformation of these major forms of ginsenoside into the more active minor forms of ginsenosides, such as Rg3, Rh2, and compound K (anti-tumor, antidiabetic, antistress, and antioxidative) [4]. Minor saponins has high pharmacological activity, however the natural availability of these saponins are very less in ginseng. Studies have been reported the success of converting major ginsenosides to the minor saponin varieties using methods such as heating [5,6], acid hydrolysis [7,8], and microbial enzymatic conversion [9-11]. Ginsenoside Rb1 has four glucose molecules with (1→2)- β-glycosidic linkages at C-3 and (1→6)-β-glycosidic linkages at C-20.
Kimchi is a traditional fermented food in Korea that accompanies many meals. The ingredients include Korean cabbage, vegetables, garlic, and other seasonings. During Kimchi fermentation, lactic acid bacteria (LAB) produce lactic acid by hydrolyzing carbohydrates (glucose and lactose). LAB is not only widely recognized, but is also well known for its physiological effects such as drug metabolism, antimutation, anticancer, gastric secretomotor activity, and radiation resistance [12-14].
In this study, we isolated β-glucosidase producing bacteria from Kimchi and tested the bio conversion ability of ginsenoside Rb1. The crude enzyme from strain 6105 was reacted with ginsenoside Rb1 to convert it into specific components. Finally, the results were confirmed with TLC and HPLC.
MATERIALS AND METHODS
Materials
Ginsenoside Rb1 was obtained from the Ginseng Genetic Resource Bank at Kyung Hee University, Korea. Standard ginsenosides including 20(S)-Rb1, 20(S)-Rd, 20(S)-Rg3, 20(S)-Rh2, and compound-K were obtained from KT&G in Daejeon, Korea. De Man, Rogosa and Sharpe (MRS) broth was purchased from Difco (Detroit, MI, USA). p-Nitrophenyl-β-D-glucopyranoside (p-NPG) and p-nitrophenol (p-NP) were purchased from Sigma Aldrich (St. Louis, MO, USA). The 60 F254 silica gel plate (Merck, Darmstadt, Germany) was used for TLC and silica gel 60 (Merck) was used for column chromatography. An HPLC (NS 3000i system, Futecs Co., Daejeon, Korea) was used with a UV detector and gradient pump.
Isolation of β-glucosidase producing bacteria from Kimchi
We collected many kinds of Kimchi from various regions in Korea, and β-glucosidase producing bacteria were isolated using a esculin agar medium. Single colonies from the plates were purified onto new plates. The isolated strains were incubated at 37℃ for 24 h in MRS agar (Difco). We suggested that the bacteria could convert the major ginsenoside Rb1 to the minor ginsenoside Rg3.
Culture conditions
One micro liter of cells were grown in 100 mL Erlenmeyer flasks containing 50 mL of MSR broth and incubated at 30℃ on a rotary shaker (160 rpm) until it reaches the absorbance 0.8 to 1.0. Later the cells were harvested and re-inoculated onto the MRS broth containing 1% (w/v) cellobiose, glucose, lactose, sucrose, hesperidin and soluble starch. Growth was monitored using spectrophotometer at O.D. 600.
Preparation of crude enzyme
Bacterial cells were removed by centrifugation at 10,000 rpm for 20 min at 4℃. The supernatant containing the extracellular β-glucosidases was used as the crude enzyme and was precipitated using acetone. The precipitates were dissolved in 20 mM sodium phosphate buffer, pH 7.0. Later, their activity was checked using 1 mM ginsenoside Rb1.
Assay of ginsenosiede Rb1 transformation activity
To determine the β-glucosidase acitivity, 50 μL of crude enzyme were added to 50 μL of 10 mM p-NPG suspended in 20 mM sodium phosphate buffer (pH 7.0). Then the mixture was incubated at 37℃ for 10 min. The reaction was stopped by adding 100 μL of 1 M Na2CO3, and the p-NP produced was measured at 405 nm by a UV spectrophotometer (Ultrospec 2100 Pro; Amersham Biosciences, Cambridge, UK). One unit of enzyme activity was defined as the amount released 1 nmol p-NP per min from p-NPG under the above reaction conditions.
Effect of temperature and pH on enzyme activity
The optimal temperature for enzyme activity was determined over a temperature ranges between 25℃ to 70℃ using the standard assay conditions. To determine the optimal pH the enzyme activity was measured over a pH range of 4.5 to 8.0. The buffers used for the assay are 20 mM acetate buffer (pH 4.5-5.5), 20 mM sodium phosphate buffer (pH 6-7.5), and 20 mM Tris-HCl buffer (pH 8.0).
Biotransformation
The reaction mixture, containing 200 μL of 1 mM ginsenoside Rb1 and 200 μL of bacterial suspension were cultured in MRS broth, was incubated for 72 h at 37℃. Then extracted twice with 200 μL of n-butanol. The n-butanol fraction of the reaction mixture was evaporated in vacuo, and the residue was dissolved in CH3OH.
Analysis of ginsenosides by thin-layer chromatography
TLC was performed using a silica gel 60 F254 plate. CHCl3-CH3OH-H2O (65:35:10, v/v/v, lower phase) mixture was used as the developing solvent for TLC. The spots on the TLC plates were detected by spraying 10% H2SO4 followed by heating at 110℃ for 5 min.
Analysis of ginsenosides by high pressure layer chromatography
HPLC was performed using a NS 3000 system (Futecs Co.), equipped with a UV detector and a gradient pump. The C18 column (YMC c18 pro, 250×4.6 mm, 5 μm) was used for this analysis with 20 μL sample volume. The mobile phase consisted of CH3CN (A) with distilled H2O (B). The solvent A/solvent B form the gradient in the ratios as follows: 15:85, 21:79, 58:42, 90:10, 90:10, 15:85, and 15:85, with run times of 0-5, 5-25, 25-70, 70-72, 72-82, 82-84, and 84-100 min, respectively, at a 1.6 mL/min flow rate. Detection was done at 203 nm.
Analysis of nuclear magnetic resonance spectrum
An 80 mL suspension of strain 6105 from the MRS broth was mixed with the same volume of 1 mM ginsenoside Rb1 and incubated at 30℃, in 160 rpm for 60 h. The reaction mixture was extracted with 150 mL of n-butanol twice and was evaporated in vacuo at 45℃. The residue was applied to the silica gel 60 column and eluted with CHCl3-CH3OH-H2O (65:35:10, v/v, lower phase). Twenty milliliter of fraction was collected, separated and the metabolite transformed from ginsenside Rb1 was dissolved in pyridine-d5 and identified by 1H-NMR and 13C-NMR using FT-NMR spectrometry (400 MHz; Varion Inova AS 400, Varion, Palo Alto, CA, USA)
RESULTS
Phylogenetic study
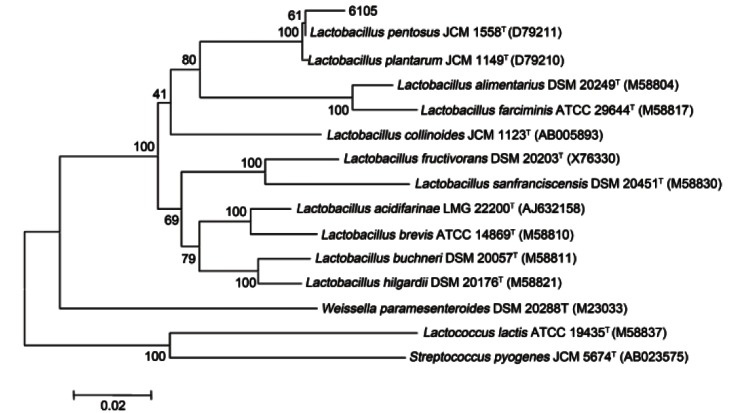
β-glucosidase producing bacteria was isolated from Kimchi using esculin-MRS agar. The 16S rRNA gene sequences of the strain 6105 were aligned with type strains found to have the closest taxonomic relationships. Strain 6105 was grouped with a Lactobacillus species, and the highest degrees of 16S rRNA gene sequence identities were to L. plantarum JCM 1149T (D79210) (99.1%), L. pentosus JCM 1558T (D79211) (99.2%) (Fig. 1).
Fig. 1. Phylogenetic tree based on 16S rRNA gene sequences showing the phylogenetic relationships between strain 6105 and related species.
Characterization of the crude enzyme of strain 6105
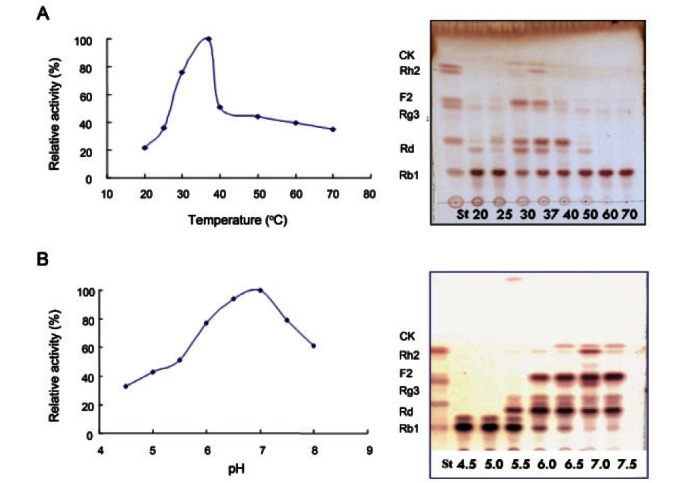
The effect of temperature and pH on the activity of β-glucosidase was shown in Fig. 2. The optimum temperature for maximal activity was 37℃. At 37℃ ginsenoside Rb1 was converted into compound K, however, the crude enzyme activity was decreased over 40℃ dramatically and not converted any compounds at 50℃ to 70℃. Under optimal temperature conditions, the crude enzyme activity was high at the pH ranges from 6.0 to 8.0. Ginsenoside Rb1 was almost hydrolyzed on pH 6.0 to 7.5 and the crude enzyme shows maximum activity at the pH 7.0 which converts ginsenoside Rb1 into ginsenoside Rd, F2 and compound K.
Fig. 2. Effect of temperature (A) and pH (B) on the transformation of ginsenoside Rb1 by crude enzyme strain 6105. CK, compound K; ST, standard.
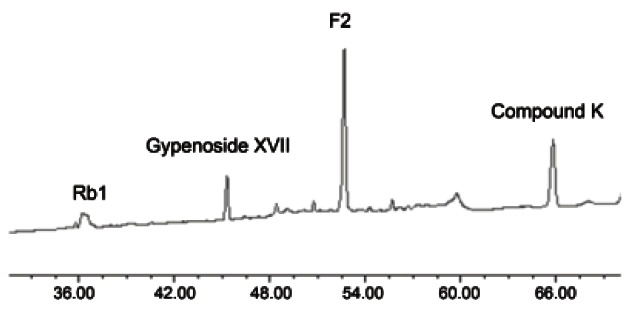
The HPLC profile of the enzymatic transformation of ginsenoside Rb1 by crude enzyme of strain 6105 is shown in Fig. 3. The identities of the peaks were established by comparison of retention time values with those of standards. In the HPLC chromatogram, the peaks with retention time 37.23, 41.68, 56.01, and 61.56 min correspond to ginsenoside Rb1, gypenoside XVII, F2, and compound K.
Fig. 3. HPLC assay of transformation of ginsenoside Rb1 to compound K by crude enzyme strain 6105.
Effect of carbon source supplements on β-glucosidase activity
The relative rates of hydrolysis of various substrates by crude enzyme are shown in Fig. 4. Strains were cultured on MRS broth and inoculated on MRS broth with 1% carbon source such as glucose, cellobiose, lactose, sucrose, hesperidin, and soluble starch to prepare crude enzyme (supernatant). The highest yields of β-glucosidase activity (0.95 U/mL) were obtained in lactose culture broth (Table 1). We hypothesized that carbon sources cause an increase β-glucosidase activity.
Fig. 4. Effects of carbon source supplementation on the transformation of ginsenoside Rb1 by crude enzyme of strain 6105. N, without elicitor; C, cellobiose; G, glucose; L, lactose; S, sucrose, H, hesperidin; T, soluble starch; St, standard; N-T, reaction with suspension culture; N’-T’, reaction with crude enzyme precipitated by acetone.
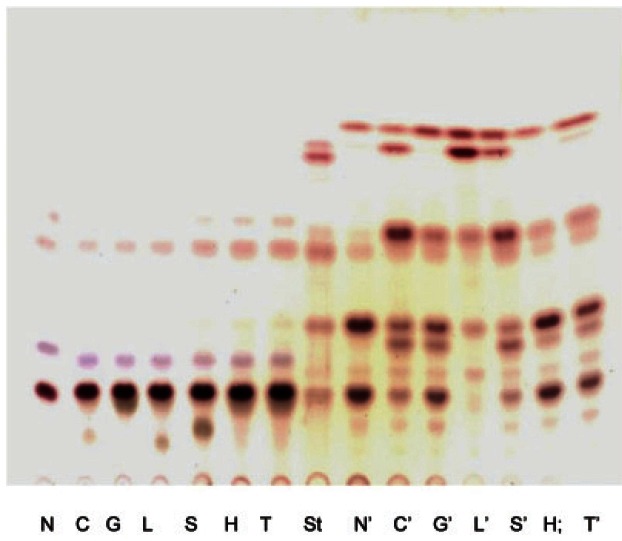
Table 1.
β-glucosidase produced by strain 6105 on various carbon sources
| β-glucosidase (U/mL) | Relative activity (%) | |
|---|---|---|
|
| ||
| Control | 0.78 | 100 |
| Cellobiose | 0.83 | 110.2 |
| Glucose | 0.82 | 108.16 |
| Lactose | 0.95 | 132.35 |
| Sucrose | 0.86 | 116.33 |
| Hesperidin | 0.59 | 65.31 |
| Soluble starch | 0.72 | 89.8 |
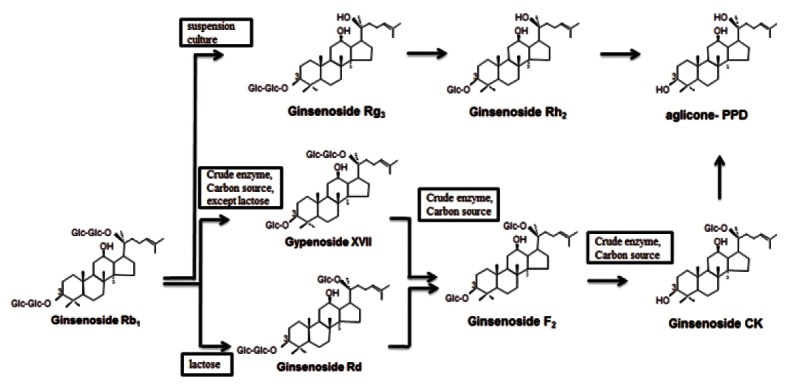
The ginsenoside Rb1 was converted in the following sequence: Rb1→Rd and gypenoside XVII→F2→compound K (Fig. 5) by the crude enzyme from strain 6105, consecutively hydrolyzing of ginsenoside Rb1.
Fig. 5. The conversion pathway of ginsenoside Rb1 using crude enzyme strain 6105.
Structural identification of metabolites
We identified the metabolites of ginsenoside Rb1 by 1H-NMR and 13C-NMR spectroscopy, because the Rf value and retention time of the metabolites were the same as the standard compound K in TLC and HPLC analysis.
Metabolite 1
In the 1H-NMR spectrum of metabolite 1, the proton signals for the H-1 of the 3-O-inner-glucopyranosyl moiety, 3-O-outer-glucopyranosyl moiety, and 20-glucopyranosyl moiety appeared respectively, at δ 4.87 ppm (1H, d, J=8.2 Hz, H-3-glc-1H’), δ 5.05 ppm (1H, d, J=7.8 Hz, H-20-glc-1H’’’), δ 5.08 ppm (1H, d, J=7.6 Hz, H-20-glc-1H’’’’), showing that the aglycon of metabolite 1 harbored three β-D-glucoses. In the 13C NMR(pyridine-d5, 100 MHz) spectrum of metabolite 1, the signals for C-2’ of the 3-inner-glucose was shifted up field, from 83.5 ppm to 76.0 ppm, but the other signals were similar to those of ginsenoside Rb1. Compared with as previous reported[15], we indicated that the metabolite is 3-O-[β-D-glucopyranosyl]-20-O-[β-D-glucopyranosyl-(6,1)-β-D-gluco-pyranosyl]-20(S)-protopanaxadiol, identical to gypenoside XVII.
Metabolite 2
In the 1H-NMR spectrum of metabolite 2, the proton signals for the H-1 of the 3-O-inner-glucopyranosyl moiety, 3-O-outer-glucopyranosyl moiety, and 20-glucopyranosyl moiety appeared respectively, at δ 4.94 ppm (1H, d, J=7.7 Hz, H-20-glc-1’’’), δ 4.87 ppm (1H, d, J=8.2 Hz, H-3-glc-1H’), δ 5.19 ppm (1H, d, J=7.4Hz, H-3-glc-1’’); Compared with as previous reported[16], metabolite 2 was identified as 3-O-[β-D-glucopyranosyl]-20(S)-protopanaxadiol, identical to ginsenoside Rd.
Metabolite 3
In the 1H-NMR spectrum of metabolite 3, the proton signals for the H-1 of the 3-O-inner-glucopyranosyl moiety and 20-glucopyranosyl moiety appeared respectively, at δ 5.20 ppm (1H, d, J=7.4Hz, H-3-glc-1’), δ 4.94 ppm (1H, d, J=7.4 Hz, H-20-glc-1’’’); for 13C-NMR (pyridine-d5, 100 MHz) data see Table 2. According to the comparison of the 13C-NMR spectrum of metabolite 3 with that of metabolite 1, the signal for the C-2 of the 3-inner-glucopyranoxyl moiety was shifted up field, from 83.2 ppm to 78.4 ppm, and other signals were similar to those of metabolite 2. Additionally, based on the fact that the anomeric carbon signals at δ 106.9 and δ 98.2 ppm showed to be similar to signals for C-1 of the 3-O-inner-glucopyranosyl moiety and 20-O-inner-glucopyranosyl moiety of metabolite 2, it was confirmed that two glucoses connected with the C-3 and C-20 sites of the aglycon moiety. Compared with as previous reported[17], metabolite 3 was identified as 3-O-[β-D-glucopyranosyl]-20-O-[β-D-glucopyranosyl]-20(S)-protopanaxadiol, identical to ginsenoside F2.
Metabolite 4
In the 1H-NMR spectrum of metabolite 4, the anomeric protone signals appeared at δ 3.42 ppm (3H, dd, 11,8, J=5.2 Hz), δ 5.18 ppm (1H, H-20-glc-1’, J=8.0 Hz), δ 5.25 ppm (24H, t, J=6.4 Hz). The 13C-NMR analysis result of metabolite is shown in Table 2. The signals for, C-24, C-25, 20-glc-6’ and 20-glc-1’ of the metabolite appeared δ 126.01 ppm, δ 130.92 ppm, δ 63.11 ppm and δ 98.33 ppm, respectively, which are similar to those for the same carbon in compound K. Compared with as previous reported [18], we indicated that the metabolite is 20-O-[β-D-glucopyranosyl]-20(S)-protopanaxadiol, identical to compound K.
DISCUSSION
This study focused on the isolation of LAB strain Lactobacillus sp. 6105 from Kimchi, finding β-glucosidase with the ability to convert ginsenonside Rb1. In previous studies we reported that the isolation of β-glucosidase producing bacteria using esculin [4,10] MRS medium. Strain 6105 converted ginsenoside Rb1 to Rg3 by live cell cultures (data not shown), however, it was shown to convert the ginsenoside Rb1 into compound K using crude enzyme. Previously, Chi and Ji [19] and Chi et al. [20] reported the transformation of major ginsenoside using food microorganisms. But low β-glucosidase activity was noted, when they incubated for transformation ginsenoside with live cells, however, crude enzyme of their bacteria shows higher activity to convert a minor saponin like compound K.
We confirmed the hydrolysis of ginsenoside Rb1. We standardized the optimal conditions of the β-glucosidase activity, and the crude enzyme of strain 6105 had the highest activity at 37℃. According to other studies, the optimal temperature was lower than those reported such as 70℃ for Paecilomyces thermophila derivative β-glucosidase [21]. An optimal temperature of 50℃ was found for Aspergilus oryzae derivative β-glucosidase [22], for Fusarium proliferatum ECU2042 derivative β-glucosidase [23], and for Candida peltata derivative β-glucosidse [24]. The 45℃ optimal temperature was found for Rhizopus japonicus derivative β-glucosidase [25] and for Bacillus subtilis natto derivative β-glucosidase [26]. Additionally, the crude enzyme of strain 6105 has optimum activity at pH 6.0 to 7.5. Comparing with others reported strain optimum pH such as P. thermophila [21], A. oryzae [22], F. proliferatum ECU2042 [23], R. japonica [25], C. peltata [24], B. subtilis natto [26], pH 6.2, 5.0, 5.0 to 6.0, 4.8 to 5.0, 5.0, 6.0, respectively.
In order to find out the maximum β-glucosidase activity, we added carbon sources such as glucose, cellobiose, lactose, sucrose, hesperidin, and soluble starch on MRS medium. Among them, the addition of lactose increased the β-glucosidase activity about 30 percent over control. On the MRS medium containing hesperidin and soluble starch, there was less activity. These results suggest that strain 6105 can increase β-glucosidase activity on the MRS medium including glucose, cellobiose, lactose, sucrose like C. peltata [24].
We found that compound K has a highly efficient function through this study. Compound K has many pharmacological activities including protection of the liver from injury [27], inhibition of the P450-mediated metabolism [28], and inhibition at the a(1G)-type of Ca2+ channel [29]. It also triggers apoptosis in activated hepatic stellate cells via caspase-3 activation [30] and potent cytotoxic activity against tumor cell lines [31]. However, minor saponins like ginsenoside F2 and compound K are found in small amounts in ginseng roots. Therefore, it is desirable to produce a greater amount of minor ginsenoside such as compound K. Our experiment demonstrates that strain 6105 is capable of producing the minor ginsenoside F2 and compound K using crude enzymes.
Acknowledgments
This research was supported by the Ministry of Knowledge Economy, Korea, under the Information Technology Research Center support program supervised by the National IT Industry Promotion Agency (NIPA-2011-C1090-1121-0003).
References
- 1.Duo D, Jin L, Chen Y. Advances and prospects of the study on chemical constituents and pharmacological activities of Panax ginseng. J Shenyang Pharm Univ. 1999;16:151–156. [Google Scholar]
- 2.Yuan CS, Wang CZ, Wicks SM, Qi LW. Chemical and Pharmacological Studies of Saponins with a Focus on American Ginseng. J Ginseng Res. 2010;34:160–167. doi: 10.5142/jgr.2010.34.3.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park JH., Kim JM, Han SB, Kim NY, Surh YJ, Lee SK, Kim ND, Park MK. A new processed ginseng with fortified activity. In: Advances in Ginseng Research – Proceedings of the 7th International Symposium on Ginseng; 1998. Korean Society of Ginseng; Seoul: 1998. pp. 146–159. [Google Scholar]
- 4.Kim MK, Lee JW, Lee KY, Yang DC. Microbial conversion of major ginsenoside Rb1 to pharmaceutically active minor ginsenoside Rd. J Microbiol. 2005;43:456–462. [PubMed] [Google Scholar]
- 5.Kwon SW, Han SB, Park IH, Kim JM, Park MK, Park JH. Liquid chromatographic determination of less polar ginsenosides in processed ginseng. J Chromatogr A. 2001;921:335–339. doi: 10.1016/s0021-9673(01)00869-x. [DOI] [PubMed] [Google Scholar]
- 6.Park EK, Choo MK, Han MJ, Kim DH. Ginsenoside Rh1 possesses antiallergic and anti-inflammatory activities. Int Arch Allergy Immunol. 2004;133:113–120. doi: 10.1159/000076383. [DOI] [PubMed] [Google Scholar]
- 7.Han BH, Park MH, Han YN, Woo LK, Sankawa U, Yahara S, Tanaka O. Degradation of ginseng saponins under mild acidic conditions. Planta Med. 1982;44:146–149. doi: 10.1055/s-2007-971425. [DOI] [PubMed] [Google Scholar]
- 8.Kim MH, Hong HD, Kim YC, Rhee YK, Kim KT, Rho J. Ginsenoside Changes in Red Ginseng Manufactured by Acid Impregnation Treatment. J Ginseng Res. 2010;34:93–97. doi: 10.5142/jgr.2010.34.2.093. [DOI] [Google Scholar]
- 9.Hasegawa H, Sung JH, Matsumiya S, Uchiyama M. Main ginseng saponin metabolites formed by intestinal bacteria. Planta Med. 1996;62:453–457. doi: 10.1055/s-2006-957938. [DOI] [PubMed] [Google Scholar]
- 10.Cheng LQ, Na JR, Kim MK, Bang MH, Yang DC. Microbial conversion of ginsenoside Rb1 to minor ginsenoside F2 and gypenoside XVII by Intrasporangium sp. GS603 isolated from soil. J Microbiol Biotechnol. 2007;17:1937–1943. [PubMed] [Google Scholar]
- 11.Choi JR, Hong SW, Kim Y, Jang SE, Kim NJ, Han MJ, Kim DH. Metabolic Activities of Ginseng and Its Constituents, Ginsenoside Rb1 and Rg1, by Human Intestinal Microflora. J Ginseng Res. 2011;35:301–307. doi: 10.5142/jgr.2011.35.3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atrih A, Rekhif N, Milliere JB, Lefebvre G. Detection and characterization of a bacteriocin produced by Lactobacillus plantarum C19. Can J Microbiol. 1993;39:1173–1179. [Google Scholar]
- 13.Klaver FA, van der Meer R. The assumed assimilation of cholesterol by Lactobacilli and Bifidobacterium bifidum is due to their bile salt-deconjugating activity. Appl Environ Microbiol. 1993;59:1120–1124. doi: 10.1128/aem.59.4.1120-1124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhee CH, Park HD. Isolation and characterization of lactic acid bacteria producing antimutagenic substance Korean Kimchi. Korean J Appl Microbiol Biotechnol. 1999;27:15–22. [Google Scholar]
- 15.Cheng LQ, Kim MK, Lee JW, Lee YJ, Yang DC. Conversion of major ginsenoside Rb1 to ginsenoside F2 by Caulobacter leidyia. Biotechnol Lett. 2006;28:1121–1127. doi: 10.1007/s10529-006-9059-x. [DOI] [PubMed] [Google Scholar]
- 16.Dong A, Ye M, Guo H, Zheng J, Guo D. Microbial transformation of ginsenoside Rb1 by Rhizopus stolonifer and Curvularia lunata. Biotechnol Lett. 2003;25:339–344. doi: 10.1023/a:1022320824000. [DOI] [PubMed] [Google Scholar]
- 17.Dou D, Wen Y, Pei Y, Chen Y, Ma Z. Active constituents reducing side-effects of prednisone acetate in leaves of Panax ginseng C.A Meyer. Zhongguo Zhong Yao Za Zhi. 1997;22:174–176, 192. [PubMed] [Google Scholar]
- 18.Sung JH, Hasegawa H, Matsumiya S, Uchiyama M. Metabolism of ginseng saponins by human intestinal bacteria. Korean J Pharmacognosy. 1995;26:360–367. [Google Scholar]
- 19.Chi H, Ji GE. Transformation of ginsenosides Rb1 and Re from Panax ginseng by food microorganisms. Biotechnol Lett. 2005;27:765–771. doi: 10.1007/s10529-005-5632-y. [DOI] [PubMed] [Google Scholar]
- 20.Chi H, Kim DH, Ji GE. Transformation of ginsenosides Rb2 and Rc from Panax ginseng by food microorganisms. Biol Pharm Bull. 2005;28:2102–2105. doi: 10.1248/bpb.28.2102. [DOI] [PubMed] [Google Scholar]
- 21.Yang S, Jiang Z, Yan Q, Zhu H. Characterization of a thermostable extracellular beta-glucosidase with activities of exoglucanase and transglycosylation from Paecilomyces thermophila. J Agric Food Chem. 2008;56:602–608. doi: 10.1021/jf072279+. [DOI] [PubMed] [Google Scholar]
- 22.Riou C, Salmon JM, Vallier MJ, Gunata Z, Barre P. Purification, characterization, and substrate specificity of a novel highly glucose-tolerant beta-glucosidase from Aspergillus oryzae. Appl Environ Microbiol. 1998;64:3607–3614. doi: 10.1128/aem.64.10.3607-3614.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su JH, Xu JH, Lu WY, Lin GQ. Enzymatic transformation of ginsenoside Rg3 to Rh2 using newly isolated Fusarim proliferatum ECU2042. J Mol Catal B Enzym. 2006;38:113–118. [Google Scholar]
- 24.Saha BC, Bothast RJ. Production, purification, and characterization of a highly glucose-tolerant novel beta-glucosidase from Candida peltata. Appl Environ Microbiol. 1996;62:3165–3170. doi: 10.1128/aem.62.9.3165-3170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SD, Seu JH. Enzymatic properties of the convertible enzyme of ginseng saponin produced from Rhizopus japonicas. Korean J Appl Microbiol Bioeng. 1989;17:126–130. [Google Scholar]
- 26.Kuo LC, Lee KT. Cloning, expression, and characterization of two beta-glucosidases from isoflavone glycoside-hydrolyzing Bacillus subtilis natto. J Agric Food Chem. 2008;56:119–125. doi: 10.1021/jf072287q. [DOI] [PubMed] [Google Scholar]
- 27.Lee HU, Bae EA, Han MJ, Kim NJ, Kim DH. Hepatoprotective effect of ginsenoside Rb1 and compound K on tert-butyl hydroperoxide-induced liver injury. Liver Int. 2005;25:1069–1073. doi: 10.1111/j.1478-3231.2005.01068.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee JH, Jeong SM, Kim JH, Lee BH, Yoon IS, Lee JH, Choi SH, Lee SM, Park YS, Lee JH, et al. Effects of ginsenosides and their metabolites on voltage-dependent Ca2+ channel subtypes. Mol Cells. 2006;21:52–62. [PubMed] [Google Scholar]
- 29.Liu Y, Zhang JW, Li W, Ma H, Sun J, Deng MC, Yang L. Ginsenoside metabolites, rather than naturally occurring ginsenosides, lead to inhibition of human cytochrome P450 enzymes. Toxicol Sci. 2006;91:356–364. doi: 10.1093/toxsci/kfj164. [DOI] [PubMed] [Google Scholar]
- 30.Park EJ, Zhao YZ, Kim J, Sohn DH. A ginsenoside metabolite, 20-O-beta-D-glucopyranosyl-20(S)-protopanaxadiol, triggers apoptosis in activated rat hepatic stellate cells via caspase-3 activation. Planta Med. 2006;72:1250–1253. doi: 10.1055/s-2006-947223. [DOI] [PubMed] [Google Scholar]
- 31.Zhou W, Feng MQ, Li JY, Zhou P. Studies on the preparation, crystal structure and bioactivity of ginsenoside compound K. J Asian Nat Prod Res. 2006;8:519–527. doi: 10.1080/10286020500208600. [DOI] [PubMed] [Google Scholar]


