Abstract
Panax ginseng has been cultivated for centuries, and nine commercial cultivars have been registered in Korea. However, these nine elite cultivars are grown in less than 10% of ginseng fields, and there is no clear authentication system for each cultivar even though their values are higher than those of local landraces. Here, we have developed 19 microsatellite markers using expressed gene sequences and established an authentication system for all nine cultivars. Five cultivars, ‘Chunpoong’, ‘Sunpoong’, ‘Gumpoong’, ‘Sunun’, and ‘Sunone’, can each be identified by one cultivar-unique allele, gm47n-a, gm47n-c, gm104-a, gm184-a (or gm129-a), and gm175-c, respectively. ‘Yunpoong’ can be identified by the co-appearance of gm47n-b and gm129-c. ‘Sunhyang’ can be distinguished from the other eight cultivars by the co-appearance of gm47n-b, gm129-b, and gm175-a. The two other cultivars, ‘Gopoong’ and ‘Cheongsun’, can be identified by their specific combinations of five marker alleles. This marker set was successfully utilized to identify the cultivars among 70 ginseng individuals and to select true F1 hybrid plants between two cultivars. We further analyzed the homogeneity of each cultivar and phylogenetic relationships among cultivars using these markers. This marker system will be useful to the seed industry and for breeding of ginseng.
Keywords: Panax ginseng, Seed industry, Breeding, EST-SSR markers, Cultivar authentication
INTRODUCTION
Ginseng (Panax ginseng Meyer) has been used and cultivated for centuries because of its positive medicinal effects. Recently, demand for ginseng is increasing because of health concerns. Ginseng is reported to offer benefits relating to diabetes, arteriosclerosis, hyperpiesia, cardiac insufficiency, cancer, stress, the immune system and sexual functions [1].
Breeding of ginseng is difficult because of its long generation time and the small number of seeds set, with only 40 seeds per plant after four years of growth [2]. For these reasons, ginseng cultivar has been bred by pure line selection among landrace populations in Korea. Up to now only nine cultivars have been officially registered with the Korea Seed and Variety Service. ‘Chunpoong’, the first cultivar registered in Korea, has orange-red fruit and good characteristics in terms of root shape and red ginseng processing. ‘Yunpoong’ has short multi-stems, red fruit, and a high yield. ‘Gopoong’ has a raddish violet stem, dark red fruit, high saponin content, and good root shape. ‘Sunpoong’ has an early germination time, a raddish violet stem, and slightly yellow roots. ‘Gumpoong’ has a green stem and yellow fruit and is good for making red ginseng. ‘Sunun’ has red fruit, a short raddish violet stem and wrinkled leaf edges. ‘Sunone’ has orange-red fruit, a raddish violet stem, a high yield and good root shape. ‘Cheongsun’ has a long green stem, red fruit, and an early germination time. ‘Sunhyang’ has a long stem, red fruit, wide oval-shaped leaves and high levels of aroma constituents [3-5]. These nine cultivars have unique and uniform phenotype characteristics and show better agricultural and processing quality than do local landraces, such as ‘Jakyung’, which has red fruit and reddish violet stems, or ‘Hwangsook’, which has yellow fruit and green stem. However, none of these elite cultivars are widely provided to ginseng farmers because of the lack of a well-organized ginseng seed industry and a proper authentication system, such as marker-aided discrimination. A reliable authentication system would promote the establishment of a ginseng seed industry and thus help ginseng production by providing superior cultivars to farmers.
Marker-assisted breeding (MAB) is established as a common and popular tool for crop breeding and seed industries. Traditional breeding requires large populations and long times for developing pure inbred lines, and involves huge phenotyping efforts. Even after development of good cultivars, it is necessary to establish a seed industry by providing stable authentication systems. MAB can reduce the effort and time needed for breeding new cultivars and establishing a seed industry [6,7]. In recent years, there have been several trials for the development of markers in ginseng using random amplified polymorphic DNA [8], restriction fragment length polymorphism [9], microsatellite [10,11], inter simple sequence repeat [12] or expressed sequence tag-simple sequence repeat (EST-SSR) [13] techniques. Microsatellites (SSRs) are distributed in all eukaryotic genomes with large amounts of polymorphism due to repeat number variation and reveal co-dominant features in gels with high reproducibility [14]. Thus, microsatellite markers have been broadly used to construct genetic maps and to authenticate cultivars in crops like rice [15], maize [16], soybean [17], and wheat [18] and in many horticultural plants [19-23].
Only a very limited number of molecular markers, and no MAB systems, have been developed in ginseng [5-10]. Recently, we described 70 expressed gene sequence-derived SSR markers, 19 of which are polymorphic among ginseng cultivars in Korea [13]. In this study, we attempted to establish a practical and reliable discrimination system for the nine cultivars using the previously described EST-SSR markers along with newly developed EST-SSR markers. The resulting system can be used for promotion of the ginseng seed industry and breeding and also for evaluation of the original cultivars using fresh or dried ginseng roots in the ginseng market.
MATERIALS AND METHODS
Plant materials and DNA extraction
Nine cultivars registered in Korea were used in this study: ‘Gumpoong’, ‘Gopoong’, ‘Yunpoong’, ‘Chunpoong’, ‘Sunpoong’, ‘Sunun’, ‘Sunone’, ‘Cheongsun’, and ‘Sunhyang’. Leaf samples from two individuals for each cultivar were collected from a research field of the Korea Ginseng Corporation in Eumseong, Korea. The leaves were frozen and ground with a mortar and pestle for DNA extraction, which was performed using modified cetyltrimethylammonium bromide method [18].
Microsatellite primer design
Previously, we developed a large number of EST-SSRs for ginseng genome mapping [13]. We used 11 of those markers in this study. Furthermore, we designed 77 EST-SSR primer pairs for the development of cultivar-specific markers that can discriminate the nine cultivars from one another. Primer pairs were designed from the flanking sequences of SSR motifs using Primer3 ver. 0.4.0 (http://frodo.wi.mit.edu/primer3) and estimated polymerase chain reaction (PCR) product sizes were 90 to 300 bp.
Polymerase chain reaction amplification and electrophoresis
PCR was performed in 25 μL reaction volumes containing the following: 20 ng genomic DNA, 10 pmole each primer, 0.5 mM dNTP, 1X Taq polymerase buffer and one unit Taq polymerase (Vivagen, Seongnam, Korea). Amplification was performed as follows: 5 min at 94℃, 35 cycles of 94℃ 20 s, 52℃ to 56℃ 20 s, 72℃ 20 s, and then 72℃ 7 min. PCR products were separated via 2% agarose gel electrophoresis and 9% polyacrylamide gel electrophoresis for identification of polymorphisms. Gels were stained with ethidium bromide and visualized with a UV lamp.
Development of cultivar-specific markers, marker combination sets and data analysis
By screening 30 polymorphic SSRs, including 11 EST-SSRs developed from our previous study [13] and 19 new markers developed in this study, we tried to select the set of the fewest polymorphic markers showing clear and reproducible band profiles that could be used to discriminate each of the nine cultivars. In 9% non-denaturing polyacrylamide gel electrophoresis, we observed duplicated bands, one expected size lower band and extra larger upper bands. Both bands showed same genotype patterns (Fig. 1) and the upper bands showed clear resolution of polymorphism for genotyping. For the phylogenetic analysis, we used genotype data for 13 of the 30 polymorphic markers. Genotype data for each individual were carefully scored and imported into PowerMarker ver. 3.0 [24] for calculation of allele frequency data, genetic diversity, and polymorphism information content (PIC) values. Phylogenetic analysis among cultivars was conducted using the unweighted pair group method with arithmetic mean (UPGMA) clustering [25] in the NTSYSpc ver. 2.1 (Exeter Software, New York, NY, USA).
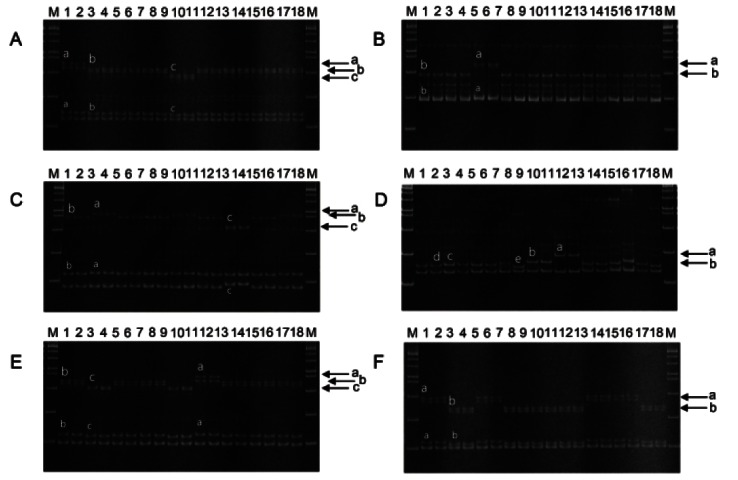
Fig. 1. Six expressed sequence tag-simple sequence repeats composing a set to authenticate nine ginseng cultivars. (A-F) are band profiles which were amplified by primer pairs gm47n, gm104, gm175, gm184, gm129, gm45n, respectively. M: 100 bp DNA ladder; Lanes: 1-2, Chunpoong; 3-4, Yunpoong; 5-6, Gumpoong; 7-8, Gopoong; 9-10, Sunpoong; 11-12, Sunun; 13-14, Sunone; 15-16, Cheongsun; 17-18, Sunhyang. Lowercase letters denote different alleles amplified by the primer pair.
RESULTS
Identification of specific markers for five cultivars
Recently, we developed 70 EST-SSR markers which were polymorphic between Panax species, and 19 of them were polymorphic between accessions of P. ginseng species [13]. Among the 19 polymorphic markers, we screened 11 low copy number markers, which generated fewer amplicons, to identify cultivar-specific markers capable of discriminating a specific cultivar from the others among the nine elite cultivars. The gm47n marker revealed three different alleles; alleles ‘a’ and ‘c’ were specific for cultivars ‘Chunpoong’ and ‘Sunpoong’, respectively, whereas allele ‘b’ was common to the other seven cultivars (Fig. 1A).
However, the other markers did not show cultivar-specific bands. Therefore, we developed more microsatellite markers, identifying 19 additional EST-SSRs showing polymorphism among ginseng cultivars in a trial of 77 EST-derived microsatellite primer pairs (Table 1). The PCR products all corresponded to their predicted sizes, ranging from 97 to 288 bp.
Table 1.
Polymorphic EST-SSR markers developed in this study
| Marker | Original sequence ID | SSR motif | Primer sequence | Ta (℃) | Estimated size | Putative gene function | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| gm99 | CN846560.1 | (TA)32 | F | AATAACCTGATTTTAAACGGTGACA | 52 | 277 | Beta glucosidase 15 |
| R | GACTATTGTAAGCGTTATCCCAAGA | ||||||
| gm104 | CN847863.1 | (GAA)7 | F | TGAATCTGGATAGATACACGACAGA | 52 | 187 | No hits |
| R | TGAAGTGTCGACAGAAGTGGTATAA | ||||||
| gm105 | DV553852.1 | (TTA)7 | F | GGTTCTTGGAGAAGAGAAAGGATAG | 52 | 288 | Jasmonate ZIM-domain protein |
| R | GATATACTTGACGGCTCTGAGAAAA | ||||||
| F | CGAAAGTGACTAACACCACTTTTCT | ||||||
| gm119 | pgcn1164 | (TA)17 | R | TGTAGTGAAACTGTGAAAGCAAGAG | 54 | 158 | Abscisic acid receptor PYL4 |
| F | CCATCCTAGCGGTCTAGTAAAGACT | ||||||
| gm123 | pgcn191 | (TA)18 | R | CACTCAGCTCAATGATATCAGCA | 52 | 169 | No hits |
| F | AAGCAAGTTGATGGAATATATGAGC | ||||||
| gm129 | pgcn2845 | (CTA)6 | R | CAGTTTTCTCTTCCTGGATATCGTA | 56 | 97 | Transcription factor WRKY4 |
| F | TTGATGAAGAAACAGACATGAAAGT | ||||||
| gm131 | pgcn2983 | (TA)10 | R | TACGTGCATTTATTTTGGCATTATT | 56 | 150 | Beta-amylase |
| F | GCCTCTTCTAGAGATCTGTCATTTG | ||||||
| gm132 | pgcn3023 | (ATTTG)6 | R | GAGATAAAAATACAGCTCCGTGACT | 56 | 133 | No hits |
| GGAGCATGACTGTCAAATCTACATA | |||||||
| gm134 | pgcn3339 | (CAG)7 | R | CTCCCGGAAAAGAGACTTAATTTAC | 56 | 130 | ELF4-like protein |
| F | ATCCCTAAGAGAAACCCAGATCTAA | ||||||
| gm137 | pgcn3646 | (TTTC)7 | R | ACACCAATCAATTTCACTTCTTGAT | 56 | 122 | Glutathione S-transferase |
| F | GTAAACTCGACCAGACAAAACCTTA | ||||||
| gm139 | pgcn3803 | (TA)10 | R | CTCATTTTTAGGCAATGGATTCTAC | 56 | 105 | No hits |
| F | GACAGAACTTCTTCATCTGCAATTT | ||||||
| gm148 | pgcn4474 | (TA)10 | R | GGCTCTCTCTTGTACCATTTAAACA | 56 | 145 | Basic blue copper protein |
| F | TTGAAGCTTACATCCTAGCAAATTA | ||||||
| gm159 | pgcn732 | (TA)7 | R | ATATAACAACAAAGCCAAGAAGCAC | 54 | 150 | PR10-2 |
| F | AGAAAACCAAATAAAGCCACACAC | ||||||
| gm160 | pgcn733 | (TA)21 | R | CACCGATAAGTCAATCCAACTATTC | 54 | 145 | Pathogenesis-related protein 10 |
| F | TTTTACTCATCTCCGTTTACACACA | ||||||
| gm166 | pgDC01027F02 | (TA)11 | R | AATTATCAGCACCCACAATTAAAAA | 54 | 123 | MADS-box protein |
| F | TCTTTAGAAAAATCTATTAGGGGTC | ||||||
| gm169 | pgDC03005G03 | (TA)10 | R | AGTAGGATCAAAATAATTACGAGGT | 56 | 194 | No hits |
| F | CCTCCAACATTATTTCAGTCTCAGT | ||||||
| gm175 | pgDC05024D08 | (TCC)9 | R | TAGTGGTAGCAGCTTAGGAGGAGTA | 56 | 123 | Hypothetical protein |
| F | CCTTGCTGCTCAAATTAATCTAAAA | ||||||
| gm176 | pgDC06005G10 | (TA)11 | R | CTCACTACTACTTGCCGCTTCTTC | 56 | 148 | Mini zinc fi nger 1 |
| F | TGGTTGACAAAGAAATTAACCAAAT | ||||||
| gm184 | pgPG04002D08 | (TTC)6 | R | GACCAAAAAGATCCGTCGTAAAG | 56 | 133 | Unnamed protein product |
EST, expressed sequence tag; SSR, simple sequence repeat.
Five of the markers have three-nucleotide SSR motifs, one has a four-nucleotide motif, and another has a five-nucleotide SSR motif. The other twelve have two-nucleotide SSR motifs, and all of them are (TA)n motifs, even though we used many primers containing other di-nucleotide motif SSRs. BLASTX searches revealed that 14 ESTs show similarity to known proteins, whereas five ESTs do not.
Among the 19 polymorphic EST-SSRs, three were cultivar-specific. The gm104 marker revealed two different alleles, with allele ‘a’ specific to the ‘Gumpoong’ cultivar and allele ‘b’ common to the other eight cultivars (Fig. 1B). The gm175 marker showed three different alleles, and allele ‘c’ was specific to the ‘Sunone’ cultivar (Fig. 1C). The gm184 marker revealed five different alleles even though alleles from b to e showed only very minor differences. Among the five alleles, gm184-a was unique to the ‘Sunun’ cultivar (Fig. 1D). Additionally, the gm129 marker showed three different alleles, with allele ‘a’ unique to ‘Sunun’ (Fig. 1E).
Authentication of ginseng cultivars by a combination of microsatellite markers
Using the cultivar-unique markers described above, ‘Chunpoong’, ‘Sunpoong’, ‘Gumpoong’, ‘Sunun’ and ‘Sunone’ could be clearly differentiated from the other cultivars. The five cultivar-specific markers were produced using four SSR primer pairs because one of them, the gm47n marker, produced two cultivar-unique bands, allele ‘a’ for ‘Chunpoong’ and allele ‘c’ for ‘Sunpoong’ (Table 2). On the other hand, ‘Sunun’ could be differentiated from the others by the appearance of either of the two ‘Sunun’-unique markers, the gm184-a and gm129-a alleles (Table 2).
Table 2.
Marker combinations for each cultivar
| Cultivar name | Original landrace | Main characteristics | Alleles for each marker | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| gm47n | gm104 | gm1841) | gm175 | gm129 | gm45n | |||
|
| ||||||||
| Chunpoong | Chunkyung | Orange-red fruit | a | b | b (d) | b | b | a |
| Yunpoong | Jakyung | Short multi-stems | b | b | b (c) | a | c | b |
| Gumpoong | Hwangsook | Yellow fruit | b | a | b (d) | b | b | a |
| Gopoong | Jakyung | High saponin content | b | b | b (c/e,e) | b | b | b |
| Sunpoong | Jakyung | Early germination | c | b | b | a | c | b |
| Sunun | Jakyung | Wrinkled leaf edges | b | b | a | b | a | b |
| Sunone | Jakyung | High yield | b | b | b (d) | c | b | a |
| Cheongsun | Chunkyung | Long green stem | b | b | b (d,b) | b | b | a |
| Sunhyang | Jakyung | High aroma constituents | b | b | b (c,d) | a | b | b |
Underlined italic letters indicate alleles required for cultivar identifi cation.
1) gm184 marker showed fi ve alleles, but there was no clear distinction among b, c, d, and e-alleles. Therefore, b, c, d, and e alleles were all regarded as b allele in this study.
We found no unique markers for ‘Yunpoong’, ‘Sunhyang’, ‘Cheongsun’ or ‘Gopoong’. To address this, we developed marker combination sets for authentication of these four cultivars. ‘Yunpoong’ could be distinguished from the other cultivars by the co-appearance of gm47n-b and gm129-c alleles, and ‘Sunhyang’ could be identified by co-appearance of gm47n-b, gm175-a, and gm129-b alleles (Table 2 and Fig. 1A, C, E). Collectively, the five cultivar-unique markers, gm47n, gm104, gm184, gm175 and gm129, could distinguish seven cultivars, ‘Chunpoong’, ‘Sunpoong’, ‘Gumpoong’, ‘Sunun’, ‘Sunone’, ‘Yunpoong’ and ‘Sunhyang’.
The gm184 marker gave rise to five alleles. However, alleles b, c, d, and e had only minor differences and could hardly be distinguished from each other. Therefore, we regarded these four alleles as single allele ‘b’ in the interest of developing a clear and reproducible identification system (Table 2 and Fig. 1D). Under this scoring system, we could not distinguish ‘Gopoong’ from ‘Cheongsun’ because they showed same genotype using the five cultivar-unique markers (Fig. 1D). Therefore, we included one more marker, gm45n, which displayed allele ‘b’ for ‘Gopoong’ and allele ‘a’ for ‘Cheongsun’ (Fig. 1F). ‘Gopoong’ and ‘Cheongsun’ could be identified from the other cultivars by co-appearance of five markers: gm47n-b + gm104-b + gm175-b + gm129-b + gm45n-b for ‘Gopoong’ and gm47n-b + gm104-b + gm175-b + gm129-b + gm45n-a for ‘Cheongsun’. By combination of these six markers, we could authenticate each of the nine ginseng cultivars (Table 2).
Blind test for cultivar identification
We utilized these markers to distinguish cultivars from 70 blind samples, which included 20 individual plants of ‘Chunpoong’ and five individual plants of seven cultivars and P. quinquefolius (Fig. 2). We could identify all individuals except for some individuals that may have been off-types or mixed genotypes.
Fig. 2. Blind identification of cultivars among 70 individuals using the marker set. Marker used: (A) gm47n, (B) gm104, (C) gm175, (D) gm184, and (E) gm45n. Deduced cultivars of the individual plants are denoted by the numbers above the lanes: 1, Chunpoong; 2, Yunpoong; 3, Gumpoong; 4, Gopoong; 5, Sunpoong; 6, Sunun; 7, Sunone; 8, Cheongsun; 9, Panax quinquefolius. Lowercase letters denote different alleles amplified by the primer pair.
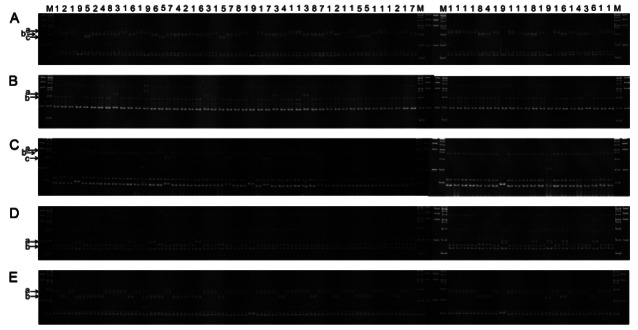
Among the 20 ‘Chunpoong’ individuals, nineteen were identical to ‘Chunpoong’ genotypes for all five markers but one individual showed heterozygous genotype with co-appearance of ‘a’ and ‘b’ for the gm47n marker. For both ‘Sunun’ and ‘Sunone’, one of five individuals showed a heterozygous genotype, gm175-b/c and gm184-a/b, respectively. Moreover, one ‘Gopoong’ and two ‘Cheongsun’ individuals showed different alleles than the major cultivar-unique allele for the gm45n marker. Overall, our authentication system was successfully applied to discriminate individuals derived from nine cultivars. However, we also ascertained heterogeneity within cultivars. In addition, P. quinquefolius individuals showed no identical genotype among five plants, but all the bands showed clear polymorphism compared to any of the ginseng cultivars in Korea.
Assessment of heterogeneity and phylogenetic relationship of cultivars
For the ‘Gopoong’, ‘Cheongsun’ and ‘Sunhyang’ cultivars, the two individuals tested showed different genotypes for the gm184 marker (Fig. 1D). In addition, ‘Sunun’, ‘Sunone’, ‘Gopoong’ and ‘Cheongsun’ showed different genotypes among the five blind samples. Even for the most elite cultivar, ‘Chunpoong’, there was one plant showing a heterozygous genotype for one of the five loci among 20 individuals in the blind sample (Fig. 2). These data indicate that the cultivars are not really homogeneous inbred lines and that some of the ginseng cultivars are actually highly heterogeneous.
Phylogenetic analysis was conducted with UPGMA clustering using genotypes of 13 SSR markers for identifying genetic relationships among 18 individuals from the nine cultivars with two plants each (Fig. 3).
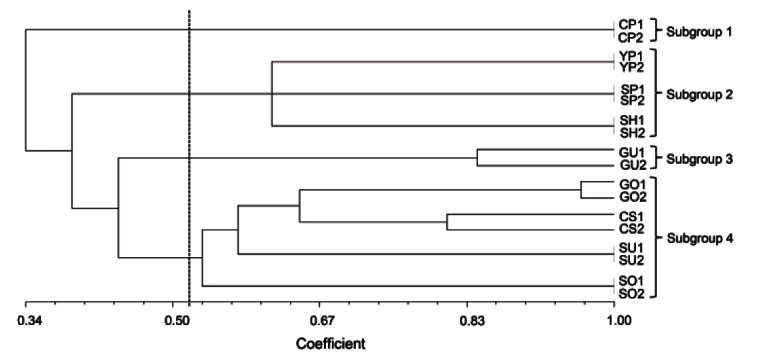
Fig. 3. Phylogenetic analysis of nine ginseng cultivars. The nine cultivars were grouped into four subgroups at a genetic coefficient value of 0.53. Analysis includes two individuals from each cultivar: CP, Chunpoong; YP, Yunpoong; SP, Sunpoong; SH, Sunhyang; GU, Gumpoong; GO, Gopoong; CS, Cheongsun; SU, Sunun; SO, Sunone.
The mean of allele number of markers was 2.8 and the major allele frequency was 0.58. The mean genetic diversity and PIC were 0.5170 and 0.4447 respectively. The average observed heterozygosity was 0.0299 (Table 3).
Table 3.
Characteristics of the markers of combination set
| Markers | No. of alleles | Genetic diversity1) | Heterozygosity2) | Polymorphism information content |
|---|---|---|---|---|
|
| ||||
| gm47n | 3 | 0.477 | 0.000 | 0.427 |
| gm104 | 2 | 0.219 | 0.000 | 0.195 |
| gm184 | 5 | 0.744 | 0.111 | 0.708 |
| gm175 | 3 | 0.531 | 0.000 | 0.468 |
| gm129 | 3 | 0.594 | 0.000 | 0.511 |
| gm45n | 2 | 0.500 | 0.000 | 0.375 |
| Average | 3 | 0.511 | 0.019 | 0.447 |
1) Genetic diversity is the probability that two randomly chosen alleles from the population are different.
2) Heterozygosity is the proportion of heterozygous individuals in the population.
For the six cultivars, ‘Chunpoong’, ‘Sunpoong’, ‘Yunpoong’, ‘Gumpoong’, ‘Sunun’, and ‘Sunone’, the two individuals tested showed the same genotype for all 13 markers. However, three cultivars, ‘Gumpoong’, ‘Gopoong’ and ‘Cheongsun’, showed differences between the two individuals with some of the markers even though they were grouped into the same clade. At a genetic coefficient value of 0.53, the nine cultivars were grouped into four subgroups (Fig. 3). ‘Chunpoong’ was placed as an independent subgroup, showing the largest divergence from the other cultivars. ‘Gumpoong’ was also independently grouped. Three cultivars, ‘Sunpoong’, ‘Yunpoong’ and ‘Sunhyang’, formed one sub-group, and four cultivars, ‘Gopoong’, ‘Sunun’, ‘Sunone’, and ‘Cheongsun’ made up another sub-group.
Selection of F1 hybrids using cultivar-specific markers
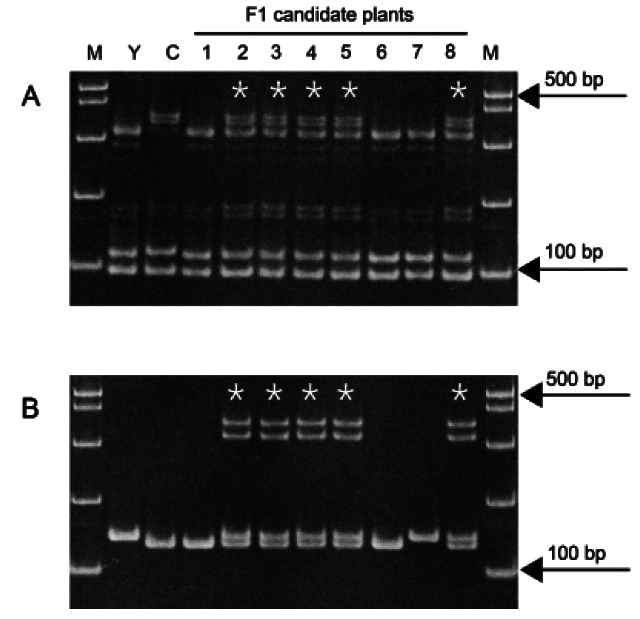
We applied these SSR markers to identify true F1 hybrid plants among eight F1 candidates from a cross of ‘Yunpoong’ and ‘Chunpoong’. These plants were grown for four years under field conditions, but it was difficult to distinguish the F1 hybrid plants from self-pollinated plants based on morphological traits. We used the gm129 and gm132 markers, which show polymorphism between ‘Chunpoong’ and ‘Yunpoong’, to identify true F1 hybrid plants. Only five of the eight candidates (plant numbers 2, 3, 4, 5, and 8) showed heterozygous band patterns, indicating that the other three plants were not successfully cross-pollinated (Fig. 4).
Fig. 4. Application of simple sequence repeat markers for the validation of F1 plants. (A,B) were amplified using primer pairs gm129 and gm132, respectively. Lanes M, Y, and C indicate 100 bp DNA ladder, ‘Yunpoong’, and ‘Chunpoong’, respectively. Lanes 1-8 indicate candidate F1 plants from a cross between ‘Yunpoong’ and ‘Chunpoong’, and true F1 plants are denoted with asterisks.
DISCUSSION
Marker-assisted selection (MAS) and marker-based cultivar identification are broadly applied in many crops, such as apple [26], mango [27], chickpea [28], peach [29], potato [30], etc. However, for ginseng there have not been sufficient molecular markers or a systematic authentication system available. Recently, some molecular markers have been developed for identification of individual ginseng cultivars which include sequence tagged site markers for ‘Chunpoong’ and ‘Gumpoong’ [31], a cleaved amplified polymorphic sequence marker for ‘Gopoong’ [32], molecular authentication of ‘Gopoong’ and ‘Gumpoong’ through internal transcribed spacer and 5.8S rDNA sequencing [33], and a single nucleotide polymorphism genotyping assay for five cultivars, ‘Chunpoong’, ‘Yunpoong’, ‘Gopoong’, ‘Gumpoong’, and ‘Sunpoong’ [34]. However, these studies were each performed based on one region of the genome and cannot discriminate each cultivar from the others, so they are difficult to utilize for practical and systematic authentication of the cultivars. To solve this problem, we developed a SSR marker combination set that can discriminate all nine registered cultivars from one another. A blind test confirmed that our marker system can successfully identify each cultivar.
Korean ginseng cultivars have been bred by selection from various local landrace populations because hybridization breeding is difficult due to the long generation time of the ginseng. Genomic study of ginseng has rarely been conducted, so breeding in ginseng has based on morphological traits. Registration of a cultivar indicates that it is a pure inbred line resulting from more than seven rounds of self-pollination in annual self-pollinating crops. However, it is hard to progress beyond seven generations of controlled self-pollination in ginseng because that would take more than 28 years. Thus, although the registered elite cultivars should be homogeneous and homozygous, it is very difficult to achieve homogeneity. We identified different levels of heterogeneity in the nine registered cultivars tested in this study. Some cultivars such as ‘Chunpoong’ and ‘Gumpoong’ showed relatively low rates of off-types, but cultivars such as ‘Cheongsun’ and ‘Gopoong’ showed high heterogeneity (Figs. 2 and 3). We expect that the cultivar authentication system developed in this study will serve to improve the purity and quality of each ginseng cultivar and thus establish a concrete seed industry.
These markers can also be successfully applied for the improvement of ginseng breeding. We could easily identify true F1 individuals among F1 candidate plants (Fig. 4). MAS would be highly beneficial for an industry facing the challenges of growing and breeding ginseng. Thus, our system should be helpful for expanding the supply of these elite cultivars, which are grown in less than 10% of ginseng fields although they are superior to the landraces, thereby promoting high quality ginseng production in Korea.
Even though the nine cultivars are derived from three landraces, ‘Hwangsook’, ‘Chungkyung’, and ‘Jakyung’ (Table 2), nine individual cultivars were clearly identified each other by the phylogenetic analysis (Fig. 3). The other six cultivars originated from landrace ‘Jakyung’ [35]. ‘Gumpoong’ was classified as an independent subgroup, which is in agreement with the breeding history because only ‘Gumpoong’ originated from landrace ‘Hwangsook’. However, ‘Chunpoong’ and ‘Cheongsun’ were grouped into different subgroups, even though they are known to be derived from the same landrace, ‘Chungkyung’. The other six ‘Jakyung’-derived cultivars were grouped into two independent subgroups. ‘Chunpoong’ seemed to be most diverged from the other cultivars, indicating that the cultivar was likely derived from a distinct individual.
In this study, we described a set of six SSR markers, developed from a total of 217 EST-SSR marker trials. Together, they composed a credible discrimination system for all nine commercial Korean ginseng cultivars. We also assessed the heterogeneity of each cultivar and found severe heterogeneity in some cultivars. This highlights the need for serious efforts aimed at improving the purity of the registered cultivars, which will help the ginseng seed industry and the ginseng industry as a whole. We determined genetic relationships among cultivars, presenting some directions for future ginseng breeding.
We expect that this marker set will be helpful for MAB in ginseng and for maintaining and improving the seed purity of each cultivar. This will support the establishment of a solid seed industry, the development of new cultivars, and the promotion of the ginseng industry.
Acknowledgments
This study was supported by the Technology Development Program for Agriculture and Forestry, Ministry for Food, Agriculture, Forestry and Fisheries, Korea (grant no. 609001) and a grant from the Next-Generation Bio-Green 21 Program (no. PJ008202) of the Rural Development Administration, Korea.
References
- 1.Nam KY. Clinical applications and efficacy of Korean ginseng. J Ginseng Res. 2002;26:111–131. doi: 10.5142/JGR.2002.26.3.111. [DOI] [Google Scholar]
- 2.Choi KT. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng C A Meyer. Acta Pharmacol Sin. 2008;29:1109–1118. doi: 10.1111/j.1745-7254.2008.00869.x. [DOI] [PubMed] [Google Scholar]
- 3.Lee SS, Lee JH, Ahn IO. Characteristics of new cultivars in Panax ginseng C.A. Meyer.; In: Korean Society of Ginseng. Proceeding of Ginseng Society Conference; 2005 Nov.; Seoul. Korean Society of Ginseng; 2005. pp. 3–18. [Google Scholar]
- 4.Ahn IO, Lee SS, Lee JH, Lee MJ, Jo BG. Comparison of ginsenoside contents and pattern similarity between root parts of new cultivars in Panax ginseng C.A. Meyer. J Ginseng Res. 2008;32:15–18. doi: 10.5142/JGR.2008.32.1.015. [DOI] [Google Scholar]
- 5.Kwon WS, Lee MG, Lee JH. Characteristics of flowering and fruiting in new varieties and lines of Panax ginseng C.A. Meyer. J Ginseng Res. 2001;25:41–44. [Google Scholar]
- 6.Collard BC, Mackill DJ. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci. 2008;363:557–572. doi: 10.1098/rstb.2007.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohan M, Nair S, Bhagwat A, Krishna TG, Yano M, Bhatia CR, Sasaki T. Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol Breed. 1997;3:87–103. [Google Scholar]
- 8.Artiukova EV, Kozyrenko MM, Reunova GD, Muzarok TI, Zhuravlev IuN. Analysis of genomic variability of planted Panax ginseng by RAPD. Mol Biol (Mosk) 2000;34:339–344. [PubMed] [Google Scholar]
- 9.Um JY, Chung HS, Kim MS, Na HJ, Kwon HJ, Kim JJ, Lee KM, Lee SJ, Lim JP, Do KR, et al. Molecular authentication of Panax ginseng species by RAPD analysis and PCR-RFLP. Biol Pharm Bull. 2001;24:872–875. doi: 10.1248/bpb.24.872. [DOI] [PubMed] [Google Scholar]
- 10.Ma KH, Dixit A, Kim YC, Lee DY, Kim TS, Cho EG, Park YJ. Development and characterization of new microsatellite markers for ginseng (Panax ginseng C. A. Meyer). Conserv Genet. 2007;8:1507–1509. [Google Scholar]
- 11.Kim J, Jo BH, Lee KL, Yoon ES, Ryu GH, Chung KW. Identification of new microsatellite markers in Panax ginseng. Mol Cells. 2007;24:60–68. [PubMed] [Google Scholar]
- 12.Bang KH, Lee JW, Kim YC, Jo IH, Seo AY, Lee JH, Kim OT, Hyun DY, Cha SW, Cho JH. Development of an ISSR-derived SCAR marker in Korean ginseng cultivars (Panax ginseng C. A. Meyer). J Ginseng Res. 2011;35:52–59. doi: 10.5142/jgr.2011.35.1.052. [DOI] [Google Scholar]
- 13.Choi HI, Kim NH, Kim JH, Choi BS, Ahn IO, Lee JS, Yang TJ. Development of reproducible EST-derived SSR markers and assessment of genetic diversity in Panax ginseng cultivars and related species. J Ginseng Res. 2011;35:399–412. doi: 10.5142/jgr.2011.35.4.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgante M, Olivieri AM. PCR-amplified microsatellites as markers in plant genetics. Plant J. 1993;3:175–182. [PubMed] [Google Scholar]
- 15.Temnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, Lipovich L, Cho YG, Ishii T, McCouch SR. Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L.). Theor Appl Genet. 2000;100:697–712. [Google Scholar]
- 16.Smith JS, Chin EC, Shu H, Smith OS, Wall SJ, Senior ML, Mitchell SE, Kresovich S, Ziegle J. An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L): comparisons with data from RFLPS and pedigree. Theor Appl Genet. 1997;95:163–173. [Google Scholar]
- 17.Rongwen J, Akkaya MS, Bhagwat AA, Lavi U, Cregan PB. The use of microsatellite DNA markers for soybean genotype identification. Theor Appl Genet. 1995;90:43–48. doi: 10.1007/BF00220994. [DOI] [PubMed] [Google Scholar]
- 18.Roder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW. A microsatellite map of wheat. Genetics. 1998;149:2007–2023. doi: 10.1093/genetics/149.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang S, Yu JK, Slabaugh B, Shintani K, Knapp J. Simple sequence repeat map of the sunflower genome. Theor Appl Genet. 2002;105:1124–1136. doi: 10.1007/s00122-002-0989-y. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Georgi LL, Zhebentyayeva TN, Reighard GL, Scorza R, Abbott AG. High-throughput targeted SSR marker development in peach (Prunus persica). Genome. 2002;45:319–328. doi: 10.1139/g01-153. [DOI] [PubMed] [Google Scholar]
- 21.Han ZG, Guo WZ, Song XL, Zhang TZ. Genetic mapping of EST-derived microsatellites from the diploid Gossypium arboreum in allotetraploid cotton. Mol Genet Genomics. 2004;272:308–327. doi: 10.1007/s00438-004-1059-8. [DOI] [PubMed] [Google Scholar]
- 22.Graham J, Smith K, MacKenzie K, Jorgenson L, Hackett C, Powell W. The construction of a genetic linkage map of red raspberry (Rubus idaeus subsp. idaeus) based on AFLPs, genomic-SSR and EST-SSR markers. Theor Appl Genet. 2004;109:740–749. doi: 10.1007/s00122-004-1687-8. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Bowman K, Choi YA, Dang PM, Rao MN, Huang S, Soneji J, McCollum T, Gmitter F. EST-SSR genetic maps for Citrus sinensis and Poncirus trifoliata. Tree Genet Genomes. 2008;4:1–10. [Google Scholar]
- 24.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 25.Sneath PH, Sokal RR. Numerical taxonomy: the principles and practice of numerical classification. W.H. Freeman; San Francisco: 1973. [Google Scholar]
- 26.Guilford P, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R. Microsatellites in Malus X domestica (apple): abundance, polymorphism and cultivar identification. Theor Appl Genet. 1997;94:249–254. [Google Scholar]
- 27.Kashkush K, Jinggui F, Tomer E, Hillel J, Lavi U. Cultivar identification and genetic map of mango (Mangifera indica). Euphytica. 2001;122:129–136. [Google Scholar]
- 28.Chowdhury MA, Vandenberg B, Warkentin T. Cultivar identification and genetic relationship among selected breeding lines and cultivars in chickpea (Cicer arietinum L.). Euphytica. 2002;127:317–325. [Google Scholar]
- 29.Aranzana MJ, Carbo J, Arus P. Microsatellite variability in peach [Prunus persica (L.) Batsch]: cultivar identification, marker mutation, pedigree inferences and population structure. Theor Appl Genet. 2003;106:1341–1352. doi: 10.1007/s00122-002-1128-5. [DOI] [PubMed] [Google Scholar]
- 30.Moisan-Thiery M, Marhadour S, Kerlan MC, Dessenne N, Perramant M, Gokelaere T, Le Hingrat Y. Potato cultivar identification using simple sequence repeats markers (SSR). Potato Res. 2005;48:191–200. [Google Scholar]
- 31.Wang H, Sun H, Kwon WS, Jin H, Yang DC. A simplified method for identifying the Panax ginseng cultivar Gumpoong based on 26S rDNA. Planta Med. 2010;76:399–401. doi: 10.1055/s-0029-1186161. [DOI] [PubMed] [Google Scholar]
- 32.Lee JW, Bang KH, Kim YC, Seo AY, Jo IH, Lee JH, Kim OT, Hyun DY, Cha SW, Cho JH. CAPS markers using mitochondrial consensus primers for molecular identification of Panax species and Korean ginseng cultivars (Panax ginseng C. A. Meyer). Mol Biol Rep. 2012;39:729–736. doi: 10.1007/s11033-011-0792-4. [DOI] [PubMed] [Google Scholar]
- 33.Kim OT, Bang KH, In DS, Lee JW, Kim YC, Shin YS, Hyun DY, Lee SS, Cha SW, Seong NS. Molecular authentication of ginseng cultivars by comparison of internal transcribed spacer and 5.8S rDNA sequences. Plant Biotechnol Rep. 2007;1:163–167. [Google Scholar]
- 34.Jo IH, Bang KH, Kim YC, Lee JW, Seo AY, Seong BJ, Kim HH, Kim DH, Cha SW, Cho YG, et al. Rapid identification of ginseng cultivars (Panax ginseng Meyer) using novel SNP-based probes. J Ginseng Res. 2011;35:504–513. doi: 10.5142/jgr.2011.35.4.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Sun H, Kwon WS, Jin H, Yang DC. Molecular identification of the Korean ginseng cultivar “Chunpoong” using the mitochondrial nad7 intron 4 region. Mitochondrial DNA. 2009;20:41–45. doi: 10.1080/19401730902856738. [DOI] [PubMed] [Google Scholar]


