Growth factor-mobilized peripheral blood progenitor cell products were collected from healthy donors and processed by elutriation. Fractions were collected and analyzed for cell count, viability, and blood cell differential. Using modified elutriation procedures, >99% of red blood cells and platelets were removed from apheresis products with high recoveries of total white blood cells and enrichment of CD34+ cells in one fraction. This process is proven to be a feasible method for the initial manipulation associated with primary blood cell therapy products and supports current good manufacturing process and tissue practice-compliant cell processing.
Keywords: Stem cell, CD34+, Immunodeficient mouse, Cellular therapy
Abstract
Cell separation by counterflow centrifugal elutriation has been described for the preparation of monocytes for vaccine applications, but its use in other current good manufacturing practice (cGMP) operations has been limited. In this study, growth factor-mobilized peripheral blood progenitor cell products were collected from healthy donors and processed by elutriation using a commercial cell washing device. Fractions were collected for each product as per the manufacturer's instructions or using a modified protocol developed in our laboratory. Each fraction was analyzed for cell count, viability, and blood cell differential. Our data demonstrate that, using standard elutriation procedures, >99% of red blood cells and platelets were removed from apheresis products with high recoveries of total white blood cells and enrichment of CD34+ cells in two of five fractions. With modification of the basic protocol, we were able to collect all of the CD34+ cells in a single fraction. The CD34-enriched fractions were formulated, labeled with a ferromagnetic antibody to CD34, washed using the Elutra device, and transferred directly to a magnetic bead selection device for further purification. CD34+ cell purities from the column were extremely high (98.7 ± 0.9%), and yields were typical for the device (55.7 ± 12.3%). The processes were highly automated and closed from receipt of the apheresis product through formulation of target-enriched cell fractions. Thus, elutriation is a feasible method for the initial manipulations associated with primary blood cell therapy products and supports cGMP and current good tissue practice-compliant cell processing.
Introduction
Hematopoietic stem and progenitor cells (HSPCs) have been widely used to provide long-lasting hematopoietic reconstitution following ablative therapy for cancer [1–8] and in gene therapy applications [9–15]. However, the inherent plasticity in CD34 differentiation and apparent paracrine effects on necrotic or ischemic tissue has generated significant nonhomologous application of this important cell source. Specifically, the clinical utility of CD34+ cells in critical limb ischemia [16–18], chronic liver disease [19], and postinfarct myocardial recovery [20–23] has been widely evaluated. With such expanded use of CD34+ cells for cellular therapy, the isolation and enrichment of these cells is of great interest to investigators for both research and clinical therapeutic development.
The most readily available source of CD34+ human HSPCs is granulocyte-colony stimulating factor (G-CSF)-mobilized peripheral blood, which is collected as an apheresis product, hematopoietic progenitor cell apheresis (HPC-A). HPC-A products typically contain 10–50 × 109 white blood cells and more than 50 × 1010 platelets and red blood cells. Platelets and red blood cells must be removed from these products prior to subsequent manipulations designed to enrich for target cell populations (e.g., CD34-cell enrichment). Additionally, the HPC-A product must be buffer-exchanged and incubated with antibody-coated magnetic beads (or fluorochrome-conjugated antibodies), then washed again to remove nonbound antibody, and formulated in the specified amount and type of buffer for cell enrichment over a magnetic cell selection device or fluorescence-activated cell sorter. Typically, washing and formulation procedures are performed manually using repeated cycles of centrifugation followed by removal of supernatant with a plasma extractor and dilution in buffered saline. This procedure is time-consuming, labor-intensive, and subject to operator-related variability. Moreover, the manipulations required to keep the system closed (repeated tubing welds to buffer bags, removal for centrifugation, and then rewelding for next buffer/wash) increase the potential for contamination or leakage of the product. Thus, we determined that a more robust system for processing HPC-A products was warranted in support of our good manufacturing practice and manufacturing operations.
Since it was first introduced in the 1970s [24–28], counterflow centrifugal elutriation (CCE) has been used extensively in research applications to separate cell products on the basis of size and density. More recently, a clinical elutriation device has been developed (Elutra; GambroBCT, Lakewood, CO, http://www.caridianbct.com) and has been successfully used to isolate monocytes from peripheral blood apheresis products for vaccine applications [29–36] and lymphocytes for adoptive immunotherapy [37]. During elutriation, platelets and red blood cells are efficiently separated from white blood cells with monocytes highly enriched in a single fraction. On the basis of these results, we began an evaluation for the use of the Elutra system as a general tool for preparation of HPC-A products for downstream processing. Our results support the implementation of this automated approach for HSPC isolation in most cell-processing laboratories.
Materials and Methods
Starting Material
HPC-A products were obtained from AllCells LLC (Emeryville, CA, http://www.allcells.com), Key Biologics LLC (Memphis, TN, http://www.keybiologics.com), or Progenitor Cell Therapy LLC (Mountain View, CA, http://www.pctcelltherapy.com). HPC-A products were collected from healthy adults following 3–4 days of G-CSF (5–10 μg/kg/day) mobilization and processed within 24 hours of collection. Informed consent was obtained for each donor by individual vendors according to vendor-specific protocols and institutional review board review.
Cell Counts
White blood cell (WBC), red blood cell (RBC), and platelet counts were obtained using an AcT 5diff CP Hematology Analyzer (Beckman Coulter, Brea, CA, http://www.beckmancoulter.com) according to the manufacturer's protocol. Cell viability was determined using the Guava Viacount Assay (Guava Technologies, Hayward, CA, http://www.guavatechnologies.com) in accordance with the manufacturer's recommendations.
Elutriation
The disposable Elutra tubing set is presterilized and is a functionally closed system that provides a means to individually connect blood, buffer, waste, and final product bags to a spinning cell separation chamber. Each Elutra set can process up to 400 ml of starting product, provided that cell count does not exceed 3 × 1010 WBCs or 7.5 ml of RBCs. Elutra protocol 1 is the standard manufacturer's protocol that separates cells into five fractions and is intended for enrichment of monocytes in fraction 5. Fraction 1 is collected using a pump speed of 37 ml/minute, 2,400 rpm. Fraction 2 is collected using a pump speed of 68 ml/minute, 2,400 rpm in approximately 975 ml. Fraction 3 is collected using a pump speed of 74 ml/minute, 2,400 rpm in 975 ml. Fraction 4 is collected using a pump speed of 103 ml/minute, 2,400 rpm in 975 ml. Fraction 5 is collected using a pump speed of 125 ml/minute at 0 rpm in approximately 300 ml. Elutra protocol 2 was developed in our laboratory in collaboration with the manufacturer and elutriates cells into three fractions. Fraction 1 is collected using a pump speed of 60 ml/minute, 2,400 rpm in approximately 1,000 ml. Fraction 2 is collected using a pump speed of 25 ml/minute, 2,400 rpm in 450 ml. Fraction 3 is collected by stopping the rotor (0 rpm) and pumping at 5 ml/minute to a final volume of 50–80 ml.
The Elutra was also used to wash the bead-labeled cells in conjunction with protocol 2. After bead labeling, the cell bag was reconnected to the original Elutra disposable set. Cells were transferred using a cell inlet flow rate of 10 ml/minute, 2,400 rpm using 200 ml of a CliniMACS (Miltenyi Biotec, Bergisch Gladbach, Germany, http://www.miltenyibiotec.com) ethylenediaminetetraacetic acid (EDTA) phosphate-buffered saline (PBS) + 0.5% human serum albumin (HSA) buffer. An additional 500-ml buffer was used to wash the cells with a gradual increase in flow rate of 2 ml/minute every 2 minutes to a maximum of 25 ml/minute. Then, the cell inlet flow rate was set to 0 ml/minute and rotor speed adjusted to 2,000 rpm to sediment the cells. The cells were then transferred to the collection bag at 5 ml/minute. Once all cells were removed from the chamber and the outlet line, the debulk pump flow rate was set to 25 ml/minute and the medium pump flow rate to 30 ml/minute to clear the line of any residual cells. The cells were collected in a maximum volume of 120 ml for loading onto the CliniMACS device.
Bag Wash by Centrifugation
G-CSF-mobilized apheresis products were diluted with three volumes of CliniMACS buffer + 0.5% HSA before magnetic labeling. Cells were pelleted by centrifugation at 200g, room temperature, acceleration 9, and deceleration 2 for 15 minutes. Samples for analysis were taken after an additional wash following CD34+ microbead labeling.
CD34+ Selection
For CD34 enrichment, elutriated cells were pelleted and then resuspended in CliniMACS/EDTA PBS + 0.5% HSA at a concentration of 6 × 108 WBCs/ml. Human immunoglobulin (ZLB Behring, Berne, Switzerland, http://www.cslbehring.com) was added at a final concentration of 1.6 mg/ml to block nonspecific binding of antibody. CliniMACS CD34 MicroBeads (Miltenyi Biotec) were added using the ratio of 7.5 ml of beads per 6 × 108 CD34+ cells and mixed well. The mixture was placed on an orbital shaker (25 rpm) and incubated for 30 minutes at room temperature. The labeled cell mixture was washed twice with 10 volumes of CliniMACS/EDTA PBS + 0.5% HSA (pelleted by centrifugation) and resuspended to a cell concentration of ≤4 × 108 cells per milliliter. Alternatively, magnetic bead-labeled cells were washed free of unbound magnetic beads by transfer to the Elutra as described above. Finally, the cells were rinsed in the debulk line at 25 ml/minute, 0 rpm in approximately 50 ml. Washed cells were selected on CliniMACS tubing set 150 using enrichment mode 3.2 according to the manufacturer's directions. Positively selected CD34+ cells were collected, and a sample was analyzed immediately. The remaining CD34-enriched cells were cryopreserved in CryoStor CS5 cell-freezing medium (BioLife Solutions, Inc., Bothell, WA, http://www.biolifesolutions.com) using a controlled rate freezer (Planer, Sunbury-on-Thames, U.K., http://www.planer.com) and stored in the vapor phase of liquid nitrogen.
Phenotypic Analysis
Antibodies used for phenotyping included CD34-PE (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com); CD3-APC-Alexa 750, CD14-fluorescein isothiocyanate (FITC), CD19-APC- Alexa 700, and CD56-PE-Texas Red (Invitrogen, Carlsbad, CA, http://www.invitrogen.com); and CD15-Alexa 647 (BioLegend, San Diego, http://www.biolegend.com). Background levels were determined with isotype-matched control antibodies, including mouse IgG1-Alexa 647 (AbD Serotec, Raleigh, NC, http://www.ab-direct.com), IgG1-APC-Alexa 700 (Invitrogen), IgG1-APC- Alexa 750 (Beckman Coulter), and IgG1-FITC and IgG1-PE (BD Biosciences).
An aliquot from each stage of the isolation process was evaluated for expression of CD34, CD3, CD14, CD15, CD19, and CD56 via flow cytometry. Each sample (3 × 105 cells) was incubated with the appropriate antibodies for 20 minutes on ice and washed three times with at least an equal volume of PBS (Irvine Scientific, Santa Ana, CA, http://www.irvinesci.com) containing 0.1% bovine serum albumin (BSA) (Sigma-Aldrich, St. Louis, http://www.sigmaaldrich.com). Flow cytometric data were collected on the Gallios cytometer (Beckman Coulter) and analyzed with FCS Express software (De Novo Software, Los Angeles, http://www.denovosoftware.com).
Mice
NOD.Cg-prkdcscid IL2rgtm1Wjl/SzJ (NSG) mice were originally obtained from Jackson Laboratory (Bar Harbor, ME, http://www.jax.org) and then bred at the Animal Resource Center at Beckman Research Institute. The 8–10-week-old healthy NSG mice were irradiated with 300 cGy. After 24 hours, the mice received either 0 or 1 million CD34+ cells via tail vein injection. Sulfamethoxazole and trimethoprim water (1:50) (Hi-Tech Pharmacal Co. Inc., Amityville, NY, http://www.hitechpharm.com) were given to the mice on the day of irradiation and continuously thereafter. All experimentations with mice were performed under protocols approved by the Institutional Animal Care and Use Committee of City of Hope National Medical Center/Beckman Research Institute.
To measure the engraftment, mice were euthanized by CO2 inhalation 8 weeks posttransplantation. Spleen and two femurs were collected and processed into single-cell suspensions. Cells were incubated with human IgG (ZLB Behring) and mouse IgG (BD Biosciences) for 20 minutes. Spleen cells were stained by anti-CD45-PC5 (BioLegend) to identify human white blood cells, anti-CD19-PE (Invitrogen) (human B cells), anti-CD4-ECD (Beckman Coulter), anti-CD8-PC7 (Invitrogen) (human T-cell subsets), and anti-CD14-APC-Alexa 750 (Invitrogen) antibodies (human monocytes) for 20 minutes and washed three times with an equal volume of PBS containing 0.1% BSA. Bone marrow cells were stained by anti-CD45-ECD (Beckman Coulter), anti-CD19-PE, anti-CD33-PC5 (BD Biosciences) (human myeloid precursors), anti-CD14-APC-Alexa 750, and anti-CD34-PC7 (Beckman Coulter) (human HSPC) antibodies for 20 minutes and washed three times with an equal volume of PBS containing 0.1% BSA. Single-color isotype controls were purchased from the same company from which the antibody was purchased, except PE isotype (BD Biosciences). Samples were analyzed by the Gallios cytometer and analyzed with FCS Express software.
Statistical Analysis
Analyses of the arithmetic mean for each group (average), distribution of values around the mean (SD), lowest value (minimum), and highest value (maximum) of cell fraction content were performed using GraphPad Prism software (LaJolla, CA, http://www.graphpad.com) using standard methods. For statistical analysis of significance, conditions were compared using an unpaired, two-tailed t test. Samples with a p value <.05 were considered significantly different.
Results
Elutriation Development
HPC-A products entering the laboratory were processed according to cell number and volume of RBCs, as outlined in Figure 1. If the total number of WBCs in the product was >3 × 1010 or the total red-cell volume exceeded 7.5 ml, the product was split and run in two separate elutriation runs. This improved separation of cells in each run and improved our ability to resolve CD34+ cells into discrete fractions. In an effort to optimize the use of CCE for processing of mobilized peripheral blood products, two elutriation protocols were tested. Initially, we used a “vendor-developed” elutriation protocol designed to enrich monocytes from apheresis products by collecting five elutriation fractions (elutriation protocol 1). Subsequently, we made incremental modifications to the buffer flow rate in an attempt to improve CD34+ cell recovery and reduce the number of fractions collected from five to three (elutriation protocol 2).
Figure 1.
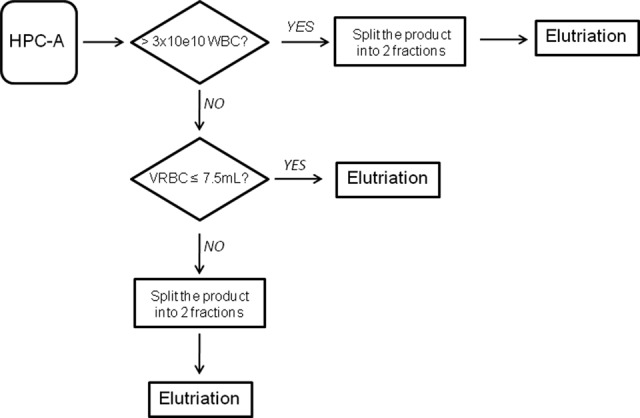
Elutriation process flowchart. Human granulocyte-colony stimulating factor-mobilized apheresis product is split into two products before elutriation if the number of WBCs is greater than 3 × 1010 or if the total volume of RBCs is greater than 7.5 ml (for protocol 1) or greater than 15 ml (for protocol 2). Abbreviations: HPC-A, hematopoietic progenitor cell apheresis; VRBC, volume of red blood cells; WBC, white blood cell.
We performed 10 elutriation runs on HPC-A from eight donors using elutriation protocol 1. An average of 4.26 × 1010 total WBCs (range, 2.48–7.19) were collected per apheresis product and processed according to total cell number and RBC counts outlined in Figure 1. An average of 2.75 × 1010 WBCs (range, 1.97–2.97) and ≤7.5 ml of RBCs were included in each processing run. Cells were loaded into the elutriation chamber and washed with Hanks' balanced salt solution (HBSS) (Lonza, Walkersville, MD, http://www.lonza.com) supplemented with 1% HSA (Grifols, Los Angeles, http://www.grifolsusa.com) at a flow rate of 37 ml/minute using a total volume of 900 ml. The supernatant from the load/wash step (F1) contained mostly platelets and red blood cells (Fig. 2A). Fractions 2, 3, and 4 (F2–F4) were eluted from the chamber by increasing buffer flow rates to 68, 74, and 103 ml/minute, respectively, and collecting 975 ml per fraction. The remaining cells in the chamber were collected by stopping the centrifugation and setting the medium flow rate to 125 ml/minute for a total collection volume of 300 ml (F5). A three-part differential count revealed that fractions 2–5 contained virtually all of the WBCs in the sample. These fractions were analyzed for lymphoid, myeloid, and CD34+ cell content.
Figure 2.
Fractionation of platelets, RBCs, and WBCs. (A): Profile of platelets, RBCs, and WBCs for protocol 1 for eight tissues. (B): Profile of platelets, RBCs, and WBCs for protocol 2 for two tissues (three elutriation runs). Abbreviations: RBC, red blood cell; WBC, white blood cell.
Our results indicate that small CD3+ or CD19+ lymphocytes (low mean forward light scatter) were contained mostly in F2 and F3 (Fig. 3A), whereas larger lymphocytes (higher mean forward light scatter) and the majority of CD34+ cells (52.7 ± 21.6%) elutriated into F4 but could also be found in F2 (three of eight tissues) and F5 (six of eight tissue) (13.8 ± 17% and 18.9 ± 13.8%, respectively). Fractions 1 and 3 did not contain significant numbers of CD34+ cells (0.7 ± 1.8% and 3.2 ± 5.3%, respectively). Fraction 5 contained a small percentage of CD15+ granulocytes and virtually all of the CD14+ monocytes. Prior to elutriation, there was an average of 210 × 106 CD34+ cells (range, 92–284) in the apheresis sample. Using protocol 1 and by combining fractions 2–5, we collected an average of 160 × 106 CD34+ cells (range, 84–214) with an average of 80% recovery (range, 54%–96%) of the CD34+ cells in the starting product (Table 1). We noted that CD34+ cells contained mostly in F2 were smaller than the CD34+ cells in F5 but did not evaluate the relationship between size and activity, as the distribution of cells in the two fractions was highly variable and we wished to capture all CD34+ in a single fraction. Cell viability was >85% in all fractions.
Figure 3.
Representative immunotype of elutriation fractions 2, 3, 4, and 5 of protocol 1 and fractions 1, 2, and 3 of protocol 2. (A): Flow cytometry plots of fractions 2, 3, 4, and 5 depicting FS versus SS, CD56 versus CD3, CD14 versus CD15, and CD34 versus CD19. (B): Flow cytometry plots of fractions 1, 2, and 3 depicting SS versus CD34. Abbreviations: FS, forward scatter; SS, side scatter.
Table 1.
Cell recovery after elutriation and purity from CD34+ selection

Comparison of the average, SD, minimum, and maximum of the percentage of CD34+ recovery and purity for protocols 1 and 2 (n = 8 and n = 3, respectively).
Modified elutriation settings were created (elutriation protocol 2) to deplete platelets, reduce cells similar in size to RBCs, and concentrate the WBCs (including all CD34+ cells) into a single fraction. The initial wash was conducted using a slightly increased flow rate (60 ml/minute vs. 37 ml/minute) followed by a line clearance at 25 ml/minute (F2), and then the rotor slowed to 2,000 rpm and all remaining cells were elutriated at 5 ml/minute to carefully control final volume. Five elutriation runs were performed on HPC-A from three donors using elutriation protocol 2. Prior to elutriation, there was an average of 319 × 106 CD34+ cells (range, 83–804). After elutriation, we collected an average of 252 × 106 CD34+ cells (range, 87–369), with an average 92% recovery (range, 75%–107%) of the CD34+ cells in the starting product using protocol 2 resulting in a single collected fraction (Table 1).
We used protocol 2, rather than protocol 1, to remove platelets and erythrocytes and enrich for the target (CD34+) leukocytes in a single fraction. Using this approach, >95% of platelets and RBCs were collected in fraction 1 along with approximately 20% of all leukocytes (Fig. 2B). Because of the high RBC content of fraction 1, we were unable to perform phenotypic analysis of lineage distribution. Three-part differential analysis, however, revealed that fraction 1 contained an approximately equal distribution of lymphocytes and neutrophils, 57% and 41%, respectively. Virtually all of the CD34+ cells were contained within fraction 3, as determined by flow cytometric analysis (Fig. 3B). The segregation of CD34− leukocytes in fraction 1 led to a modest fold increase in CD34+ cell frequency in fraction 3 (1.85 ± 0.28). The enrichment of the frequency of CD34+ cells in a single fraction from HPC-A products starting with low CD34 contents (<0.5%) potentially improves the yield and purity of CD34+ cells during subsequent magnetic bead collection. In subsequent studies, protocol 2 was used prior to CD34+ enrichment by magnetic column purification.
Comparison of Elutriation to Bag Washing
Having established optimized elutriation conditions, we wished to determine the utility of elutriation compared with standard methods of cell washing (bag-wash). HPC-A products were elutriated as described above or washed with three volumes of HBSS, and cells were pelleted by centrifugation in the HPC-A collection bag. A plasma extractor was used to remove supernatant, and cells were resuspended in CliniMACS buffer with 0.5% HSA. Analysis of the final washed, pooled products demonstrated a significant reduction of the number of remaining platelets and red blood cells in the elutriated samples versus the bag-wash samples (Fig. 4). This resulted in products that formed fewer clumps and more solid pellets upon subsequent centrifugation.
Figure 4.

Comparison of RBCs and platelets after bag wash versus elutriated wash. Total red blood cell counts (n = 5) (A) and platelet counts (n = 4) (B) in samples in bag-washed and elutriated hematopoietic progenitor cell apheresis samples. Significance: p < .01 and p < .001, as indicated. Abbreviation: RBC, red blood cell.
In subsequent studies, we evaluated the performance of HPC-A products processed by elutriation or by traditional bag washing methods prior to CD34+ enrichment by magnetic column purification. We first compared the two elutriation protocols to determine whether there were differences in yield and purities of CD34+ cells between the two procedures. Prior to selection, there was an average of 283.3 × 106 CD34+ cells (range, 77–869) in each washed HPC-A product. An average of 142.3 × 106 CD34+ cells (range, 32–406) were obtained from the column for a 54.2% average yield (range, 41%–81%) through the CliniMACS selection process. The CD34+ cells had a purity of 99% (range, 97%–99%). No differences were seen in the purity or yield of CD34+ cells isolated using either protocol (Table 1). We then compared the yields and purities of magnetically enriched CD34+ cells derived from bag-washed and elutriated HPC-A products. CD34+ cells isolated from the bag-washed product had an average purity of 96.8 ± 2%, whereas CD34+ cells from elutriated products had a significantly higher average purity of 98.9 ± 0.7% (p < .0053) (Fig. 5A). CD34+ recovery was 48.7 ± 12.6% in the bag-washed population and 54.2 ± 11.7% in the elutriated population and was not significantly different (Fig. 5B).
Figure 5.

CD34 selection results. (A):CD34+ purity following bag centrifugation (n = 12) versus Elutra processing (n = 13). (B): Same as (A) for CD34+ recovery.
Engraftment with Enriched CD34+ HSPCs
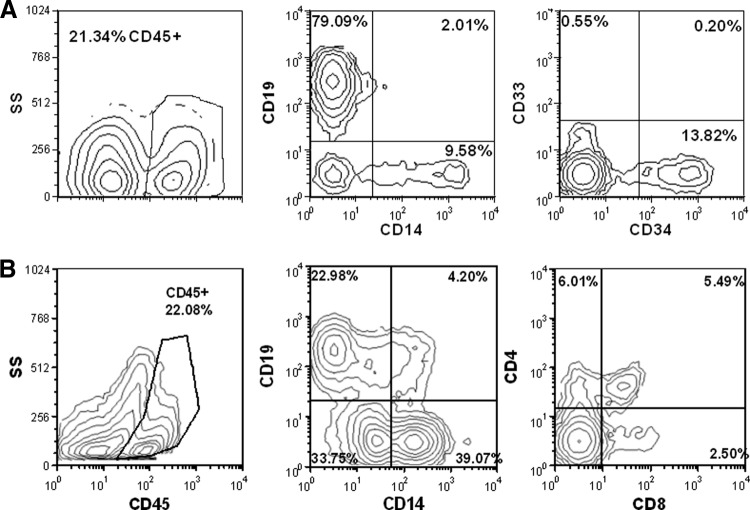
We wished to evaluate the potency of the CD34+ HSPCs processed using the revised elutriation method. Cohorts of 8-week-old NSG mice were irradiated with 300 cGy and then injected via the tail vein with either 5 × 105 or 1 × 106 CD34+ HSPCs or PBS. Eight and a half weeks after transplant, the mice were evaluated for the extent of human cell engraftment and lineage distribution. We observed engraftment of human (CD45+) cells in the bone marrow and spleen of five of five mice transplanted with CD34+ HSPCs (average, 12.2; range, 2–36). An example of multilineage engraftment of blood, bone marrow, and spleen is shown (Fig. 6). The bone marrow contained predominantly CD19+ B cells with evidence of CD14+ monocytes as well as CD34+/CD33− progenitors (Fig. 6A). Conversely, the predominant population of cells in the spleen were CD14+ monocytes, but CD4+ and CD8+ T lymphocytes and CD19+ B cells were also present (Fig. 6B). Taken together, these data demonstrate CD34+ cells isolated according to revised elutriation protocols retain multilineage in vivo engrafting capability with maintenance of the progenitor cell compartment in the bone marrow and thus retain the biological properties of hematopoietic stem cells required for blood replacement therapies.
Figure 6.
Hematopoietic stem and progenitor cells (HSPCs) engrafted on NOD.Cg-prkdcscid IL2rgtm1Wjl/SzJ mouse. (A): Example of human cell engraftment in the bone marrow of a mouse 8.5 weeks after transplantation with HSPCs. (B): Analysis of spleen of same mouse. All subsequent analysis of lineage distribution is among the CD45+ population. Antibodies identify specific human cell lineages, as described in Materials and Methods. Abbreviation: SS, side scatter.
Discussion
The isolation of hematopoietic stem and progenitor cells from peripheral blood apheresis products (HPC-A) has become a common procedure for a steadily increasing number of applications. Many laboratories have been able to perform pilot studies using manual methods for washing and preparing products for downstream processing steps, such as CD34+ HSPC enrichment. However, the introduction of an automated process for upfront processing that allows for strictly defined standardized procedures with highly reproducible results would enhance outcomes and support larger scale clinical trials and commercial use of such procedures.
We (and others) previously reported on the use of the Cytomate (Nexell, Irvine, CA) device to wash HPC-A products prior to downstream processing steps [38–43]. Unfortunately, this device is no longer manufactured, and thus we sought an alternative for HPC-A processing. We wished to avoid processes and devices that involved open steps (pipetting), repeated procedures (centrifugation), and/or operator variability in bag processing (expressing plasma or buffer). The Elutra was identified as an ideal device, as it was made for processing blood products (a descendant of the Cobe Spectra [Terumo, Lakewood, CO, http://www.terumobct.com] series of blood devices) and had been successfully used for clinical manufacturing of dendritic cell products. Service and support of the device was provided by the manufacturer, who has a significant presence in the blood cell-processing field. Therefore, we were likely to be able to obtain devices and disposables for the foreseeable future.
We have developed a series of standard operating procedures for counterflow centrifugal elutriation of HPC-A products using the Elutra along with disposable tubing and processing sets that eliminate the need for open manipulations. Minor modification to fluid flow rates and rotor speeds allowed us to identify conditions that result in the isolation of virtually all of the CD34+ HSPC in a single fraction. As a result, the HPC-A preparation process was shortened by 45–60 minutes, and we completely eliminated the need for centrifugation during product washing and formulation. This resulted in significant savings in terms of labor and capital equipment requirements. Moreover, increasing the frequency of CD34+ cells in the preselected product improves average yield and purity when CD34+ cell frequencies in the original HPC-A product are <0.5% (D.D., unpublished observation). Since we wished to determine cell yields and purities at full scale, we did not split products to compare the performance of each process on the same sample and evaluated products from 12 separate donors for the bag process and 13 for elutriation. When the HPC-A product did exceed the capacity of the processing chamber (3 × 1010 WBC), we processed with sequential elutriation runs to establish the feasibility of handling large products in this fashion. This latter issue may be readily addressed by using a larger capacity device, such as the K-Sep 400 elutriation device (KBI Biopharma Inc., Durham, NC, http://www.kbibiopharma.com).
After elutriation, we wished to further enrich for CD34+ cells by magnetic bead selection using the CliniMACS device. Typically, cells are labeled with magnetic beads in a blood bag, and then the unbound beads are removed through a series of buffer additions, centrifugation, and supernatant removal. At each step, cell bags are welded to buffer bags, filled with buffer, removed, centrifuged, and welded to a waste bag; the supernatant is removed using a plasma extractor; and the process is repeated. This series of manipulations is time-consuming, labor-intensive, and potentially susceptible to contamination from repeated tubing welding and removal steps required for the wash. Additionally, in some cases, the bags rupture during centrifugation, leading to product loss. In order to reduce the number of manipulations required to wash away the nonbound beads, we also used the Elutra to perform the postincubation wash steps in a single closed cycle with the same disposable set as was used to wash the initial product. This resulted in an equivalent yield of cells comparable to the bag wash method in considerably less time with fewer operator interventions. Subsequent magnetic purification of elutriated cells produced a population of CD34+ HSPCs with significantly high purity and greater reproducibility. This is a critical step, as higher purities of CD34+ cells means fewer T cells and a lower overall chance for graft-versus-host disease when these cells are used in allogeneic transplantation protocols.
The process has been conducted by several members of the laboratory staff with essentially equivalent results, suggesting that it is not operator-dependent. We have not observed any inherent loss of viability, defects, or reduction in hematopoietic potential following these procedures. However, in order to ensure that we had not adversely affected the cells or mistakenly eliminated a fraction of cells required for engraftment, we evaluated the in vivo hematopoietic potential of elutriated, CD34-selected HSPCs. Our results demonstrate that these cells are suitable for engraftment and are able to maintain the CD34+ bone marrow compartment for at least 8 weeks following transplant. This serves as a functional demonstration of maintenance of homologous activity of the cells and addresses regulatory requirements for the potency of such products.
Conclusion
Thus, although elutriation devices have been commercialized for monocyte enrichment, they are easily adapted to optimize the collection of CD34+ cells in a single fraction. The methods are robust and can be automated to eliminate variability from site to site. CD34+ HSPCs isolated by this process are routinely used in our laboratories for preclinical process development and clinical materials manufacturing and meet most (if not all) regulatory requirements for phase I/II clinical investigations.
Acknowledgments
This research was supported by grants from the California Institute for Regenerative Medicine (Grants DR1-01490 and TR2-01771, to D.D.). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of CIRM or any other agency of the State of California. We thank Andrea Jeworowski (formerly at Caridian BCT) and Linda A. Taylor (Caridian BCT) for their support in the process development of this project.
Author Contributions
C.-A.T., M.T.-C., and A. Gu: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; A. Gardner: conception and design, collection and assembly of data; H.V. and L.-F.C.: collection and assembly of data; A.R.: conception and design; A.A.: administrative support; D.D.: conception and design, data analysis and interpretation, manuscript writing, financial support, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Scheding S, Brugger W, Mertelsmann R, et al. Peripheral blood stem cells: In vivo biology and therapeutic potential. Stem Cells. 1994;12(suppl 1):203–210. doi: 10.1002/stem.5530120717. discussion 211. [DOI] [PubMed] [Google Scholar]
- 2.Berenson RJ, Shpall EJ, Auditore-Hargreaves K, et al. Transplantation of CD34+ hematopoietic progenitor cells. Cancer Invest. 1996;14:589–596. doi: 10.3109/07357909609076903. [DOI] [PubMed] [Google Scholar]
- 3.Johnson RJ, Owen RG, Smith GM, et al. Peripheral blood stem cell transplantation in myeloma using CD34 selected cells. Bone Marrow Transplant. 1996;17:723–727. [PubMed] [Google Scholar]
- 4.Handgretinger R, Greil J, Schürmann U, et al. Positive selection and transplantation of peripheral CD34+ progenitor cells: Feasibility and purging efficacy in pediatric patients with neuroblastoma. J Hematother. 1997;6:235–242. doi: 10.1089/scd.1.1997.6.235. [DOI] [PubMed] [Google Scholar]
- 5.Somlo G, Sniecinski I, Odom-Maryon T, et al. Effect of CD34+ selection and various schedules of stem cell reinfusion and granulocyte colony-stimulating factor priming on hematopoietic recovery after high-dose chemotherapy for breast cancer. Blood. 1997;89:1521–1528. [PubMed] [Google Scholar]
- 6.Handgretinger R, Lang P, Schumm M, et al. Isolation and transplantation of autologous peripheral CD34+ progenitor cells highly purified by magnetic-activated cell sorting. Bone Marrow Transplant. 1998;21:987–993. doi: 10.1038/sj.bmt.1701228. [DOI] [PubMed] [Google Scholar]
- 7.Gandhi M, Jestice H, Scott M, et al. A comparison of CD34+ cell selected and unselected autologous peripheral blood stem cell transplantation for multiple myeloma: A case controlled analysis. Bone Marrow Transplant. 1999;24:369–375. doi: 10.1038/sj.bmt.1701938. [DOI] [PubMed] [Google Scholar]
- 8.Vescio R, Schiller G, Stewart AK, et al. Multicenter phase III trial to evaluate CD34(+) selected versus unselected autologous peripheral blood progenitor cell transplantation in multiple myeloma. Blood. 1999;93:1858–1868. [PubMed] [Google Scholar]
- 9.Barese CN, Dunbar CE. Contributions of gene marking to cell and gene therapies. Hum Gene Ther. 2011;22:659–668. doi: 10.1089/hum.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yannaki E, Papayannopoulou T, Jonlin E, et al. Hematopoietic stem cell mobilization for gene therapy of adult patients with severe β-thalassemia: Results of clinical trials using G-CSF or plerixafor in splenectomized and nonsplenectomized subjects. Mol Ther. 2012;20:230–238. doi: 10.1038/mt.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papanikolaou E, Georgomanoli M, Stamateris E, et al. The new self-inactivating lentiviral vector for thalassemia gene therapy combining two HPFH activating elements corrects human thalassemic hematopoietic stem cells. Hum Gene Ther. 2012;23:15–31. doi: 10.1089/hum.2011.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaspar HB, Cooray S, Gilmour KC, et al. Hematopoietic stem cell gene therapy for adenosine deaminase-deficient severe combined immunodeficiency leads to long-term immunological recovery and metabolic correction. Sci Transl Med. 2011;3:97ra80. doi: 10.1126/scitranslmed.3002716. [DOI] [PubMed] [Google Scholar]
- 13.Frittoli MC, Biral E, Cappelli B, et al. Bone marrow as a source of hematopoietic stem cells for human gene therapy of β-thalassemia. Hum Gene Ther. 2011;22:507–513. doi: 10.1089/hum.2010.045. [DOI] [PubMed] [Google Scholar]
- 14.DiGiusto DL, Krishnan A, Li L, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sumiyoshi T, Holt NG, Hollis RP, et al. Stable transgene expression in primitive human CD34+ hematopoietic stem/progenitor cells, using the Sleeping Beauty transposon system. Hum Gene Ther. 2009;20:1607–1626. doi: 10.1089/hum.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Procházka V, Gumulec J, Jalůvka F, et al. Cell therapy, a new standard in management of chronic critical limb ischemia and foot ulcer. Cell Transplant. 2010;19:1413–1424. doi: 10.3727/096368910X514170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolvenbach R, Kreissig C, Cagiannos C, et al. Intraoperative adjunctive stem cell treatment in patients with critical limb ischemia using a novel point-of-care device. Ann Vasc Surg. 2010;24:367–372. doi: 10.1016/j.avsg.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Murphy MP, Lawson JH, Rapp BM, et al. Autologous bone marrow mononuclear cell therapy is safe and promotes amputation-free survival in patients with critical limb ischemia. J Vasc Surg. 2011;53:1565–1574. doi: 10.1016/j.jvs.2011.01.074. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pai M, Spalding D, Xi F, et al. Autologous bone marrow stem cells in the treatment of chronic liver disease. Int J Hepatol. 2012;2012:307165. doi: 10.1155/2012/307165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quyyumi AA, Waller EK, Murrow J, et al. CD34(+) cell infusion after ST elevation myocardial infarction is associated with improved perfusion and is dose dependent. Am Heart J. 2011;161:98–105. doi: 10.1016/j.ahj.2010.09.025. [DOI] [PubMed] [Google Scholar]
- 21.He H, Cao J, Wang D, et al. Gene-modified stem cells combined with rapid prototyping techniques: A novel strategy for periodontal regeneration. Stem Cell Rev. 2010;6:137–141. doi: 10.1007/s12015-009-9110-0. [DOI] [PubMed] [Google Scholar]
- 22.Losordo DW, Henry TD, Davidson C, et al. Intramyocardial, autologous CD34+ cell therapy for refractory angina. Circ Res. 2011;109:428–436. doi: 10.1161/CIRCRESAHA.111.245993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmet H, Porapakkham P, Porapakkham P, et al. Short- and long-term outcomes of intracoronary and endogenously mobilized bone marrow stem cells in the treatment of ST-segment elevation myocardial infarction: A meta-analysis of randomized control trials. Eur J Heart Fail. 2012;14:91–105. doi: 10.1093/eurjhf/hfr148. [DOI] [PubMed] [Google Scholar]
- 24.Abrahamsen TG, Carter CS, Read EJ, et al. Stimulatory effect of counterflow centrifugal elutriation in large-scale separation of peripheral blood monocytes can be reversed by storing the cells at 37 degrees C. J Clin Apher. 1991;6:48–53. doi: 10.1002/jca.2920060110. [DOI] [PubMed] [Google Scholar]
- 25.Stevenson HC. Isolation of human mononuclear leukocyte subsets by countercurrent centrifugal elutriation. Methods Enzymol. 1984;108:242–249. doi: 10.1016/s0076-6879(84)08087-3. [DOI] [PubMed] [Google Scholar]
- 26.Persidsky MD, Ling NS. Separation of platelet-rich plasma by modified centrifugal elutriation. J Clin Apher. 1982;1:18–24. doi: 10.1002/jca.2920010106. [DOI] [PubMed] [Google Scholar]
- 27.Contreras TJ, Jemionek JF, French JE, et al. Human granulocyte isolation by continuous flow centrifugation leukapheresis and counterflow centrifugation elutriation (CFCL/CCE) Transfusion. 1979;19:695–703. doi: 10.1046/j.1537-2995.1979.19680104095.x. [DOI] [PubMed] [Google Scholar]
- 28.Ito Y, Suaudeau J, Bowman RL. New flow-through centrifuge without rotating seals applied to plasmapheresis. Science. 1975;189:999–1000. doi: 10.1126/science.1220011. [DOI] [PubMed] [Google Scholar]
- 29.Micklethwaite KP, Garvin FM, Kariotis MR, et al. Clinical-scale elutriation as a means of enriching antigen-presenting cells and manipulating alloreactivity. Cytotherapy. 2009;11:218–228. doi: 10.1080/14653240802702160. [DOI] [PubMed] [Google Scholar]
- 30.Perseghin P, D'Amico G, Dander E, et al. Isolation of monocytes from leukapheretic products for large-scale GMP-grade generation of cytomegalovirus-specific T-cell lines by means of an automated elutriation device. Transfusion. 2008;48:1644–1649. doi: 10.1111/j.1537-2995.2008.01756.x. [DOI] [PubMed] [Google Scholar]
- 31.You Y, Xia LH, Zhang C, et al. Clinical observation of selected CD34(+) cell autologous transplantation in non-Hodgin lymphoma: Report of 5 cases [in Chinese] Zhonghua Yi Xue Za Zhi. 2007;87:3127–3129. [PubMed] [Google Scholar]
- 32.Lemarie C, Sugaye R, Kaur I, et al. Purification of monocytes from cryopreserved mobilized apheresis products by elutriation with the Elutra device. J Immunol Methods. 2007;318:30–36. doi: 10.1016/j.jim.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 33.Kasow KA, Sims-Poston L, Eldridge P, et al. CD34(+) hematopoietic progenitor cell selection of bone marrow grafts for autologous transplantation in pediatric patients. Biol Blood Marrow Transplant. 2007;13:608–614. doi: 10.1016/j.bbmt.2007.01.074. [DOI] [PubMed] [Google Scholar]
- 34.Erdmann M, Dörrie J, Schaft N, et al. Effective clinical-scale production of dendritic cell vaccines by monocyte elutriation directly in medium, subsequent culture in bags and final antigen loading using peptides or RNA transfection. J Immunother. 2007;30:663–674. doi: 10.1097/CJI.0b013e3180ca7cd6. [DOI] [PubMed] [Google Scholar]
- 35.Schwanke U, Nabereit A, Moog R. Isolation of monocytes from whole blood-derived buffy coats by continuous counter-flow elutriation. J Clin Apher. 2006;21:153–157. doi: 10.1002/jca.20077. [DOI] [PubMed] [Google Scholar]
- 36.Huang W, Tan W, Zhong Q, et al. Development of a gene therapy based bone marrow purging system for leukemias. Cancer Gene Ther. 2005;12:873–883. doi: 10.1038/sj.cgt.7700848. [DOI] [PubMed] [Google Scholar]
- 37.Hiwase DK, White DL, Powell JA, et al. Blocking cytokine signaling along with intense Bcr-Abl kinase inhibition induces apoptosis in primary CML progenitors. Leukemia. 2010;24:771–778. doi: 10.1038/leu.2009.299. [DOI] [PubMed] [Google Scholar]
- 38.Zinno F, Landi F, Aureli V, et al. Positive immunomagnetic CD34(+) cell selection in haplo-identical transplants in beta-thalassemia patients: Removal of platelets using an automated system. Cytotherapy. 2010;12:60–66. doi: 10.3109/14653240903348301. [DOI] [PubMed] [Google Scholar]
- 39.Giancola R, Olioso P, Di Riti M, et al. Evaluation of an automated closed fluid management device for processing expanded cytokine-induced killer cells to use in immunotherapy programs for cancer. Transfusion. 2008;48:629–639. doi: 10.1111/j.1537-2995.2007.01587.x. [DOI] [PubMed] [Google Scholar]
- 40.Mullally A, Ritz J. Beyond HLA: The significance of genomic variation for allogeneic hematopoietic stem cell transplantation. Blood. 2007;109:1355–1362. doi: 10.1182/blood-2006-06-030858. [DOI] [PubMed] [Google Scholar]
- 41.Lemarie C, Calmels B, Malenfant C, et al. Clinical experience with the delivery of thawed and washed autologous blood cells, with an automated closed fluid management device: CytoMate. Transfusion. 2005;45:737–742. doi: 10.1111/j.1537-2995.2005.04126.x. [DOI] [PubMed] [Google Scholar]
- 42.Perotti CG, Del Fante C, Viarengo G, et al. A new automated cell washer device for thawed cord blood units. Transfusion. 2004;44:900–906. doi: 10.1111/j.1537-2995.2004.03389.x. [DOI] [PubMed] [Google Scholar]
- 43.Calmels B, Houz é P, Hengesse JC, et al. Preclinical evaluation of an automated closed fluid management device: Cytomate, for washing out DMSO from hematopoietic stem cell grafts after thawing. Bone Marrow Transplant. 2003;31:823–828. doi: 10.1038/sj.bmt.1703905. [DOI] [PubMed] [Google Scholar]