This study demonstrates that fibroblasts from patients with ataxia-telangiectasia (A-T) can be reprogrammed as bona fide induced pluripotent stem cells (iPSCs), albeit at a reduced efficiency. It is shown that iPSCs can be generated from a chromosomal instability syndrome and that these cells can be used to discover early developmental consequences of ATM deficiency, such as altered mitochondrial function, that may be relevant to A-T pathogenesis and amenable to therapeutic intervention.
Keywords: Experimental models, Gene expression, Reprogramming, iPS
Abstract
Pluripotent stem cells can differentiate into every cell type of the human body. Reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) therefore provides an opportunity to gain insight into the molecular and cellular basis of disease. Because the cellular DNA damage response poses a barrier to reprogramming, generation of iPSCs from patients with chromosomal instability syndromes has thus far proven to be difficult. Here we demonstrate that fibroblasts from patients with ataxia-telangiectasia (A-T), a disorder characterized by chromosomal instability, progressive neurodegeneration, high risk of cancer, and immunodeficiency, can be reprogrammed to bona fide iPSCs, albeit at a reduced efficiency. A-T iPSCs display defective radiation-induced signaling, radiosensitivity, and cell cycle checkpoint defects. Bioinformatic analysis of gene expression in the A-T iPSCs identifies abnormalities in DNA damage signaling pathways, as well as changes in mitochondrial and pentose phosphate pathways. A-T iPSCs can be differentiated into functional neurons and thus represent a suitable model system to investigate A-T-associated neurodegeneration. Collectively, our data show that iPSCs can be generated from a chromosomal instability syndrome and that these cells can be used to discover early developmental consequences of ATM deficiency, such as altered mitochondrial function, that may be relevant to A-T pathogenesis and amenable to therapeutic intervention.
Introduction
Ataxia-telangiectasia (A-T) is a rare autosomal recessive genetic disorder characterized by chromosomal instability, progressive neurodegeneration, a high risk of cancer, and immunodeficiency. ATM, the gene defective in A-T, was initially localized to chromosome 11q22–23 [1] and then cloned by positional cloning [2]. ATM is a Ser/Thr protein kinase and a member of the phosphatidylinositol 3-kinase-related protein kinase family. ATM is recruited to the sites of DNA double-strand breaks (DSBs) by the Mre11-Rad50-Nbs1 complex, where, in the presence of other DNA damage response proteins, it is activated and subsequently phosphorylates as many as 700 substrates involved in DNA repair and cell cycle checkpoint activation [3, 4]. Biton et al. used human embryonic stem cells (hESCs) to demonstrate that ATM is nuclear and that it responds to DNA DSBs [5]. More recently, ATM was shown to be vital for the coordination of cell cycle control of pluripotent stem cells after ionizing radiation (IR) in G2 but not G1 [6]. Interestingly, interference with ATM activity in hESCs using the specific ATM inhibitor KU55933 [7] or bacterial artificial chromosome (BAC)-mediated gene knockout [8] suggested that ATM may be dispensable for repair of DSBs and genomic stability in hESCs. Although Atm-deficient mice recapitulate some of the cellular defects observed in A-T, including radiosensitivity, immunodeficiency, high incidence of cancer, and defective germ cell development [9, 10], other A-T-related defects, such as neuronal degeneration, are not evident in Atm-deficient mice, highlighting the need for a human A-T model system. Induced pluripotent stem cells (iPSCs) from individuals with A-T therefore present an opportunity to elucidate the role of ATM in the pluripotent context, to study A-T pathogenesis, and to create relevant patient-specific cell platforms for drug screening.
To date, the generation of iPSCs from DNA damage and chromosome instability syndromes without prior gene correction has not been reported. We found that reprogramming of A-T fibroblasts into iPSCs was indeed inefficient. Here we report on the generation and characterization of bona fide iPSCs from a family with A-T and show that these cells recapitulate important aspects of the A-T phenotype, including deregulation of molecular pathways previously associated with ATM, as well as gene expression changes in the pentose phosphate and mitochondrial oxidative phosphorylation pathways. These findings provide novel insights into early developmental consequences of ATM deficiency that may contribute to A-T pathogenesis. We also show that A-T iPSCs are capable of generating functional neurons and thus offer a potential model system to investigate the neurodegeneration associated with this disorder.
Materials and Methods
Generation of iPSCs
Primary fibroblasts were isolated from dermal punch biopsies collected from patients with ataxia-telangiectasia attending the A-T Clinic, University of Queensland Centre for Clinical Research. Biopsies were dissected into small pieces and incubated under coverslips in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Grand Island, NY, http://www.invitrogen.com) with 12% fetal calf serum (FCS) until fibroblasts grew out. Primary human fibroblasts were harvested with TrypLE select (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) and expanded in DMEM (Gibco) with 15% FCS until cryopreservation at passage 2. Fibroblasts were transduced with lentiviral constructs carrying OCT4/IRES/SOX2 and KLF4/IRES/c-MYC (Adgene plasmid 21162: pSIN4-EF2-O2S and plasmid 21163: pSIN4-EF2-N2L) [11]. After transduction, >50,000 A-T fibroblasts were allowed to recover for 24–48 hours before being transferred to mouse embryonic fibroblast (MEF) feeder plates (36,000 cells per cm2). Transduced A-T fibroblasts were stepwise transferred from DMEM with 15% FCS to 100% hESC culture medium over a period of 4 days at 25% per day, as this was shown to greatly increase their survival and proliferation. In accordance with a recent attempt to standardize nomenclature across hESCs and iPSCs [12], we have named these lines UQ0001i-ATh47.x and UQ0002i-AT34.y, where UQ refers to the institution in which they originated, the subsequent four-digit number refers to the order in which they were generated, i denotes iPSC origin, and A-T or ataxia-telangiectasia heterozygote (A-Th) nomenclature was as previously developed for naming A-T-cell lines, followed by internal patient identifier and clone number (x or y). All work was carried out with informed consent from patients under the approval of the University of Queensland Human Research Ethics Committee (HREC/09/QRCH/103).
Cell Culture Conditions
hESCs and iPSCs were grown in knockout serum replacement (KOSR) hESC culture medium (80% DMEM Ham's F-12 medium [Gibco], 20% KnockOut Serum Replacement [Gibco], 2 mM l-glutamine [Gibco], 1% nonessential amino acids [Gibco], 0.1 mM 2-mercaptoethanol, and 50–100 ng/ml basic fibroblast growth factor) (Invitrogen) at 37°C at 5% CO2 and high humidity. Cells were maintained on MEF feeder layers supplied by the Australian Stem Cell Centre. For experimentation, cells were cultured in feeder-free conditions on Matrigel (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com) in MEF-conditioned hESC culture medium. Cells were passaged as previously described [13] before being replating at a seeding ratio of between 1:2 and 1:6. hESC medium was replaced daily, and cells were split at approximately 80% confluence on days 6–7.
Screening of Clones
iPSC colonies were picked at early (2 weeks) and late (5 weeks) time points and subcultured clonally on MEFs in organ culture dishes. Clones were screened for expression of TRA-1-60 and Hoechst dye efflux, transgene persistence by reverse transcription-polymerase chain reaction (RT-PCR), stem cell marker expression, methylation status at Oct4/Nanog promoters, and karyotypic stability via G-band analysis (>15 metaphases analyzed per sample) by a commercial genotyping service (Sullivan Nicolaides Pathology, Brisbane, QLD, Australia, http://www.snp.com.au). Transgene silencing in selected clones was later confirmed by quantitative RT-PCR.
Teratoma Formation
iPSCs grown on MEFs were collected by collagenase IV treatment, and approximately 2 × 106 iPSCs resuspended in 50 μl of DMEM/Ham's F-12 medium supplemented with 30% Matrigel were injected into hind limb muscles of methoxyflurane-anesthetized 6-week-old immune-compromised SCID mice (CB17-SCID mice from the Animal Resources Centre [ARC], Canning Vale, WA, Australia, http://www.arc.wa.gov.au). After 8–10 weeks, teratomas were dissected and fixed in 4% paraformaldehyde. Samples were embedded in paraffin, stained with hematoxylin and eosin, and examined for the presence of representatives of the three germ layers by an independent pathologist. All mouse procedures were conducted under local ethical guidelines and after gaining permission from the local animal ethics committee (University of Queensland, Brisbane, QLD, Australia).
Bisulfite Sequencing
Live iPSCs were sorted by flow cytometry for TRA-1-60, and genomic DNA was isolated. One to 2 μg of DNA was bisulfite-converted using the EpiTect Bisulfite kit (Qiagen, Hilden, Germany, http://www.qiagen.com) before PCR of Oct4/Nanog promoter regions and cloning into the PCR2.1 vector. Clones were screened and selected for sequencing (primers are listed in supplemental online Table 8). Synthetically hypermethylated HeLa cells and H9 hESCs were also included as controls.
Irradiation
A 60Co source irradiator was used to deliver 2 Gy of IR to the cells. Cells were returned to the incubator to recover to the appropriate time point before harvesting/fixation with 4% paraformaldehyde, lysate preparation, or processing for fluorescence-activated cell sorting (FACS).
Terminal Deoxynucleotidyl Transferase dUTP Nick-End Labeling Assay
Following IR or mock dose, cells were washed once in phosphate-buffered saline (PBS) and harvested with cell dissociation buffer (Gibco). The terminal deoxynucleotidyl transferase dUTP nick-end labeling assay was used to determine apoptosis according to the in situ cell death detection kit (Roche Diagnostics, Basel, Switzerland, http://www.roche-applied-science.com).
Immunoblotting
Cell extracts were prepared as previously described [14]. Proteins were separated using SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose using Towbin's buffer and 100 V for 1 hour at 4°C prior to immunoblotting. Membranes were blocked with PBS blocking buffer containing 5% skim milk and 0.05% Tween 20 for 1 hour at room temperature and incubated for 16 hours with antibodies to SMC1 (1 μg/ml), SMC1p5957 (1 μg/ml), KAP-1 (1 μg/ml), or KAP-1pS824 (1 μg/ml) (rabbit polyclonals; Novus Biologicals, Littleton, CO, http://www.novusbio.com); anti-rabbit p53 (2 μg/ml) or anti-mouse p53 Ser15 (2 μg/ml) (Cell Signaling Technology, Beverly, MA, http://www.cellsignal.com); or anti-rabbit Chk2 (2 μg/ml), Chk2 pT68 (3 μg/ml) (Abcam, Cambridge, U.K., http://www.abcam.com), anti-rabbit ATM pSer1981 (2 μg/ml) (Rockland Immunochemicals, Gilbertsville, PA, http://rockland-inc.com), or anti-ATM (2 μg/ml) (mouse monoclonals; GeneTex, San Antonio, TX, http://www.genetex.com) diluted in blocking buffer at the indicated dilutions. Following washing in PBS buffer containing 0.05% Tween 20, anti-mouse horseradish peroxidase (1 μg/3 ml) (Millipore, Billerica, MA, http://www.millipore.com) and anti-rabbit (1 μg/5 ml) (Rockland Immunochemicals) secondary antibodies diluted in blocking buffer were used to reveal antibody binding. Secondary antibody cross-reactivity was visualized using ECL (PerkinElmer Life and Analytical Sciences, Waltham, MA, http://www.perkinelmer.com).
Immunostaining
For immunostaining cells were washed in PBS and fixed in 4% paraformaldehyde for 15 minutes at 4°C. For nuclear staining, samples were permeabilized in 0.1% Triton X-100 at room temperature for 10 minutes before blocking with 10% goat serum and incubation with the relevant antibodies overnight at 4°C. Antibodies and dilutions used were Oct4 (2.5 μg/ml), SSEA-4 (1/100), TRA-1-60 (2.5 μg/ml), Nanog (1/400), TRA-1-80 (3.8 μg/ml) (all from Millipore), or Oct4 (1 μg/ml) (Santa Cruz Biotechnology Inc., Santa Cruz, CA, http://www.scbt.com). ATM antibody preparation is described in [15]. Following washing with PBS (three times for 5 minutes at room temperature) the secondary antibodies goat anti-mouse IgG1, goat anti-mouse IgG2B, goat anti-mouse IgM, or donkey anti-rabbit IgG (Alexa Fluor) (2 μg/ml) were used to reveal cross-reactivity. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) or Hoechst. This preparation minus the addition of primary antibody was used to confirm specificity of staining.
G2/M Checkpoint Analysis
Activation of the G2/M checkpoint was determined by histone H3 phosphorylation [16]. Immunohistochemistry for the mitosis-specific marker phosphorylated histone H3 (serine 10) (H3s10; P-Histone H3 S10; Cell Signaling Technology; 0.5 μg/ml) was performed 2 hours after 2 Gy of ionizing radiation or mock dose. The mitotic index was derived by counting the proportion of immune-positive cells for H3s10 staining divided by the number of nuclei stained by DAPI. General linear models were performed to assess for differences in the proportions for the three different groups (A-T, control iPSCs, and H9) and the irradiation states (irradiated and not irradiated). Analyses were performed on the mean arcsine square-root proportions, and the results were then back-transformed for presentation. All results presented were back-transformed to the original scale. No significant difference was found between control iPSCs and H9s, so we pooled these data to compare with A-T iPSCs. An average of >650 events were quantified form each condition in three independent experiments. Slides were imaged using an Olympus BX61 microscope (Olympus, Tokyo, http://www.olympus-global.com).
Radioresistant DNA Synthesis
Radioresistant DNA synthesis was determined by DNA fiber labeling as described previously [17]. Briefly, A-T and control iPSCs were pulsed for 15 minutes with 50 μM chlorodeoxyuridine and washed, followed by exposure to mock/2 Gy of radiation prior to a second pulse for 15 minutes with 50 μM iododeoxyuridine. DNA fibers were made following the approach as previously outlined. Ongoing initiations and new replication forks were visualized via immunofluorescent microscopy after staining with rat monoclonal anti-bromodeoxyuridine (anti-BrdU) (Abcam) (13.3 μg/ml) and mouse monoclonal anti-BrdU (BD Biosciences) (1/8). Secondary antibodies were goat anti-rat Alexa 488-conjugated secondary (Invitrogen) (6.7 μg/ml) and donkey anti-mouse Alexa 594-conjugated secondary (6.7 μg/ml). Two-by-two factorial analysis of variance was performed to assess differences in the proportions of the two different genotypes (A-T vs. control) and the irradiated states (irradiated and not irradiated). Analyses were performed on the arcsine square-root proportions, and the results (means and confidence intervals) were then back-transformed for presentation. Analyses showed that there were no differences between the two substudies, and all analyses were performed on the combined data set, not taking into account the substudies. All results presented from analyses were back-transformed to the original scale. More than 700 events were quantified from each condition in two independent experiments. Slides were imaged using a Zeiss LSM 710 confocal microscope (Carl Zeiss, Jena, Germany, http://www.zeiss.com).
Neuronal Differentiation
iPSC cultures were grown for 5 days after passage prior to commencement of differentiation and changed directly into KOSR hESC medium supplemented with 5 μM dorsomorphin (Stemgent, Cambridge, MA, https://www.stemgent.com) and 10 μM SB431542 for the first 6 and 12 days of differentiation, respectively, with medium changes every 2 days. The KOSR hESC medium was gradually replaced with Neurobasal medium (Gibco) (with N2B27 supplements used according to the manufacturer's specifications) (Gibco) with 25%, 50%, 75%, and 100% N2B27 Neurobasal medium in KOSR hESC medium on days 4, 6, 8, and 10 respectively. Neurospheres were formed on day 6 of differentiation by 10 minutes of incubation in 1 mg/ml collagenase IV (Gibco) at 37°C and dislodging of large pieces of colonies by use of a cell scraper and P1000 pipette. Neuralized colony fragments were seeded into Ultra-Low Cluster plates (Corning Costar, Acton, MA, http://www.corning.com/lifesciences), where they aggregated into tight spheres. On day 12 of differentiation, neurospheres were seeded onto Matrigel (BD Biosciences)-coated plates, and N2B27 Neurobasal medium was subsequently changed every 3–4 days as neurons grew out from the sphere borders. Cultures were passaged every week by cell dissociation buffer (Sigma-Aldrich, St. Louis, http://www.sigmaaldrich.com) at a ratio of 1:2 to 1:3, eventually leading to the complete dissociation of neurosphere aggregates. Four- to 6-week-old cultures were subsequently matured by the addition of 20 ng/ml brain-derived neurotrophic factor and 20 ng/ml glial cell line-derived neurotrophic factor (R&D Systems Inc., Minneapolis, http://www.rndsystems.com), 200 nM ascorbic acid, and 0.5 mM dibutyryl-cAMP (Sigma-Aldrich) for 2 weeks with medium changes every 2 days to replenish growth factors.
Calcium Imaging in Neuronal Cultures
Calcium imaging was performed using the FLIPR TETRA High Throughput Cellular Screening System (Molecular Devices, Sunnyvale, CA, http://www.moleculardevices.com), essentially as described by Vetter and Lewis [18]. Neurons were seeded in a black-walled 386-well culture format, loaded with a calcium-sensitive fluorescent dye, and depolarized with 50 mM KCl.
RNA Isolation, cDNA Synthesis, PCR, and Quantitative PCR
RNA was isolated using the RNeasy isolation kit (Qiagen). Concentration and 260/280 ratios were quantified using a NanoDrop 1000 spectrophotometer (NanoDrop, Wilmington, DE, http://www.nanodrop.com) before synthesis of cDNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, http://www.bio-rad.com) according to the manufacturer's specifications (500 ng). RT-PCR was performed for OCT4 (657 base pairs [bp]) and SOX2 (498 bp) transgenes according to the following conditions: 94°C for 5 minutes; 32 cycles of 94°C for 1 minute, 58°C for 30 seconds, and 72°C for 1 minute; 72°C for 7 minutes. RT-PCR was performed for KLF4 (563 bp) and c-MYC (350 bp) transgenes and for GAPDH (152 bp) according to the following conditions: 94°C for 5 minutes; 32 cycles of 94°C for 1 minute, 57°C for 30 seconds, and 72°C for 1 minute; and 72°C for 7 minutes. Primers are listed in supplemental online Table 8. Quantitative PCR (qPCR) was performed for the OCT4 transgene, endogenous OCT4, and β-Actin using a C1000 thermal cycler (Bio-Rad) using Ssofast evagreen qPCR mix (Bio-Rad) according to the following conditions: 95°C for 3 minutes; 30 cycles of 95°C for 10 seconds and 60°C for 30 seconds. Expression data were calculated using the ΔΔCt method.
Expression Analysis
RNA was harvested from TRA-1-60 FACS-sorted hESCs (MEL1) and control (UQ0001i-control1), A-T heterozygote (UQ0001i-ATh47.1), and homozygote iPSCs (UQ0002i-AT34.7), as well as from nonsorted parental fibroblasts. Total RNA was isolated from each FACS-sorted iPSC line and from unsorted fibroblast cell samples using the RNeasy Mini Kit (Qiagen). The total RNA (and A260/A280 ratio) was then quantified using a NanoDrop 1000 spectrophotometer. Total RNA (100 ng) was subjected to reverse transcription, second-strand cDNA synthesis, and in vitro transcription using the TotalPrep RNA Amplification Kit (Illumina Inc., San Diego, http://www.illumina.com). cRNA was hybridized to Illumina HT12 v4 BeadChip microarrays. The raw expression data were normalized using quantile normalization and without background correction, using the lumi R/Bioconductor package (version 2.4.0) [19]. Only probes passing the Illumina detection threshold were included in the expression analysis; a probe passed the Illumina detection score if it had a detection p value of ≤.01 in at least 75% of cell lines in the same group; these criteria resulted in 20,593 probes being retained. All statistical analyses were performed using R, version 2.13.2. All probes were mapped using the annotation package illuminaHumanv4.db (version 1.10.0) available from Bioconductor. The expression data are available for download from Stemformatics (http://www.stemformatics.org) and GEO (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE35347.
Heatmaps were constructed using the gplots R/Bioconductor package (version 2.10.1) where agglomerative hierarchical clustering was used on the basis of a measure of dissimilarity 1 − R, where R represents the Pearson correlation coefficient between any two gene expression profiles and ranges from −1 to 1. Probes mapping to multiple gene symbols were filtered to ensure a one-to-one mapping between probe and gene symbol; the probe with the most significant p value assessing the significance of differential expression between A-T and control iPSCs was retained and represented in the resulting heatmap. The p values were generated using the R/Bioconductor package limma (version 3.8.3) and adjusted for multiple testing using the Benjamini-Hochberg method [20]. The PluriNet gene list, as identified by Müller et al., was originally downloaded from http://www.stemcellmatrix.org and consisted of 299 gene symbols, listed in supplemental online Table 9 [21]. Pathway analysis was performed using the R/Bioconductor package attract (version 1.4.0) [22], where pathways were defined using the Kyoto Encyclopedia of Genes and Genomes (KEGG) [23]. The comparison of expression between mitochondrial genes in A-T and control iPSCs was based on genes represented in MitoCarta, a curated list of genes known to be involved in mammalian mitochondrial function. We used attract to compute a p value representing the enrichment of genes showing differential expression across the seven cell types for which expression analysis was present. We found that the mitochondrial gene list had a statistically significant p value of 9.47 × 10−19, and we were able to decompose this pathway into four groups of genes showing distinct patterns of correlated expression.
Results
Generation and Characterization of A-T Homozygote and A-T Heterozygote iPSCs
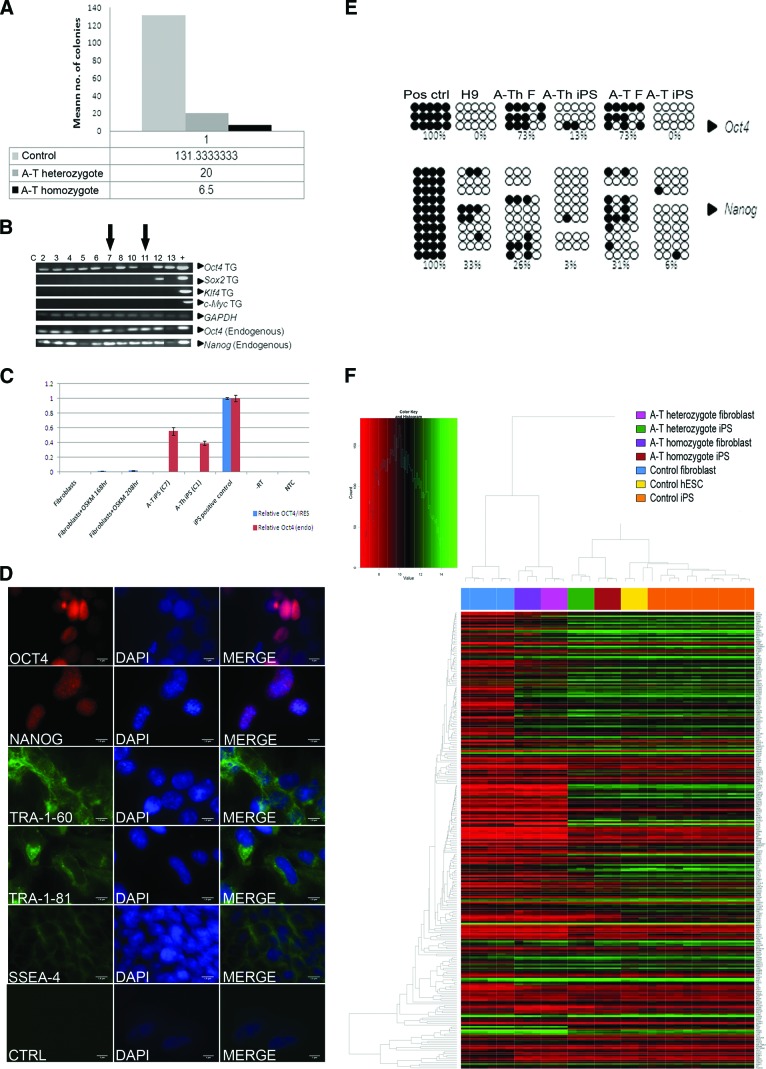
Skin fibroblast cultures were established from punch biopsies from A-T heterozygote and A-T homozygote patients (mother and daughter). Mutation analysis revealed two frameshift deletions (7004delCA and 7886delTATTA) predicted to result in truncated and thus unstable ATM protein [24]. In order to optimize conditions and reduce the risk of chromosomal instability, we selected early passage (passage <5) fibroblasts for reprogramming. Following transduction with OCT4/IRES/SOX2 and KLF4/IRES/c-MYC lentivirus, we stepwise adapted the cells to KOSR hESC culture medium over the first 4–5 days of iPSC generation, since direct replacement with KOSR hESC culture medium was found to lead to extensive death of the A-T fibroblasts. Control cells from healthy, unrelated individuals were also reprogrammed in parallel. After 2 weeks, transduced A-T patient fibroblasts gave rise to hESC-like colonies (supplemental online Fig. 1A). Although our data show that it was possible to reprogram A-T fibroblasts, the efficiency was markedly reduced to 4% compared with controls (Fig. 1A). It is also of note that reprogramming efficiency was reduced to 15% for the A-T heterozygote, indicating a potential role for ATM in reprogramming. Thirteen colonies from the ATM homozygote patient that expressed the TRA-1-60 stem cell surface marker and exhibited Hoechst dye efflux [25] (supplemental online Fig. 1B) were expanded for further analysis. Eleven of these clones displayed a stable pluripotent stem cell phenotype and could be culture expanded. Two of the A-T homozygote clones (C7 and C11) and one clone (C1) of the heterozygote samples were selected for further analysis as these showed SOX2, KLF4, and c-MYC transgene silencing after 13 weeks of culture with some persistence of OCT4 transgene (Fig. 1B). A-T iPSC clones 7 and 11 robustly expressed endogenous OCT4 and NANOG, as shown by RT-PCR (OCT4) (Fig. 1B). Following 30 weeks of culture, quantification of OCT4 transgene expression by qPCR in A-T iPSC clone 7 and A-Th iPSC clone 1 revealed that exogenous OCT4 expression was undetectable (C7) or present in trace amounts (C1), relative to controls (Fig. 1C). These clones further exhibited clearly detectable expression of the pluripotency markers TRA-1-60, TRA-1-81, SSEA4, NANOG, and OCT4 by immunofluorescence (Fig. 1D; supplemental online Fig. 1C). Immunoblotting failed to detect ATM protein in protein extracts from the patient fibroblasts (Fig. 2A). Flow cytometric analysis of TRA-1-60 expression further exemplifies that the A-T iPSCs (clone 7 shown) exhibit robust and uniform expression of this pluripotency marker in the majority of the population (supplemental online Fig. 1D). In keeping with these observations, CpG islands in the OCT4 and NANOG promoter regions in the A-T iPSCs (clone 7) and A-Th-iPSCs (clone 1) were found to be hypomethylated as compared with the parental fibroblasts (Fig. 1E). Heterozygote and control clones were screened similarly as described above and conformed to the same criteria (data shown where applicable).
Figure 1.
Generation of A-T heterozygote and homozygote induced pluripotent stem cells (iPSCs). (A): The mean number of colonies following reprogramming of control (n = 3), A-T heterozygote (n = 3), and A-T homozygote (n = 2) fibroblasts. The figure shows pooled data from more than one experiment. (B): Polymerase chain reaction (PCR) analysis of transgene expression and endogenous expression of NANOG and OCT4 in homozygote and heterozygote A-T iPSCs. Arrows indicate the two clones (C7 and C11) with the greatest levels of transgene silencing. Positive controls were lentiviral plasmids and H9 hESC cDNA. (C): Quantitative PCR data show that the transgene was detectable in transduced fibroblasts at 168 and 208 hours post-transduction, but not in control fibroblasts. Transgene OCT4 was not detected in A-T iPS (C7) but was present in trace amount in A-Th iPS (C1) after 30 weeks of passage. Results were normalized to a control iPS clone that expressed transgene robustly. Both A-T iPS (C7) and A-Th iPS (C1) strongly expressed endogenous OCT4. Error bars show SEM from three technical replicates; data are from one experiment. Primers for β-ACTIN were used as a control. (D): A-T iPSC (C7) colonies expressed the following pluripotency markers: OCT4, NANOG, Tra-1-80, Tra-1-61, SSEA4. DAPI stained the nuclei. Scale bars = 1 μm. (E): Bisulfite sequencing of CpG islands in the NANOG and OCT4 promoters in nuclear DNA from heterozygote A-T fibroblasts (A-Th-F) and heterozygote A-T iPSCs (A-Th-iPS C1) and homozygote A-T fibroblasts (A-T-F) and A-T iPSCs (A-T-iPS C7). Methylated HeLa DNA (positive control) and H9 hESC DNA (negative control) are also shown. Open circles represent unmethylated dinucleotides, and closed circles represent methylated dinucleotides. (F): Hierarchical clustering and heatmap comparison of the 299 Plurinet genes between control fibroblasts, homozygote A-T fibroblasts, heterozygote A-T fibroblasts, heterozygote A-T iPSCs (C1), homozygote A-T iPSCs (C7), H9 hESCs, and control iPSCs. Abbreviations: A-T, ataxia-telangiectasia; A-Th, ataxia-telangiectasia heterozygote; C, clone; CTRL, control; DAPI, 4′,6-diamidino-2-phenylindole; F, fibroblast; hESC, human embryonic stem cell; iPS, induced pluripotent stem; Pos, positive; TG, transgene.
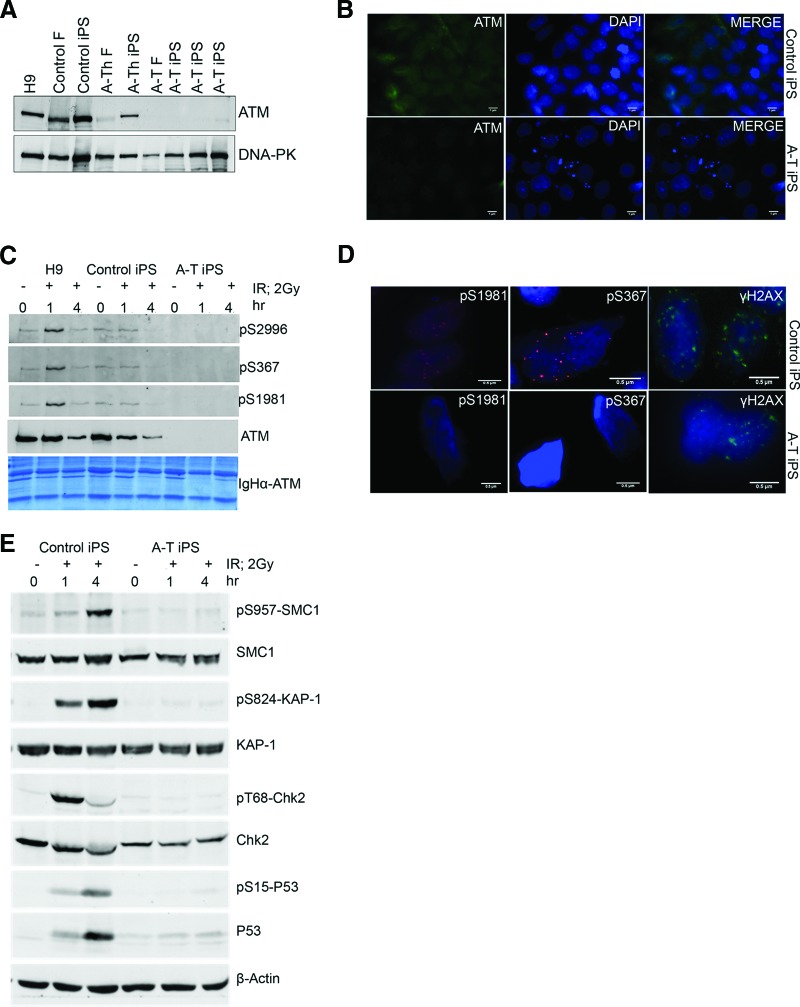
Figure 2.
Ionizing radiation failed to activate ATM signaling in A-T induced pluripotent stem cells (iPSCs). (A): Western blot showing expression of ATM in (lanes from left to right) H9 human embryonic stem cells (hESCs), control fibroblasts, control iPSCs, heterozygote A-T fibroblasts, heterozygote A-T iPSCs, A-T homozygote fibroblasts, and homozygote A-T iPSCs. DNA-protein kinase was used as a loading control. (B): Immunofluorescent detection of ATM protein in control iPSCs (top) and A-T iPSCs (bottom). DAPI stained the nuclei. Secondary antibody staining alone revealed no cross-reactivity (not shown). Scale bars = 1 μm. (C): Western blot analysis of ATM autophosphorylation sites S2996, S367, and S1981 in immunoprecipitated ATM protein from H9 hESCs, control iPSCs and A-T iPSCs following irradiation with 2 Gy of IR for 0, 1, and 4 hours. A Coomassie-stained gel of the ATM immunoprecipitate was used as a loading control. (D): Immunofluorescent detection of ATM autophosphorylation sites S1981 and S367 and γH2AX foci in control iPSCs (top) and A-T iPSCs (bottom) after IR (2 Gy, 1 hour). Scale bars = 0.5 μm. (E): Western blot analysis of phosphorylation of ATM downstream targets SMC1 (S957), KAP1 (S824), and Chk2 (T68) and p53 stabilization in control and A-T iPSCs following 0, 1, and 4 hours of IR (2 Gy). Abbreviations: A-T, ataxia-telangiectasia; A-Th, ataxia-telangiectasia heterozygote; DAPI, 4′,6-diamidino-2-phenylindole; F, fibroblast; iPS, induced pluripotent stem; IR, ionizing radiation.
iPSCs from both the A-T homozygote (clones 7 and 11) and their A-T heterozygote parent (clone 1) formed teratomas when injected into SCID mice (one mouse per line, with each mouse developing a teratoma) comprising tissue types from all three germ layers (endoderm, mesoderm and ectoderm), indicating pluripotential trilineage differentiation (supplemental online Fig. 1E), as did control iPSCs (supplemental online Fig. 1E). A subset of the 11 clones that displayed the characteristics of pluripotent cells were examined for gross karyotypic abnormalities (supplemental online Fig. 2). Although 2 of the 11 clones that displayed long-term self-renewal developed chromosomal abnormalities, 5 A-T iPSC clones displayed normal karyotypes between passage 11 and passage 16, and this was maintained out to passage 31 in the case of one A-T clone (clone 7) (supplemental online Table 1). To further validate the A-T iPSCs, we compared the transcriptome of A-T iPSC clone 7 with control iPSCs (UQ0001i-control1) and hESCs (MEL1). Principal component analysis revealed the similarity in gene expression between A-T iPSCs, control iPSCs, and hESCs, and this was further reinforced by hierarchical clustering of the 299 pluripotency-associated genes of the PluriNet [21] (Fig. 1F). We conclude from these analyses that the phenotype and gene expression profiles of A-T homozygote (and UQ0002i-AT34.7) and heterozygote (UQ0001i-ATh47.1) iPSC lines are consistent with a fully reprogrammed iPSC phenotype.
Defective DNA Damage Response in A-T iPSCs
ATM is a cellular DNA damage sensor that coordinates the cell cycle through damage-response checkpoints and mediates DNA repair to preserve genomic integrity. ATM has also been implicated in additional roles such as regulation of metabolic activity and response to reactive oxygen species (ROS) [24, 26]. There is still little consensus regarding the role of ATM in human pluripotent stem cells, and we therefore investigated several A-T-associated phenotypes in the A-T iPSCs.
The A-T patient in this study possesses frameshift mutations in both ATM alleles; accordingly neither the A-T fibroblasts (Fig. 2A) nor the A-T iPSCs (Fig. 2A) showed expression of ATM protein. This was confirmed by immunofluorescent imaging, revealing predominantly nuclear localization of ATM in control iPSCs and undetectable ATM in A-T iPSCs (Fig. 2B). Consequently, after exposure to radiation (2 Gy, 1 hour) and immunoprecipitation with ATM, phosphospecific antibodies that detect S367, S1981, or S2996 phosphorylation sites, ATM autophosphorylation was not detected, whereas autophosphorylation was clearly detected in control iPSCs (Fig. 2C). In agreement with these data, immunofluorescent staining failed to detect ionizing radiation-induced foci in irradiated A-T iPSCs (ATM pS1981 or ATM pS367), whereas clear nuclear localization of ATM pS367 and ATM pS1981 was detected in irradiated control iPSCs (Fig. 2D). γH2AX foci were present in both controls and A-T iPSCs, indicating that DNA repair was actively proceeding in the absence of ATM protein (Fig. 2D). Once ATM is activated it phosphorylates a large number of substrates in multiple pathways [3, 14, 27], including proteins involved in cell cycle control (e.g., p53 and Chk2), cell survival (SMC1), and maintaining chromatin structure (Kap1). In control iPSCs, all of these substrates were phosphorylated and p53 was stabilized in response to radiation exposure, but this did not occur in A-T iPSCs (Fig. 2E). We conclude that ionizing radiation-induced ATM-dependent signaling is defective in A-T iPSCs.
Defective Cell Cycle Checkpoint Activation in A-T iPSCs
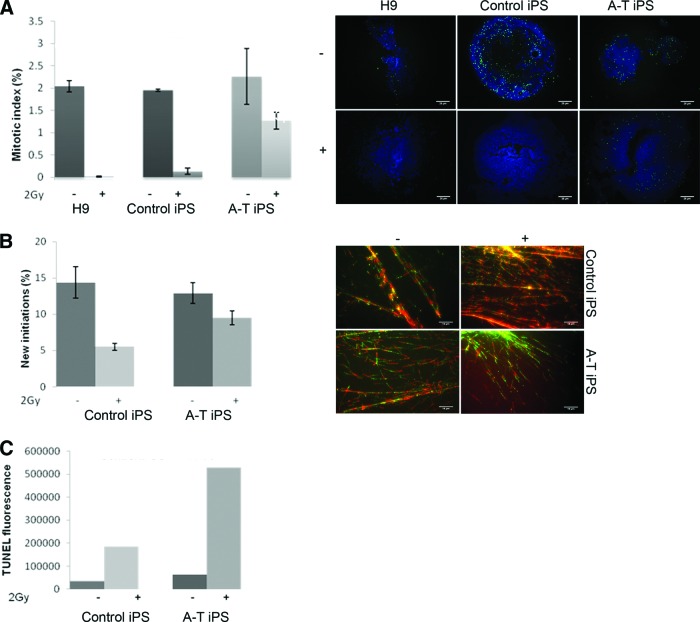
Another cellular hallmark of A-T is defective cell cycle checkpoint activation in response to DNA DSBs [28]. We therefore measured G2/M delay using histone H3 phosphorylation after exposure to 2 Gy of radiation. Control iPSCs showed characteristic inhibition of entry into mitosis 2 hours after exposure to 2 Gy, similar to H9 hESCs (Fig. 3A), whereas A-T iPSCs exhibited a greatly reduced inhibition of mitotic entry after the same dose of radiation. Exposure of cells to radiation leads to rapid inhibition of DNA synthesis, and this is used to determine the intra-S-phase checkpoint [17]. When somatic A-T-cells are irradiated they exhibit radioresistant DNA synthesis or a defective S-phase checkpoint [29]. The S-phase checkpoint was determined using a DNA fiber assay where the extent of DNA synthesis is expressed as a percentage of new initiations relative to total elongations in irradiated and nonirradiated cells [14]. DNA synthesis was inhibited by 60% in control iPSCs, whereas in A-T iPSCs DNA synthesis was inhibited only by 20%, demonstrating the presence of radioresistant DNA synthesis in A-T iPSCs (Fig. 3B). Because hypersensitivity to ionizing radiation is a well-established characteristic of somatic A-T cells [30, 31], we next examined the incidence of apoptosis in A-T and control iPSCs 24 hours after exposure to 2 Gy of radiation. As shown in Figure 3C, radiation-induced apoptosis was approximately threefold higher in A-T iPSCs than in control iPSCs (supplemental online Fig. 3 gives FACS plots). Figure 3C further shows that even under standard culture conditions, A-T iPSCs exhibited a twofold load of apoptosis relative to controls. In somatic cells, ATM activation by DNA DSBs and subsequent signaling through p53 represents a major pathway for induction of apoptosis [32], implying that loss of ATM activity may confer resistance to apoptosis. In contrast our data show an increased sensitivity to spontaneous and radiation-induced apoptosis.
Figure 3.
A-T induced pluripotent stem cells (iPSCs) exhibited cell cycle defects and hypersensitivity after ionizing radiation. (A): H3s10 immunostaining of cells following 2 Gy of ionizing radiation (IR) or mock dose was quantified. A-T iPSCs showed persistent staining relative to controls (top panels, H9 human embryonic stem cells and control iPSCs shown), indicative of a failure to arrest at the G2/M checkpoint (>600 events were quantified from each condition in three independent experiments; error bars show SEM; p < .05). Scale bars = 25 μm. (B): A-T iPSCs exhibited radioresistant DNA synthesis following 2 Gy of IR (>700 events were quantified from each condition in two independent experiments; error bars show SEM; p < .05). Scale bars = 16 μm. (C): Flow cytometric quantification of TUNEL staining of control and A-T iPSCs 24 hours after mock or 2 Gy of IR. Populations were gated to exclude cellular debris and autofluorescence. Positive control was DNase-treated iPSCs (histograms shown in supplemental online Fig. 3) (n = 1). Abbreviations: A-T, ataxia-telangiectasia; iPS, induced pluripotent stem; TUNEL, terminal deoxynucleotidyl transferase dUTP nick-end labeling.
Bioinformatic Analysis of Transcriptome Changes in A-T iPSCs
In addition to its role in responding to DNA DSB repair, ATM has also been implicated in a range of other cellular processes, and its role in nonirradiated pluripotent cells has remained largely unexplored. We therefore examined in detail the transcriptome of unchallenged A-T iPSCs in culture using a combinatorial bioinformatics approach using the Genego and attract bioinformatic analysis tools. Whereas Genego identifies pathways within significantly differentially expressed genes on the basis of existing knowledge (on the basis of KEGG annotation) the attract pathway analysis tool examines the entire data set and identifies and amplifies new coordinately regulated gene sets that are relevant to the mechanisms underlying particular phenotypes [22].
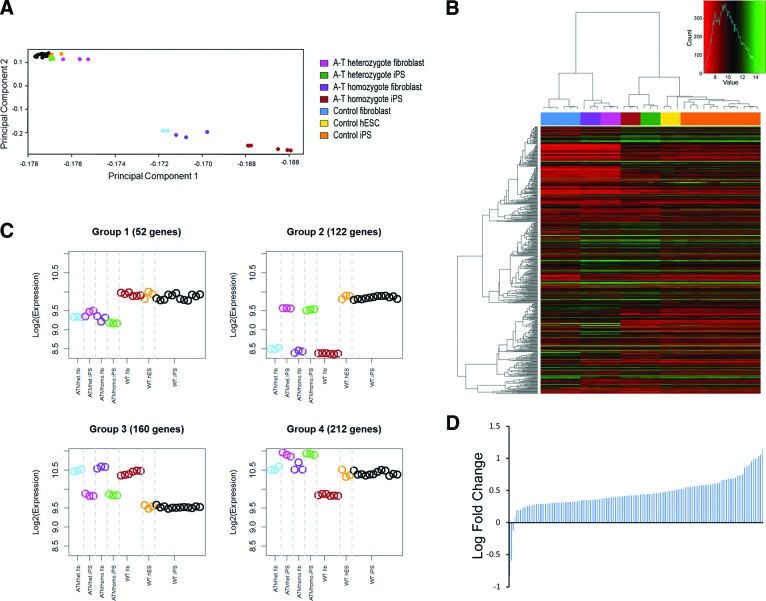
Principal component analysis of the entire data set (Fig. 4A) showed that A-T homozygote, A-T heterozygote, wild-type iPSCs, and hESCs clustered together closely but away from the fibroblast samples, which did show clustering based on genotype. These data indicate that the large transcriptome differences that exist between A-T fibroblasts and wild-type fibroblasts are largely resolved after reprogramming. Nevertheless 7,921 genes showed differential expression between A-T and wild-type iPSCs (p < .05) (supplemental online Table 2). Genego analysis of this cohort of genes identified pathways previously associated with ATM, such as cell cycle control, DNA damage, and apoptosis (supplemental online Table 3). When we used attract to analyze the entire data set, we identified not only these known pathways but also oxidative phosphorylation and pentose phosphate pathway as significantly altered (supplemental online Fig. 4). This led us to analyze the expression of 1,080 genes, previously identified in the MitoCarta database [33], that are directly or indirectly associated with mitochondrial function within the 7,921 genes that were differentially expressed (p < .05) between A-T iPSCs and controls. Remarkably, 464 of 1,080 of these mitochondria-associated genes were identified within the A-T iPSCs differentially expressed gene list (heatmap shown in Fig. 4B), suggesting that mitochondrial function is a significantly altered pathway in nonirradiated A-T iPSCs (supplemental online Table 5 shows list of upregulated and downregulated genes). Using attract we were further able to attribute these mitochondrial gene expression differences to either cell state (i.e., induced pluripotent stem + embryonic stem vs. fibroblasts) or genotype (i.e., A-T homozygous + heterozygous vs. wild-type), identifying four groups (supplemental online Fig. 5 shows statistical analyses) of mitochondrial activity. In two of these groups (groups 2 and 3) ATM deficiency showed no difference, whereas group 1 consisted of conserved ATM-centric changes that were apparent in both fibroblasts and pluripotent cells (Fig. 4C). Most interestingly, group 4 identified 212 mitochondria-associated genes that were specifically altered in A-T iPSCs but not in fibroblasts (supplemental online Table 6 shows gene lists of all four groups). Further curation of this list selecting for genes either encoded by mtDNA or imported into mitochondria shows that 140 of 143 truly mitochondrial genes were upregulated in A-T iPSCs (Fig. 4D). The majority of these 143 genes are either components of the respiratory chain, involved in assembly and import of respiratory chain complexes, mitochondrial metabolite transporters, mitochondrial ribosomal proteins, or tRNA synthetases (supplemental online Table 7 shows an annotated gene list). These gene expression changes are therefore consistent with an upregulation of mitochondrial biogenesis in human pluripotent cells in the absence of ATM.
Figure 4.
Bioinformatic analysis of A-T induced pluripotent stem cells (iPSCs) revealed mitochondrial gene expression changes. (A): Principal component analysis of the entire gene expression data sets of A-T heterozyogote fibroblasts and iPSCs, A-T homozygote fibroblasts and iPSCs, control fibroblasts and iPSCs, and MEL1 hESCs. (B): Heatmap comparison of 546 mitochondria-associated genes (MitoCarta) between A-T heterozyogote fibroblasts and iPSCs, A-T homozygote fibroblasts and iPSCs, control fibroblasts and iPSCs, and MEL1 hESCs. (C): On the basis of a gene list of mitochondria-associated genes, the (MitoCarta) attract analysis identified four groups of mitochondria-associated gene expression showing distinct patterns of correlated expression depending on cell type or genotype. (D): Expression of 141 genes expressed in mitochondria derived from group 4 (iPSC-specific changes in A-T iPSCs from [B]). Graph indicates log fold change of group 4 genes compared with control iPSCs and MEL1 hESCs. Abbreviations: A-T, ataxia-telangiectasia; ATMhet, heterozyogote; ATMhomo, homozygote; hES, human embryonic stem; hESC, human embryonic stem cell; iPS, induced pluripotent stem; WT, wild-type.
Directed Differentiation of A-T iPSCs into Functional Neurons
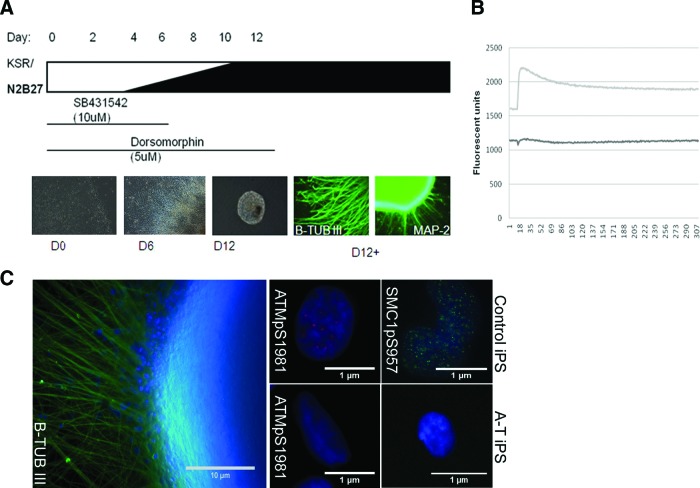
We next differentiated A-T iPSCs (UQ0002i-AT34.7) and control iPSCs (UQ0001i-control1) into neuronal progenitors using a modified version of the dual SMAD inhibition protocol previously described [34] (Fig. 5A). A-T iPSCs readily differentiated into βIII-TUBULIN and MAP-2-expressing cells with a neuronal morphology that, after 2 weeks of culture in neuronal maturation medium, displayed electrophysiological activity in the form of calcium spikes following KCl depolarization, as shown by Ca2+ imaging performed using the FLIPR TETRA High Throughput Cellular Screening System [18] (Fig. 5B). When irradiated, these cells also exhibited a defective DNA damage response, as shown by the absence of ionizing radiation-induced foci, relative to controls (ATM S1981 and SMC1 S957) (Fig. 5C).
Figure 5.
Directed differentiation of A-T induced pluripotent stem cells (iPSCs) into mature neurons. (A): Schematic representation of neural induction protocol involving stepwise addition of N2B27 neurobasal medium and small molecules SB431542 and dorsomorphin for the first 6 and 12 days, respectively. Neurospheres were generated on day 6 of induction and plated after day 12, giving rise to colonies with neuronal projections and morphologies that were βIII-TUBULIN- and MAP-2-positive. (B): Depolarization-induced calcium transients in control and A-T iPSC-derived neuronal cultures following in vitro maturation. A representative experiment is shown. (C): Immunofluorescent detection of βIII-TUBULIN and radiation-induced (2 Gy, 1 hour) foci (ATM pS1981 and SMC1 pS957) in control iPSCs and A-T iPSCs. 4′,6-Diamidino-2-phenylindole stained the nuclei. Scale bars = 10 μm (left panel), 1 μm (right panels). Abbreviations: A-T, ataxia-telangiectasia; B-TUBIII, βIII-TUBULIN; D, day; iPS, induced pluripotent stem.
Discussion
Emerging evidence suggests that the DNA damage pathways are activated early during reprogramming and may pose a barrier to iPSC generation. The generation of iPSCs from chromosomal instability syndromes has indeed proven to be difficult without gene manipulation. Raya et al. showed that somatic cells from patients with the rare recessive chromosomal instability disorder Fanconi anemia could be reprogrammed to pluripotency to generate patient-specific iPSCs only after correction of the defective gene with cDNA [35]. We have shown that this is not necessary, by demonstrating for the first time that fibroblasts from patients with A-T, a syndrome characterized by genome instability, can be reprogrammed to pluripotency and meet all the established criteria for bona fide iPSCs. The efficiency of reprogramming A-T fibroblasts to iPSCs was approximately 4% of that seen with controls, with heterozygotes of intermediate efficiency. This is in keeping with the observation that reprogramming of mouse Atm-deficient tail-tip fibroblasts occurs with efficiency less than 2% of that of wild-type fibroblasts [36]. During the process of reprogramming, cells may be less tolerant of the presence of DNA damage where p53 may play an important role in removing these cells by inducing apoptosis [37]. It is likely that the accumulation of DNA DSBs during reprogramming of A-T fibroblasts renders these cells more susceptible to apoptosis or makes them otherwise unavailable for reprogramming. Short telomeres also contribute to the barrier of cell reprogramming imposed by p53 [38]. This is also significant for A-T since fibroblasts and lymphoblastoid cells from these patients are characterized by abnormally short telomeres [39, 40]. We show that these iPSCs recapitulate key features of the A-T cellular phenotype, including radiation-induced cell cycle defects (radioresistant DNA synthesis/reduced delay at G2/M), which is supported by numerous transcriptional changes in DNA damage and cell cycle-related processes. A-T iPSCs also show an increased sensitivity to spontaneous and radiation-induced apoptosis. A similarly increased cell death phenotype was previously reported by Ivanov et al. [41] in human melanoma cells treated with ATM small interfering RNA (siRNA).
iPSCs from Atm-deficient mice were previously found to accumulate abnormal genome structures with continuing passage [36], whereas hESCs genetically modified with a BAC-based homologous recombination system to knock out ATM were devoid of gross genomic abnormalities after extended culture [8]. In our study we were also able to culture karyotypically normal human A-T iPSCs for more than 20 weekly passages. Our data are thus in agreement with recent reports showing that ATM-independent homologous repair and recombination is the principal mediator of DNA damage in pluripotent stem cells [42]. This notion is further supported by a 10-fold elevation of Rad51 foci in embryonic stem cells (ESCs) compared with differentiated astrocytes [7] and by the fact that ATM signaling proceeds in a heterochromatin-dependent manner, whereas ESCs are largely euchromatic [43].
Our pathway and phenotypic analysis of A-T iPSCs supports the prediction that ATM plays a role in the DNA damage response and cell cycle control in pluripotent cells and confirms the observations made by Momcilovic et al., who used the ATM inhibitor KU55933 to show that ATM function is essential for induction of G2 arrest in irradiated hESCs [44]. The A-T iPSCs further behave similarly to hESCs in which ATM has been abrogated by disruption of the gene [8] or by siRNA knockdown [5].
It is becoming increasingly clear that in addition to its nuclear role in DSB repair, cytosolic ATM also functions as an ROS sensor [24, 26] and can be associated with mitochondria [45]. The attract pathway analysis of A-T iPSCs and fibroblasts uncovered extensive differences in the regulation of oxidative phosphorylation genes, supporting a defect at the level of the mitochondrion specifically in pluripotent A-T-cells. Previous reports suggested that loss of ATM function in somatic cells caused reduced mitochondrial function and/or abnormal homeostasis [46–48]. It was recently demonstrated that ATM-mediated mitochondrial defects are highly cell context-dependent and that in thymocytes from Atm-deficient mice there is in fact an increase in mitochondrial number and mitochondrial ROS production due to a defect in mitophagy [45]. In strong agreement with these data, our gene expression analysis shows that in addition to a dramatic increase in mitochondrion-associated transcripts, mRNA expression of HMGB1 and HSPB1 [49, 50], two important regulators of mitophagy, is specifically reduced in A-T iPSCs but not in A-T fibroblasts, suggesting that mitophagy may be specifically impaired in ATM-deficient iPSCs. A failure to clear mitochondria with low membrane potential, reduced ATP production, elevated ROS production, and consequently increased mtDNA mutations is consistent with the increased oxidative stress reported in A-T-cells [46, 51, 52], the increased mtDNA mutations reported in tissues of A-T patients [47], and the observation that cerebellar Purkinje cell death and neurological deficits in Atm-deficient mice can be rescued by antioxidants [53–55]. It is not unlikely that in response to the increased oxidative stress in A-T-cells, possibly arising from mitochondrial dysfunction, carbohydrate metabolism is rerouted from glycolysis to the pentose phosphate pathway. Our gene expression data indeed identified the pentose phosphate pathway as a significantly altered and revealed that G6PD is downregulated in A-T iPSCs, in agreement with a recent report showing that ATM activates the pentose phosphate pathway by stimulating complex formation between heat shock protein 27 and G6PD to increase G6PD activity [56]. Of additional interest, we observed significant upregulation of USMG5, a gene recently implicated in human inherited ataxia and Purkinje cell degeneration [57]. If one accepts that iPSCs are equivalent to hESCs derived from the inner cell mass, it appears reasonable to hypothesize that increased mitochondrial biogenesis accompanied by defective mitophagy may lead to an accumulation of defective high ROS producing stem and progenitor cells and that this DNA damage response-independent function of ATM could contribute to the pathology of A-T.
Our data demonstrating that ATM deficiency does not disrupt gross neuronal differentiation suggests that iPSCs may be able to be used for screening of drugs aimed at promoting survival of cell types affected in A-T, such as Purkinje cells in at the cerebellum. Generation of mature cerebellar Purkinje cells from mouse embryonic stem cells was only recently described [58, 59] and provides proof of concept that selective preparation of Purkinje neurons or committed Purkinje neuron precursors from A-T iPSCs may be possible.
Conclusion
In summary, we have demonstrated for the first time the generation of iPSCs from a patient with a chromosomal breakage disorder, in this case A-T, without the requirement for correcting the genetic defect in advance. We have shown that A-T iPSCs are pluripotent and provide further evidence implicating roles for ATM in DNA damage-dependent signaling in pluripotent cells. In addition to this, we have shown that it is possible to maintain a number of clones, one to high passage, that were stable in culture. Extensive transcriptional profiling and analysis of mutant and wild-type human iPSCs and parental fibroblasts showed gene expression changes in human pluripotent stem cells and allowed the identification of DNA damage, cell cycle regulation, pentose phosphate metabolism, and mitochondrial oxidative phosphorylation defects as early ATM-mediated phenotypes. It was possible to generate functionally active neurons using published protocols, providing a tool that may be used to further the understanding of this disease.
Acknowledgments
We gratefully acknowledge the Stemcore facility for cell culture support. We also thank Louise Marquart (Queensland Institute of Medical Research) and Katia Nones (Queensland Centre for Medical Genomics) for expert technical support and the BrAshA-T Foundation and the Australian National Health and Medical Research Council for financial support. We thank Aine Farrell and Jian Sun for their assistance with tissue culture.
Author Contributions
S.N.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; M.G., S.K., and R.G.: collection and assembly of data; J.C.M.: data analysis and interpretation, manuscript writing; C.A.W.: data analysis and interpretation, final approval of manuscript; M.L.: conception and design, financial support, provision of study material/patients, data analysis and interpretation, manuscript writing, final approval of manuscript; E.W.: conception and design, financial support, provision of study material/patients, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Gatti RA, Berkel I, Boder E, et al. Localization of an ataxia-telangiectasia gene to chromosome 11q22–23. Nature. 1988;336:577–580. doi: 10.1038/336577a0. [DOI] [PubMed] [Google Scholar]
- 2.Savitsky K, Sfez S, Tagle DA, et al. The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. Hum Mol Genet. 1995;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- 3.Bensimon A, Schmidt A, Ziv Y, et al. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal. 2010;3:rs3. doi: 10.1126/scisignal.2001034. [DOI] [PubMed] [Google Scholar]
- 4.Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 5.Biton S, Gropp M, Itsykson P, et al. ATM-mediated response to DNA double strand breaks in human neurons derived from stem cells. DNA Repair (Amst) 2007;6:128–134. doi: 10.1016/j.dnarep.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Momcilovic O, Choi S, Varum S, et al. Ionizing radiation induces ataxia telangiectasia mutated-dependent checkpoint signaling and G(2) but not G(1) cell cycle arrest in pluripotent human embryonic stem cells. Stem Cells. 2009;27:1822–1835. doi: 10.1002/stem.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams BR, Golding SE, Rao RR, et al. Dynamic dependence on ATR and ATM for double-strand break repair in human embryonic stem cells and neural descendants. PLoS One. 2010;5:e10001. doi: 10.1371/journal.pone.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song H, Chung SK, Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 2010;6:80–89. doi: 10.1016/j.stem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Elson A, Wang Y, Daugherty CJ, et al. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc Natl Acad Sci USA. 1996;93:13084–13089. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavin MF. Ataxia-telangiectasia: From a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 11.Yu J, Hu K, Smuga-Otto K, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luong MX, Auerbach J, Crook JM, et al. A call for standardized naming and reporting of human ESC and iPSC lines. Cell Stem Cell. 2011;8:357–359. doi: 10.1016/j.stem.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 14.Gatei M, Jakob B, Chen P, et al. ATM protein-dependent phosphorylation of Rad50 protein regulates DNA repair and cell cycle control. J Biol Chem. 2011;286:31542–31556. doi: 10.1074/jbc.M111.258152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kozlov S, Gueven N, Keating K, et al. ATP activates ataxia-telangiectasia mutated (ATM) in vitro. Importance of autophosphorylation. J Biol Chem. 2003;278:9309–9317. doi: 10.1074/jbc.m300003200. [DOI] [PubMed] [Google Scholar]
- 16.Hans F, Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- 17.Parra I, Windle B. High resolution visual mapping of stretched DNA by fluorescent hybridization. Nat Genet. 1993;5:17–21. doi: 10.1038/ng0993-17. [DOI] [PubMed] [Google Scholar]
- 18.Vetter I, Lewis RJ. Characterization of endogenous calcium responses in neuronal cell lines. Biochem Pharmacol. 2010;79:908–920. doi: 10.1016/j.bcp.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 19.Du P, Kibbe WA, Lin SM. lumi: A pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Method. 1995;57:289–300. [Google Scholar]
- 21.Müller FJ, Laurent LC, Kostka D, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mar JC, Matigian NA, Quackenbush J, et al. attract: A method for identifying core pathways that define cellular phenotypes. PLoS One. 2011;6:e25445. doi: 10.1371/journal.pone.0025445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watters DJ. Oxidative stress in ataxia telangiectasia. Redox Rep. 2003;8:23–29. doi: 10.1179/135100003125001206. [DOI] [PubMed] [Google Scholar]
- 25.Chan EM, Ratanasirintrawoot S, Park IH, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 26.Guo Z, Kozlov S, Lavin MF, et al. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 27.Bensimon A, Aebersold R, Shiloh Y. Beyond ATM: The protein kinase landscape of the DNA damage response. FEBS Lett. 2011;585:1625–1639. doi: 10.1016/j.febslet.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, Ashley T, Brainerd EE, et al. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 29.Houldsworth J, Lavin MF. Effect of ionizing radiation on DNA synthesis in ataxia telangiectasia cells. Nucleic Acids Res. 1980;8:3709–3720. doi: 10.1093/nar/8.16.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen PC, Lavin MF, Kidson C, et al. Identification of ataxia telangiectasia heterozygotes, a cancer prone population. Nature. 1978;274:484–486. doi: 10.1038/274484a0. [DOI] [PubMed] [Google Scholar]
- 31.Taylor AM, Harnden DG, Arlett CF, et al. Ataxia telangiectasia: A human mutation with abnormal radiation sensitivity. Nature. 1975;258:427–429. doi: 10.1038/258427a0. [DOI] [PubMed] [Google Scholar]
- 32.Morgan SE, Kastan MB. p53 and ATM: Cell cycle, cell death, and cancer. Adv Cancer Res. 1997;71:1–25. doi: 10.1016/s0065-230x(08)60095-0. [DOI] [PubMed] [Google Scholar]
- 33.Pagliarini DJ, Calvo SE, Chang B, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morizane A, Doi D, Kikuchi T, et al. Small-molecule inhibitors of bone morphogenic protein and activin/nodal signals promote highly efficient neural induction from human pluripotent stem cells. J Neurosci Res. 2011;89:117–126. doi: 10.1002/jnr.22547. [DOI] [PubMed] [Google Scholar]
- 35.Raya A, Rodriguez-Piza I, Guenechea G, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinoshita T, Nagamatsu G, Kosaka T, et al. Ataxia-telangiectasia mutated (ATM) deficiency decreases reprogramming efficiency and leads to genomic instability in iPS cells. Biochem Biophys Res Commun. 2011;407:321–326. doi: 10.1016/j.bbrc.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 37.Marión RM, Strati K, Li H, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marion RM, Strati K, Li H, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 39.Pandita TK. ATM function and telomere stability. Oncogene. 2002;21:611–618. doi: 10.1038/sj.onc.1205060. [DOI] [PubMed] [Google Scholar]
- 40.Vaziri H. Critical telomere shortening regulated by the ataxia-telangiectasia gene acts as a DNA damage signal leading to activation of p53 protein and limited life-span of human diploid fibroblasts. A review. Biochemistry (Mosc) 1997;62:1306–1310. [PubMed] [Google Scholar]
- 41.Ivanov VN, Zhou H, Partridge MA, et al. Inhibition of ataxia telangiectasia mutated kinase activity enhances TRAIL-mediated apoptosis in human melanoma cells. Cancer Res. 2009;69:3510–3519. doi: 10.1158/0008-5472.CAN-08-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan J, Robert C, Jang YY, et al. Human induced pluripotent cells resemble embryonic stem cells demonstrating enhanced levels of DNA repair and efficacy of nonhomologous end-joining. Mutat Res. 2011;713:8–17. doi: 10.1016/j.mrfmmm.2011.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodarzi AA, Noon AT, Deckbar D, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Momcilovic O, Knobloch L, Fornsaglio J, et al. DNA damage responses in human induced pluripotent stem cells and embryonic stem cells. PLoS One. 2010;5:e13410. doi: 10.1371/journal.pone.0013410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valentin-Vega YA, Maclean KH, Tait-Mulder J, et al. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119:1490–1500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ambrose M, Goldstine JV, Gatti RA. Intrinsic mitochondrial dysfunction in ATM-deficient lymphoblastoid cells. Hum Mol Genet. 2007;16:2154–2164. doi: 10.1093/hmg/ddm166. [DOI] [PubMed] [Google Scholar]
- 47.Eaton JS, Lin ZP, Sartorelli AC, et al. Ataxia-telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis. J Clin Invest. 2007;117:2723–2734. doi: 10.1172/JCI31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu X, Wan S, Lyu YL, et al. Etoposide induces ATM-dependent mitochondrial biogenesis through AMPK activation. PLoS One. 2008;3:e2009. doi: 10.1371/journal.pone.0002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang D, Kang R, Livesey KM, et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab. 2011;13:701–711. doi: 10.1016/j.cmet.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang R, Livesey KM, Zeh HJ, 3rd, et al. Metabolic regulation by HMGB1-mediated autophagy and mitophagy. Autophagy. 2011;7:1256–1258. doi: 10.4161/auto.7.10.16753. [DOI] [PubMed] [Google Scholar]
- 51.Heiss EH, Schilder YD, Dirsch VM. Chronic treatment with resveratrol induces redox stress- and ataxia telangiectasia-mutated (ATM)-dependent senescence in p53-positive cancer cells. J Biol Chem. 2007;282:26759–26766. doi: 10.1074/jbc.M703229200. [DOI] [PubMed] [Google Scholar]
- 52.Patel AY, McDonald TM, Spears LD, et al. Ataxia telangiectasia mutated influences cytochrome c oxidase activity. Biochem Biophys Res Commun. 2011;405:599–603. doi: 10.1016/j.bbrc.2011.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barlow C, Dennery PA, Shigenaga MK, et al. Loss of the ataxia-telangiectasia gene product causes oxidative damage in target organs. Proc Natl Acad Sci USA. 1999;96:9915–9919. doi: 10.1073/pnas.96.17.9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Browne SE, Roberts LJ, 2nd, Dennery PA, et al. Treatment with a catalytic antioxidant corrects the neurobehavioral defect in ataxia-telangiectasia mice. Free Radic Biol Med. 2004;36:938–942. doi: 10.1016/j.freeradbiomed.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 55.Chen P, Peng C, Luff J, et al. Oxidative stress is responsible for deficient survival and dendritogenesis in purkinje neurons from ataxia-telangiectasia mutated mutant mice. J Neurosci. 2003;23:11453–11460. doi: 10.1523/JNEUROSCI.23-36-11453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cosentino C, Grieco D, Costanzo V. ATM activates the pentose phosphate pathway promoting anti-oxidant defence and DNA repair. EMBO J. 2011;30:546–555. doi: 10.1038/emboj.2010.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim J, Hao T, Shaw C, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 58.Muguruma K, Nishiyama A, Ono Y, et al. Ontogeny-recapitulating generation and tissue integration of ES cell-derived Purkinje cells. Nat Neurosci. 2010;13:1171–1180. doi: 10.1038/nn.2638. [DOI] [PubMed] [Google Scholar]
- 59.Tao O, Shimazaki T, Okada Y, et al. Efficient generation of mature cerebellar Purkinje cells from mouse embryonic stem cells. J Neurosci Res. 2010;88:234–247. doi: 10.1002/jnr.22208. [DOI] [PubMed] [Google Scholar]