This study investigated the differences between the differentiation potentials of three human embryonic stem cell (hESC) lines, four retrovirally derived human induced pluripotent stem cell (hiPSC) lines, and one hiPSC line derived with the nonintegrating Sendai virus technology. Cells differentiated from hESCs and hiPSCs showed functional similarities and similar expression of genes characteristic of specific cell types. However, some differences between individual cell lines were also detected, which could be explained by residual activity of viral transgenes and the reactivation of several transgenes in retrovirally derived hiPSC lines. These findings highlight the need for integration-free reprogramming technologies, resulting in transgene-free iPSCs, which could also be potentially therapeutically applicable, unlike the retrovirally derived cells used in this study.
Keywords: Embryonic stem cells, Pluripotent stem cells, Hepatocyte differentiation, Cardiac, Neural differentiation, Retinal pigmented epithelium, Differentiation
Abstract
Functional hepatocytes, cardiomyocytes, neurons, and retinal pigment epithelial (RPE) cells derived from human embryonic stem cells (hESCs) or human induced pluripotent stem cells (hiPSCs) could provide a defined and renewable source of human cells relevant for cell replacement therapies, drug discovery, toxicology testing, and disease modeling. In this study, we investigated the differences between the differentiation potentials of three hESC lines, four retrovirally derived hiPSC lines, and one hiPSC line derived with the nonintegrating Sendai virus technology. Four independent protocols were used for hepatocyte, cardiomyocyte, neuronal, and RPE cell differentiation. Overall, cells differentiated from hESCs and hiPSCs showed functional similarities and similar expression of genes characteristic of specific cell types, and differences between individual cell lines were also detected. Reactivation of transgenic OCT4 was detected specifically during RPE differentiation in the retrovirally derived lines, which may have affected the outcome of differentiation with these hiPSCs. One of the hiPSC lines was inferior in all directions, and it failed to produce hepatocytes. Exogenous KLF4 was incompletely silenced in this cell line. No transgene expression was detected in the Sendai virus-derived hiPSC line. These findings highlight the problems related to transgene expression in retrovirally derived hiPSC lines.
Introduction
Human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), collectively termed human pluripotent stem cells (hPSCs), are considered a renewable source of cells for regenerative medicine because of their potential to differentiate into all cell types found in the adult human body [1]. hESCs are derived from the inner cell mass of developing embryos [2], whereas hiPSCs are reprogrammed from somatic cells [3, 4]. hiPSCs share several characteristics with hESCs, including similar morphology, expression of pluripotency markers, and the ability to differentiate into definitive cell lineages [5–8]. Initial studies have suggested that fully reprogrammed iPSCs are indistinguishable from ESCs [3, 4, 9]. More comprehensive studies have revealed that particularly early passage iPSCs show differences in their gene expression profile, but continued propagation tends to increase the similarity between hESCs and iPSCs [10, 11].
Recent studies have revealed that iPSCs maintain differential DNA methylation patterns as a sign of incomplete reprogramming [12, 13]. The possible consequences of this “epigenetic memory” still remain unknown. Some recent studies indicate that the origin of iPSCs is relevant for their differentiation capacity. iPSCs derived from retinal pigment epithelial (RPE) cells have a high tendency for pigmentation [14], and reprogramming of cardiac fibroblasts produces more cardiomyocytes than fibroblasts from other sources [15]. Although hiPSCs in general seem to differentiate into specific lineages as efficiently as hESCs, there are several examples of incomplete pluripotent differentiation capacity, possibly reflecting their epigenetic barriers [11, 16].
Most studies comparing the properties of hESCs and hiPSCs have focused on their undifferentiated phenotype, their uncontrolled differentiation capacity in embryoid bodies or teratomas, or their differentiation toward a single specific lineage [6, 17–19]. However, many extended protocols have been developed for the differentiation of specific derivatives of all germ lines, such as hepatocytes (endoderm), cardiomyocytes (mesoderm), or neurons and retinal cells (ectoderm). The rationale of our study was to systematically compare the capability of the same hiPSC and hESC lines to develop into functional cell types following the protocols optimized by researchers dedicated to their respective line of differentiation. We studied four hiPSC and three hESC lines for their ability to differentiate into functional hepatocyte-like cells (HLCs), beating cardiomyocytes, neurons forming active neuronal networks, and highly pigmented mature RPE cells. Cell lines hiPSC1 to hiPSC4 were derived in two different laboratories, from neonatal and adult fibroblasts using retroviral vectors. One of the cell lines was derived using NANOG, OCT4, SOX2, and LIN28, whereas the other cell lines were derived by overexpressing OCT4, SOX2, KLF4, and c-MYC. In addition, the hiPSC5 line was derived with an integration-free Sendai-viral system. All of the cell lines were adapted in the same culture conditions prior to each differentiation protocol to avoid variation caused by different culture environments. Our results did not point to a systematic difference in the differentiation efficiency between hiPSC and hESC lines, except for one cell line with incomplete transgene silencing. Reactivation of transgenes was occasionally observed, especially with a long RPE differentiation protocol. These observations raise concerns related to the use of integrating reprogramming methods.
Materials and Methods
Ethical Issues
The Institute of Biomedical Technology has an approval of National Authority for Medicolegal Affairs Finland to study human embryos (Dnro1426/32/300/05), as well as the support of the Ethical Committee of Pirkanmaa Hospital District to derive, culture, and differentiate hESC lines from surplus human embryos (R05116) and to produce new hiPSC lines (R08070). The generation of hiPSC lines in Biomedicum Stem Cells Center Helsinki was approved by the Coordinating Ethics Committee of the Helsinki and Uusimaa Hospital District (Nro 423/13/03/00/08).
Cell Lines and Cell Culture
Three hESC lines (H7 [hESC1; WiCell Research Institute, Madison, WI, http://www.wicell.org], FES29 [hESC2] [20], and Regea 08/023 [hESC3] [21]) and five hiPSC lines (FiPS5-7 [hiPSC1] [22], UTA.00112.hFF [hiPSC2] [23], A116 [hiPSC3] [supplemental online Fig. 1], UTA.01006.WT [hiPSC4] [23], and Hel24.3 [hiPSC5] [supplemental online Fig. 1]) were used in this study. The hiPSC lines FiPS5-7 and UTA.00112.hFF were derived from human foreskin fibroblasts (hFFs) (CRL-2429; American Type Culture Collection, Manassas, VA, http://www.atcc.org), and the hiPSC lines A116, UTA.01006.WT, and Hel24.3 were derived from adult skin fibroblasts. FiPS5-7 (hiPSC1) was reprogrammed with NANOG, OCT4, SOX2, and LIN28 and the other hiPSC lines with OCT4, SOX2, KLF4, and c-MYC. The cell lines used in this study are presented in supplemental online Table 1. Details of hiPSC reprogramming conditions are provided in the supplemental online Materials and Methods.
Differentiation Protocols
Pluripotent stem cell lines were differentiated into hepatocyte-like cells, cardiomyocytes, neural cells, and RPE cells. Detailed methods of differentiation and characterization are provided in the supplemental online Materials and Methods.
The efficiency of hepatic differentiation was evaluated by studying the expression of OCT4, SOX17, FOXA2, AFP, and Albumin at day (d) 7, d14, and d21 by quantitative polymerase chain reaction (qPCR) analysis and by studying the expression of OCT4, FOXA2, SOX17, AFP, and albumin with immunocytochemistry. The definitive endoderm induction was analyzed at d7 by flow cytometry for CXCR4+ cells, and the functionality of the differentiated hepatocyte-like cells was studied by albumin secretion measured with an enzyme-linked immunosorbent assay.
Cardiac differentiation was characterized by studying the expression of Nanog, OCT4, SOX17, Brachyury T, and NKX2.5 at time points d0, d3, d6, d13, and d30 by qPCR and by studying the expression of α-actinin, Troponin T, connexin-43, and ventricular myosin heavy chain (MHC) with immunocytochemistry. The efficiency of cardiac differentiation was evaluated by immunocytochemical analysis of cytospin samples on day 20 and counting the number of beating areas in the end of differentiation on day 30. The functionality of the cardiomyocytes was analyzed using the microelectrode array (MEA) platform.
Neural differentiation was evaluated at the 4- and 8-week time points by studying the expression of OCT4, Musashi, Neurofilament-68 (NF-68), and glial fibrillary acidic protein (GFAP) by qPCR and by studying the expression of OCT4, EpCAM, Nestin, microtubule-associated protein 2 (MAP-2), GFAP, brain lipid-binding protein (BLBP), chondroitin sulfate proteoglycan (NG2), and galactocerebroside (GalC) by immunocytochemistry. The morphological analysis was performed with time-lapse imaging. The spontaneous functionality of developing neuronal networks was characterized using MEA.
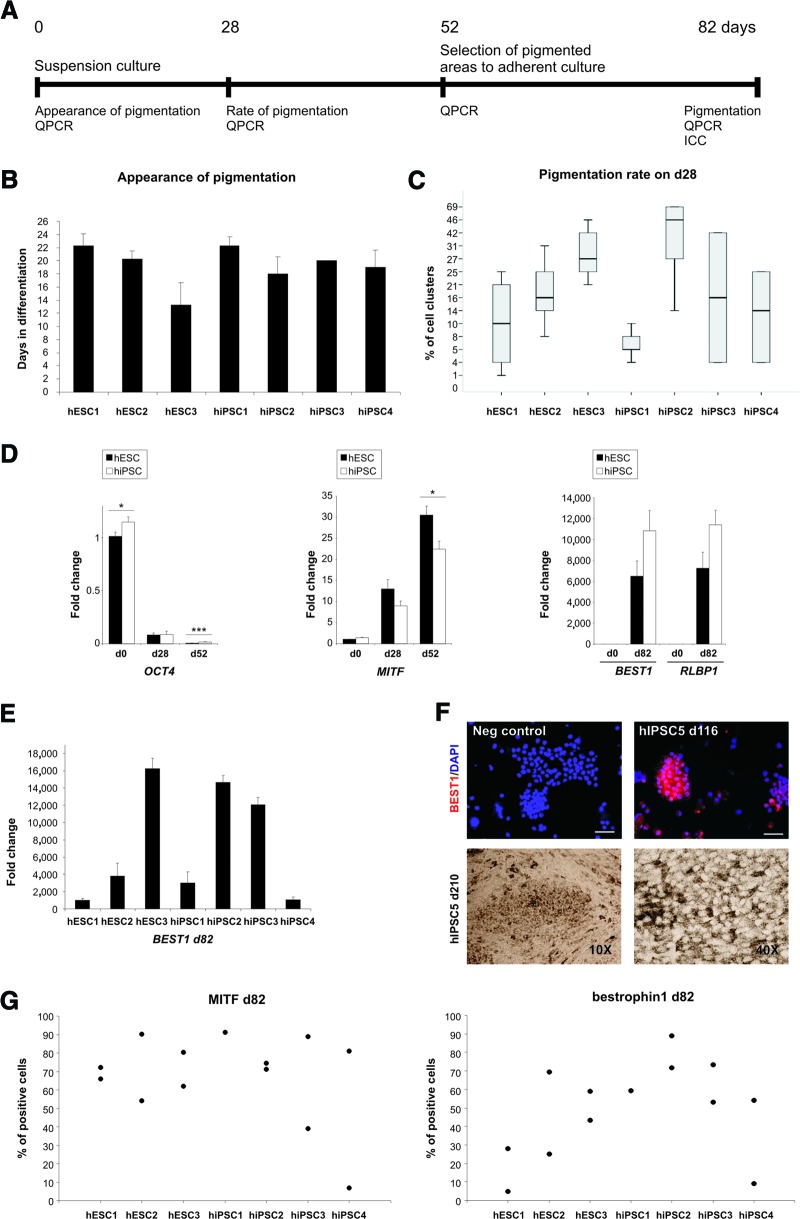
To evaluate putative RPE cell differentiation, the appearance of the first pigmented cells was followed daily and recorded. The percentage of pigment-containing cell aggregates from the total amount of aggregates was counted on day 28 ± 1 of the differentiation. The expression of OCT4, MITF, BEST1, and RLBP1 was analyzed by qPCR from d0, d28, d52, and d82 of RPE differentiation. The expression of OCT4, MITF and bestrophin-1 proteins was quantified with cytospin analysis on day 82 or on day 116.
Statistical Analysis
Statistical analysis between two groups was performed with the unpaired Student's t test or Mann-Whitney U test according to the sample set. In the case of multiple groups, one-way analysis of variance and the Tukey post hoc test were used. A p value of <.05 was considered statistically significant.
Results
Transgene Silencing
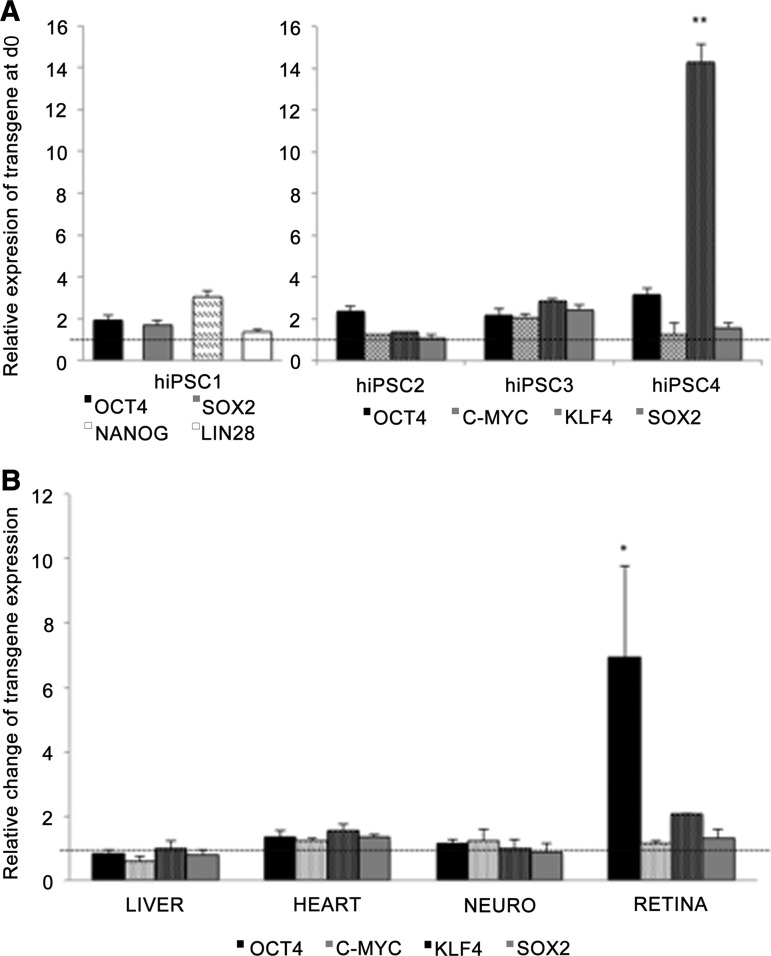
hiPSC lines hiPSC1 [22], hiPSC2 [23], and hiPSC4 [23] were independently established by retroviral infection (OCT4, SOX2, c-MYC, and KLF4 or OCT4, SOX2, NANOG, and LIN28) and characterized as described previously [22, 23]. hiPSC3 and hiPSC5 (supplemental online Fig. 1) lines were separately characterized for this study. Relative transcriptional levels of the transgenes were studied by qPCR before and at the end of each differentiation protocol in lines hiPSC1–hiPSC4. The results revealed constant expression of exogenous KLF4 in hiPSC4 at d0, whereas transgenes in other cell lines were silenced (Fig. 1A; supplemental online Fig. 2A). Transgene expression in general was not significantly induced by the differentiation protocols, with one remarkable exception. Levels of exogenous OCT4 mRNA were systematically increased at the end of the long-term RPE differentiation protocol in all retrovirally derived hiPSC lines (Fig. 1B; supplemental online Fig. 2B), and OCT4+ cells could be detected by immunocytochemistry after 82 days of RPE differentiation (supplemental online Fig. 3B). In addition, exogenous LIN28 and NANOG mRNA levels were markedly increased during the RPE differentiation in hiPSC1, the only cell line derived by overexpression of these factors (supplemental online Fig. 2B). When the Sendai-virally derived hiPSC5 line was differentiated into RPE cells, no reactivation of transgene expression was detected (supplemental online Fig. 3A, 3B).
Figure 1.
Transgene silencing. (A): Quantitative polymerase chain reaction (qPCR) analysis for expression of the transgenes OCT4, SOX2, NANOG, LIN28, c-MYC, and KLF4 at the onset of differentiation (d0). The data are shown as the average (±SEM) relative value from four independent experiments. The value 1 indicates total silencing of transgenes. One-way analysis of variance with Tukey post hoc test was used for statistical analysis. **, p < .01. (B): qPCR analysis for activation of transgene expression during each differentiation protocol. The value 1 indicates no change in transgene expression. *, p < .05. Abbreviations: d, day; hiPSC, human induced pluripotent stem cell; NEURO, neural differentiation.
Definitive Endoderm Differentiation
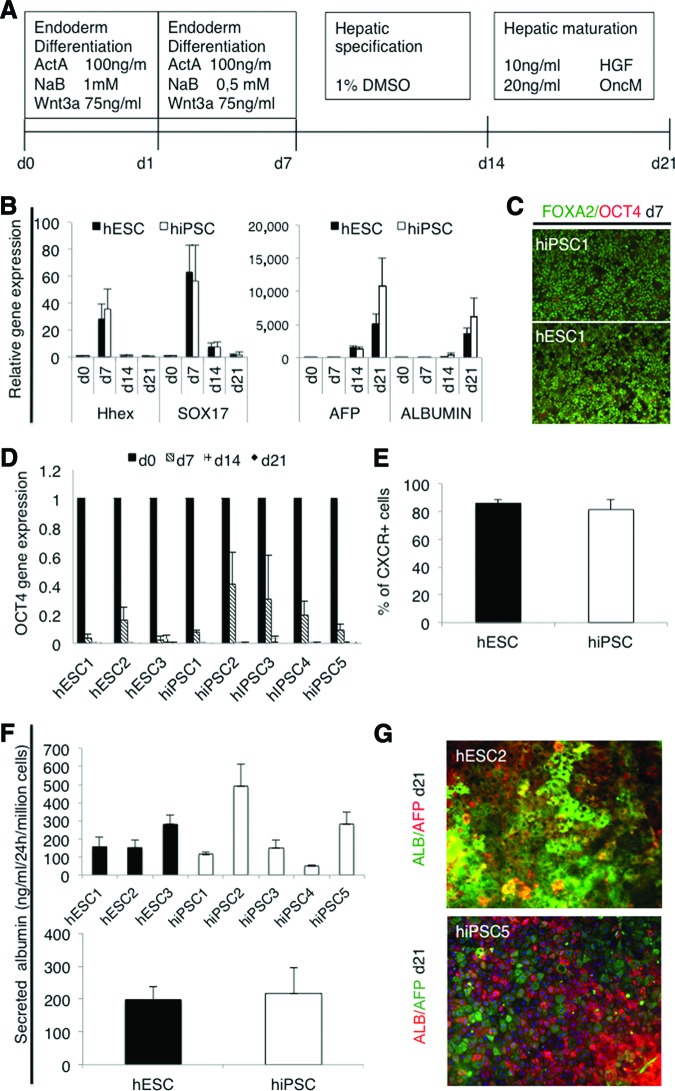
Hepatocyte differentiation protocol consists of three stages, slightly modified from that described by Hay et al. [24] (Fig. 2A). The first stage directs the cells from pluripotent cells into committed definitive endoderm (DE) cells. In this stage, after 7 days from the onset of induction, all the cell lines had lost their embryonic stem-like small, round, and dense morphology and the cells were growing as homogeneous monolayers. qPCR analysis showed marked upregulation of the anterior definitive endoderm genes SOX17 and Hhex in all lines at day 7 (Fig. 2B; supplemental online Fig. 4A). During differentiation, the expression of OCT4 decreased in all cell lines and became undetectable by day 14. The process was somewhat slower in hiPSCs than hESCs (Fig. 2D). There was no change in the expression level of the extraembryonic endoderm gene SOX7 (data not shown). In immunocytochemical analysis more than 90% of the cells were positive for definitive endoderm marker FOXA2, and very few if any OCT4+ cells could be found (Fig. 2C; supplemental online Fig. 5A). The percentage of CXCR4+ cells as analyzed by flow cytometry varied between 65% and 96% between all the lines (supplemental online Fig. 5B), and there were no significant difference between hESC (n = 3) and hiPSC (n = 4) lines in group comparison (Fig. 2E). These results suggest that the hESC and hiPSC lines used in this study differentiated into definitive endoderm stage with equal efficiency.
Figure 2.
Hepatocyte differentiation. (A): Schematic presentation of the protocol used to differentiate human pluripotent stem cells into hepatocyte-like cells. (B): Quantitative polymerase chain reaction (qPCR) analysis for expression of the genes marking key stages of differentiation, first into definitive endoderm cells (SOX17, Hhex) and then into hepatocyte-like cells (AFP, Albumin). The columns show the average fold change from at least two independent experiments ± SEM for each line. (C): Representative immunostaining after 7 days of differentiation, demonstrating that almost all cells expressed nuclear FOXA2 as a sign of definitive endoderm differentiation and very few cells still expressed the pluripotency marker OCT4. (D): qPCR analysis for expression of the pluripotency gene OCT4 at d0, d7, d14, and d21. The columns show the average fold change from at least two independent experiments ± SEM, demonstrating the rapid downregulation in both hESC and hiPSC lines. (E): Fluorescence-activated cell sorting analysis of cells expressing the endoderm marker CXCR4 at d7. Columns represent the average for hESCs (n = 6) and hiPSCs (n = 8). (F): Albumin secretion into the medium by the differentiated cells. In the upper graph, each cell line is presented separately (two or three repeated experiments). Lower graph shows the comparison between iPSCs (n = 10) and hESCs (n = 6). (G): Representative immunostaining after 21 days of differentiation of hESC2 and hiPSC5. Hepatocyte markers ALB and AFP are shown. Magnification, ×20. Abbreviations: AFP, α-fetoprotein; ALB, albumin; d, day; DMSO, dimethyl sulfoxide; hESC, human embryonic stem cell; HGF, hepatocyte growth factor; hiPSC, human induced pluripotent stem cell.
Hepatocyte Differentiation
The resulting DE cells were then differentiated into HLCs by 7-day culture in medium supplemented with 1% dimethyl sulfoxide (stage 2) and by a final maturation step in medium supplemented with hepatocyte growth factor and Oncostatin M for a further 7 days [24] (Fig. 2A). During this time the cells displayed morphological changes from a spiky shape to a polygonal shape. On day 21, the cultures contained foci exhibiting features of human hepatocytes, including a typical polygonal shape with distinct round nuclei, and many of the cells were binuclear (supplemental online Fig. 5C). Only hiPSC4-derived cells failed to develop a distinct hepatocyte-like morphology (supplemental online Fig. 5C). In qPCR analysis, AFP was highly upregulated at day 14 and Albumin at day 21 (Fig. 2B; supplemental online Fig. 4A).
The HLCs derived from hiPSC2 expressed the highest levels of AFP and Albumin, whereas the expression of these hepatocyte-specific markers was nearly undetectable with hiPSC4-derived cells. Variation in the differentiation efficiency, as measured by albumin expression, was detected between hESCs and iPSCs, but the differences were not statistically significant in a group comparison. The hepatocyte-specific functionality of the differentiated cells was analyzed by albumin secretion assay. The results correlated well with qPCR data; there was no overall difference in albumin secretion rate between the hESC1–hESC3 lines and the hiPSC1–hiPSC5 lines in a group comparison, although there was variation between the individual cell lines (Fig. 2F), particularly because hiPSC4 failed to develop into albumin-secreting cells. Taken together, both the highest and the lowest levels of differentiation were observed in the hiPSC lines, whereas there was less variation among the hESC lines.
Cardiomyocyte Differentiation
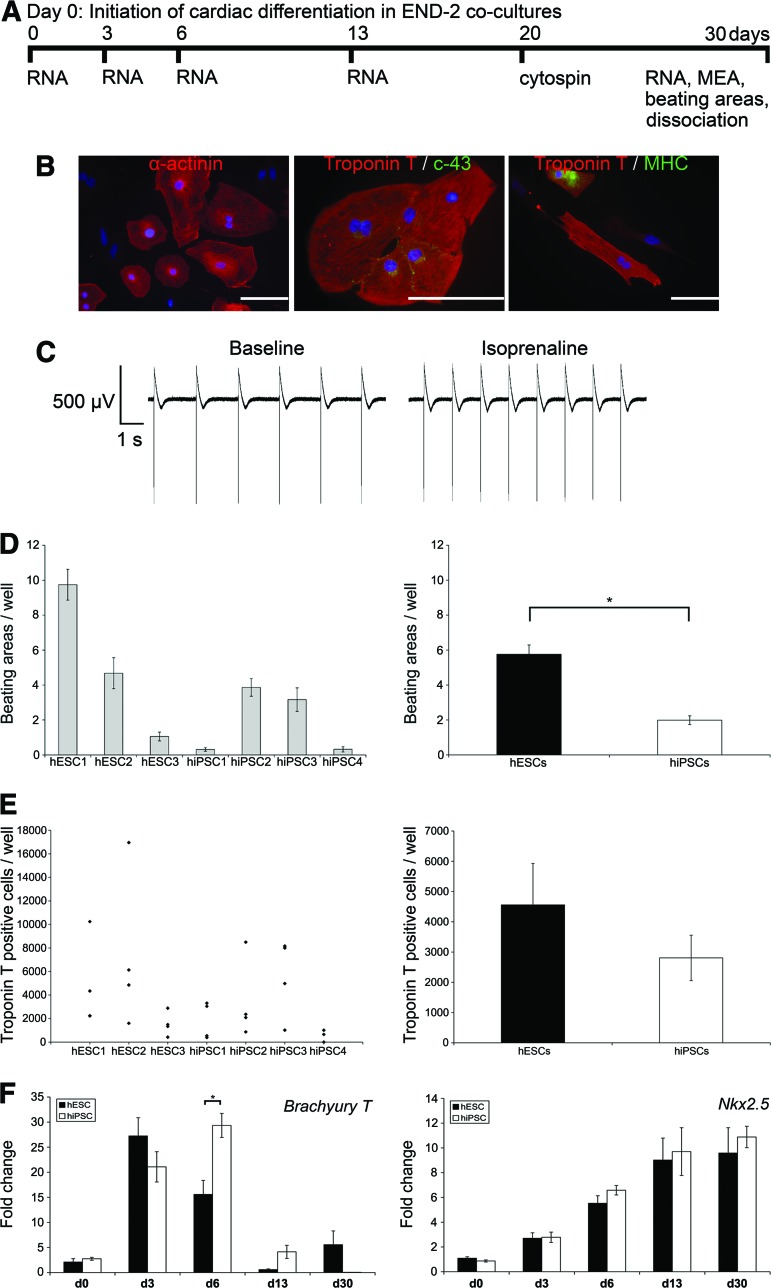
hPSC lines were differentiated into cardiomyocytes using the END-2 coculture method [25]. The progression of cardiac differentiation was monitored and cells analyzed as shown in Figure 3A. All four hiPSC and three hESC lines differentiated into beating cardiomyocytes, but the differentiation efficiency was variable. In addition, the cardiac differentiation efficiency varied between separate differentiation experiments within the same cell line. All cell lines formed compact structures in END-2 coculture except hiPSC4, which tended to form more cystic structures than the other cell lines. Cardiomyocytes derived from the cell lines expressed α-actinin, Troponin T, connexin-43, and MHC in immunocytochemical stainings (Fig. 3B). The electrical activity of cardiomyocytes was monitored with MEA measurements. The normal beating rate of the cell clusters was measured and the beating rate was increased by adding the β-adrenergic agonist isoprenaline to MEA chambers. All hESC- and hiPSC-derived cardiomyocytes beat and gave a signal on MEA and thus can be considered functional cardiomyocytes (Fig. 3C).
Figure 3.
Cardiomyocyte differentiation. (A): Schematic presentation of the cardiac differentiation protocol and experimental design. (B): Cardiomyocytes derived from all human pluripotent stem cell (hPSC) lines expressed α-actinin, connexin-43, and ventricular myosin heavy chain proteins. Representative images of hiPSC2 line. Scale bars = 100 μm. (C): All hPSC lines gave a signal on the MEA platform, and the beating rate was increased by isoprenaline (80 nM). Shown are representative images of the hiPSC3 line. (D): The number of beating areas in one well with each hPSC line (left) and in hESC and hiPSC groups (right). Error bars show the SEM. *, p < .001. (E): Scatter plots (left) show the number of Troponin T-positive cells in one well, and the columns (right) show the Troponin T-positive cells found in the hESC and hiPSC groups. Error bars show the SEM. (F): Results of the gene expression analysis on Brachyury T and Nkx2.5 genes at the d0, d3, d6, d13, and d30 time points during cardiac differentiation. The expression of genes was compared between hESC and hiPSC lines. In hiPSC lines the highest expression of Brachyury T was detected on day 6, and it was significantly higher in hiPSC lines than in hESC lines (*, p < .003). Error bars show the SEM. Abbreviations: c-43, connexin-43; d, day; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; MEA, microelectrode array; MHC, myosin heavy chain; RNA, quantitative polymerase chain reaction samples.
Quantitative immunocytochemical analysis was performed on cells cultured in END-2 cocultures on day 20, and beating areas were counted at the end of differentiation on day 30. The number of beating areas was highly variable between separate differentiation experiments within the same cell line. As a group, hESCs formed more beating areas than hiPSCs (p < .001, Fig. 3D). hESC1 had the most efficient cardiac differentiation efficiency, whereas hiPSC1 and hiPSC4 had the least efficient cardiac differentiation and the lowest number of beating areas (Fig. 3D). The results from the quantitative immunocytochemical analysis detecting cardiac Troponin T-positive cells were in accordance with the number of beating areas (Fig. 3E). In the hESC1 line, the beating areas were smaller than in other cell lines, but there were more beating areas. This may explain the difference between quantitative immunocytochemical results and the number of beating areas detected.
The expression of pluripotency markers NANOG and OCT4 was highest on day 0 and descended during cardiac differentiation (supplemental online Fig. 6A, 6B). The expression of endodermal SOX17 was the highest on day 3 in all hiPSC and hESC lines (supplemental online Fig. 6C). The expression of Brachyury T was also the highest on day 3 with the hESC lines (Fig. 3F). Interestingly, with hiPSC lines the highest expression of Brachyury T was detected on day 6, and it was significantly higher in hiPSC lines than in hESC lines (p = .003), suggesting a slower tempo of cardiac differentiation. The expression of Nkx2.5 ascended evenly during differentiation in all hiPSC and hESC lines.
Neural Differentiation
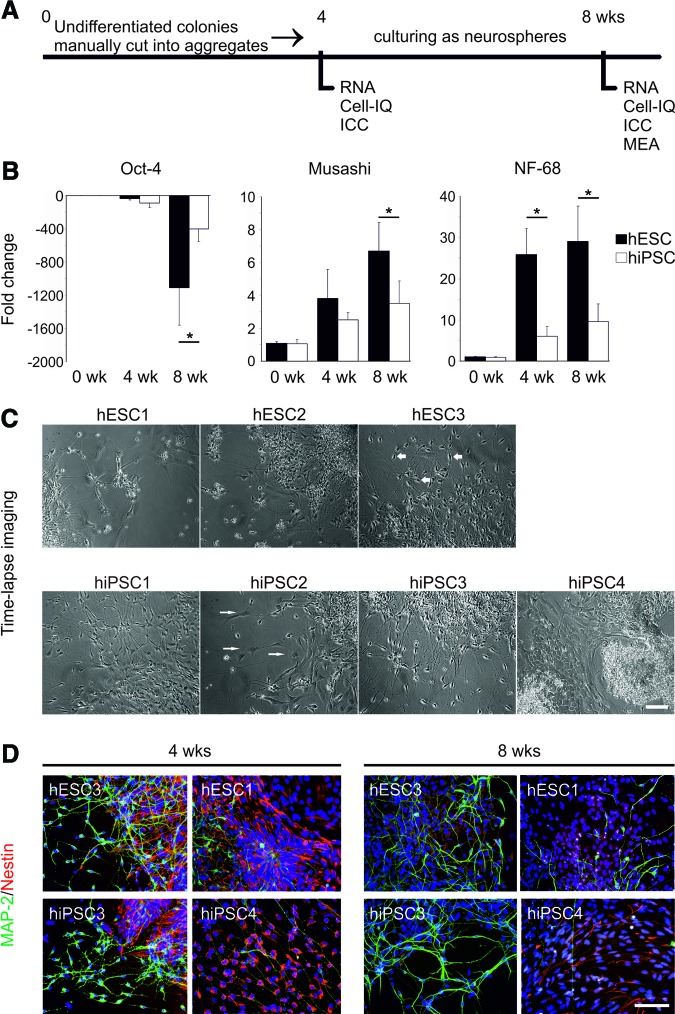
The neural differentiation protocol and the analyses used in this study are summarized in Figure 4A. Cell lines displayed clear differences when differentiated and cultured as neurospheres in neural differentiation medium. Neurospheres usually develop into firm cell aggregates within 2 weeks. The hiPSC3-derived neurospheres, however, showed persistent growth of unwanted cystic structures up to 4 weeks of differentiation (supplemental online Fig. 7). When the cysts were repeatedly manually removed from the cultures, the cyst formation declined. In addition to hiPSC3, the hiPSC2 line produced relatively fast-growing and less firm neurospheres. The growth of hiPSC4-derived neurospheres was weaker than that in the other lines. Neurospheres derived from hESC lines showed less variation during differentiation and culture.
Figure 4.
Neuronal differentiation. (A): Schematic presentation of the neural differentiation protocol and experimental design. (B): Results of the gene expression analysis of OCT4, Musashi, and NF-68 at the 0-, 4-, and 8-week time points. The expressions of the genes were compared between hESC and hiPSC lines. Columns represent an average of hESC (n = 3) and hiPSC (n = 4) lines ± SEM. Musashi week 8, p = .034; NF-68 weeks 4 and 8, p = .002 and p = .01, respectively. *, p < .05, Mann-Whitney U test. (C): Morphologies of the cells derived from different cell lines at the 8-week time point. hESC3-, hiPSC1-, and hiPSC3-derived cells displayed mostly neuronal morphology (thick arrows), whereas other cell lines produced cells with flat epithelial cell-like morphology (thin arrows). Scale bar = 100 μm. (D): Immunocytochemical characteristics of the differentiated cells. Neural precursor cell marker Nestin (red) and neural marker MAP-2 (green) were both detected in all the populations derived from hESC and hiPSC lines. Cell cultures derived from hESC3 and hiPSC3 lines were detected with high amounts of MAP-2-positive cells at both time points, whereas clearly fewer MAP-2-positive cells could be detected from the cultures of hESC1 and hiPSC4. The number of Nestin-positive cells decreased within all the cell lines from 4 to 8 weeks. Scale bar = 100 μm. Abbreviations: Cell-IQ, time-lapse imaging; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; ICC, immunocytochemistry; MAP-2, microtubule-associated protein 2; MEA, microelectrode array; NF-68, Neurofilament-68; RNA, quantitative polymerase chain reaction samples; wk, week.
According to qPCR analysis, the expression of OCT4 was strongly downregulated within every cell line during the neural differentiation at 8 weeks (Fig. 4B). The downregulation was, however, significantly stronger in neurospheres derived from hESC than from hiPSC lines (p < .01). The expression of neural precursor cell marker Musashi and neural marker NF-68 increased during differentiation, and both were significantly higher in neurospheres derived from hESCs than from hiPSC lines (Musashi week 8, p = .034; NF-68 weeks 4 and 8, p = .002 and p = .01, respectively) (Fig. 4B). Expression of glial marker GFAP was undetectable in 0 and 4 weeks, and it was expressed at a low level in every cell line after 8 weeks of differentiation. Line-specific expression of OCT4, Musashi, and NF-68 is shown in supplemental online Figure 4.
The cells were monitored with time-lapse imaging at 4 and 8 weeks during the neural differentiation. Quantitative analysis of time-lapse imaging data was performed by Cell-IQ analysis software (Chip-Man Technologies Ltd., Tampere, Finland) [26], but the accurate neuronal cell number could not be reliably determined because of confluence of the cultures (supplemental online Fig. 7). Qualitative analysis of the imaged data showed that hESC3 and hiPSC3 produced very pure neuronal populations in 8 weeks, whereas hiPSC4 was clearly the weakest cell line for neural differentiation, producing a lot of flat epithelial-like cells (Fig. 4C). The cells in hiPSC1-derived cultures were also mostly neuronal, but more cells with non-neuronal morphology were detected compared with hESC3- and hiPSC3-derived cultures.
Immunocytochemical staining supported the results of the time-lapse imaging analysis (Fig. 4D). The highest levels of MAP-2-positive cells were detected within hESC3- and hiPSC3-derived cultures. In hiPSC4-derived cultures only single cells positive for MAP-2 could be detected. The number of Nestin-positive cells decreased in all lines from 4 weeks to 8 weeks. No OCT4, CD326, GFAP, or BLBP was detected in any cell lines, indicating that no undifferentiated cells, astrocytes, or radial glial cells were present in the cultures. Only single cells positive for NG2 or GalC could be detected at 8 weeks, indicating the presence of few oligodendrocyte precursor cells in the cultures (supplemental online Fig. 7).
Electrophysiological Properties of Neuronal Networks
As previously described [27], the first form of electrical activity detected from the neuronal networks was single spikes, whereas the mature neuronal networks displayed bursts containing multiple spikes simultaneously on several electrodes (supplemental online Fig. 8). The cell lines with the highest neural differentiation efficiencies based on morphological and immunocytochemical characterizations (hESC3 and hiPSC3) displayed burst-activity within 3 weeks in MEA culture. The spontaneously active bursting neuronal networks were routinely recorded from hESC3- and hiPCS3-derived cultures. The neuronal networks formed by the other cell lines displayed activities varying from single spikes to bursts.
RPE Differentiation
The hESC and hiPSC1–hiPSC5 lines were differentiated into RPE cells according to a previously reported protocol, which is based on spontaneous differentiation in EB-like cultures [8]. The differentiation protocol and analyses are summarized in Figure 5A. The RPE differentiation potential of the cell lines was studied by monitoring the appearance of the first pigmented cells emerging in the cultures. In addition, the percentage of cell clusters containing pigmented cells was counted on day 28 after initiation of differentiation.
Figure 5.
Retinal pigment epithelial (RPE) differentiation. (A): Schematic representation of the RPE differentiation protocol and experimental design. (B): Appearance of the first pigmented cells in the cultures at the beginning of RPE differentiation. Columns are representing an average of two to four independent experiments (n) ± SEM. Shown are hESC1 (n = 4), hESC2 (n = 4), hESC3 (n = 3), hiPSC1 (n = 4), hiPSC2 (n = 3), hiPSC3 (n = 2), and hiPSC4 (n = 3). (C): Rate of pigmentation on differentiation day 28. Box plots show the sample minimum, lower quartile, median, upper quartile, and sample maximum of two to four independent experiments. The number of experiments/total number of cell clusters counted were as follows: hESC1, 4/674; hESC2, 3/448; hESC3, 3/339; hiPSC1, 3/656; hiPSC2, 3/427; hiPSC3, 2/445; and hiPSC4, 2/704. (D): QPCR analysis for expression of genes marking key stages of human pluripotent stem cell differentiation (OCT4) into pigment-producing cells (MITF) and subsequently into RPE-like cells (BEST1 and RLBP1). The columns show the average fold change from at least two independent experiments ± SEM. *, p < .05; ***, p < .001. Statistical analyses were performed with independent samples t test or Mann-Whitney U test according to the sample set. (E): QPCR analysis for expression of BEST1 on d82 in each analyzed cell line. The columns show the average fold change from at least two independent experiments ± SEM. (F): Top row: Bestrophin-1 (BEST1) staining for cytospin samples collected from hiPSC5-derived RPE cells. Bottom row: pigmented cells derived from hiPSC5 at d210 in passage 2. (G): Expression of MITF and bestrophin-1 proteins on d82. Scatter plots show the percentage of positive cells from one or two independent experiments. The total number of cells counted were as follows for MITF/bestrophin-1: hESC1, 326/274; hESC2, 351/337; hESC3, 466/454; hiPSC1, 184/202; hiPSC2, 579/518; hiPSC3, 355/402; and hiPSC4, 288/269. Abbreviations: d, day; hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; ICC, immunocytochemistry; Neg, negative; QPCR, quantitative polymerase chain reaction.
All the examined cell lines produced pigmented cells on average within 22 days after initiation of differentiation (Fig. 5B). hESC lines produced pigmented cells on average 2 days earlier than hiPSC lines. The first pigmented cells were detected on day 13 (hESC3). On day 28, the highest proportion of pigmented cell clusters in hESC lines was detected in hESC3 (31%) and the lowest in hESC1 (11%) (Fig. 5C). Of the hiPSC lines, the best performer was hiPSC2 (43%) and the weakest hiPSC1 (6%) (Fig. 5C). In a groupwise comparison, none of the differences between the hESC and hiPSC lines were statistically significant.
During differentiation, the expression of endogenous OCT4 decreased in all cell lines. However, it was higher in the hiPSC than in the hESC lines on day 0 (p = .03) and day 52 (p < .001) (Fig. 5D). Expression of the selected differentiation markers MITF, BEST1, and RLBP1 increased in all cell lines during differentiation. In a comparison of hESC and hiPSC lines, the pigment cell marker MITF was higher in hESC on day 52 (p = .011) (Fig. 5D). The more RPE-specific markers BEST1 and RLBP1 appeared higher on d82 in the hiPSC lines, but the differences were not statistically significant (Fig. 5D). The Sendai-virally derived hiPSC5 line was characterized only partially, but it also differentiated into pigmented epithelium with cobblestone morphology and bestrophin-1 immunoreactivity (Fig. 5F). At the protein level, all cell lines expressed MITF and bestrophin-1 proteins. The highest proportion of MITF-positive cells in hESC lines was detected in hESC2 (72%) and the lowest in hESC1 (69%). The results were less reproducible in the hiPSC lines (Fig. 5G). Bestrophin-1-positive cells tended to be more abundant in the hiPSC than the hESC lines (Fig. 5G), and the results correlated with BEST1 gene expression and also with the rate of pigmentation.
When analyzed comprehensively, it appears that the hESC lines (particularly hESC1 and hESC3) displayed variable propensities for mesodermal versus ectodermal differentiation. The same cell lines differentiated consistently more efficiently in the ectodermal (neuronal and RPE) directions or mesodermal (cardiac) direction. However, none of the induced pluripotent stem cell (iPSC) lines showed such preferential differentiation capacity (Table 1).
Table 1.
Differentiation potential of individual human pluripotent stem cell lines
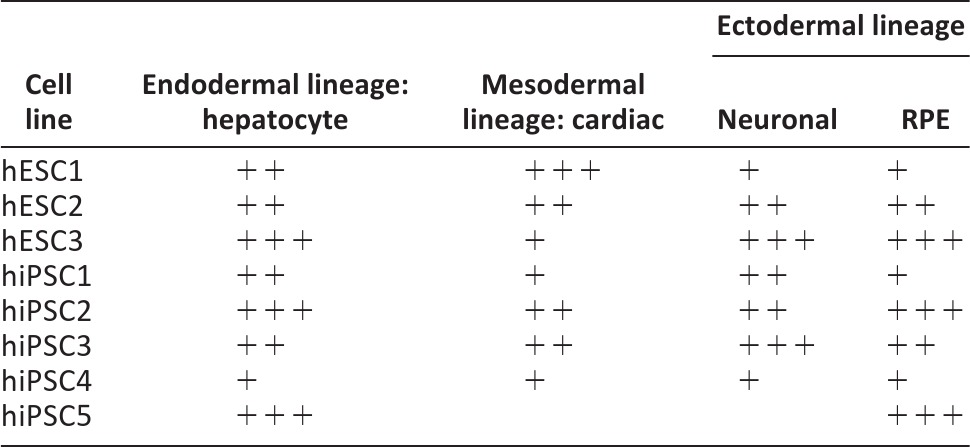
Shown is the differentiation efficiency of the cell lines based on the rate of albumin secretion (hepatocyte), number of beating cardiomyocytes (cardiac), morphological and immunocytochemical criteria (neural), and rate of pigmentation (retinal pigment epithelial). + indicates lower differentiation potential than average, ++ indicates average differentiation capacity, and +++ indicates excellent differentiation capacity.
Abbreviations: hESC, human embryonic stem cell; hiPSC, human induced pluripotent stem cell; RPE, retinal pigment epithelial.
Discussion
We studied the differentiation capacity of three hESC and five hiPSC lines. The four retrovirally derived hiPSC lines were characterized in detail using four well-established differentiation protocols and specific functional assays. Through this approach, we hope to elucidate the true variability between human pluripotent stem cell lines with respect to their most important characteristic: the ability to develop into physiologically functional cell types.
To our knowledge, our study is the first to use four separate extended differentiation protocols into derivatives of all germ layers in a systematic comparison of hPSC lines. In this study most of the cell lines showed no differentiation preference toward any specific cell lineages but rather showed more or less differentiation potential toward all different cell types produced. Only two of the cell lines (hESC1 and hESC3) had consistently more differentiation potential toward specific lineages. hESC1 differentiated well into beating cardiomyocytes and poorly into ectodermal lineages, and hESC3 had the best ectodermal differentiation capacity but produced few beating areas in cardiomyocyte differentiation. One reason for these differences may be the fact that the cardiac (END2 coculture) and neuronal (EB formation) protocols in this study are based more on the spontaneous differentiation of the cells than the hepatocyte protocol, which is based on guidance by specific growth factors. It is likely that the genetic background of the cells plays a more crucial role in the former than the latter situation.
Analysis of transgene expression showed that KLF4 was incompletely silenced in hiPSC4 (Fig. 1A; supplemental online Fig. 2A), suggesting that this cell line was only partially reprogrammed. The retroviral transgenes are usually silenced as a late event of the reprogramming process [28] because of the activation of DNA [29] and histone methyltransferases [12]. This process, however, is often incomplete, resulting in partially reprogrammed cell lines [9, 30, 31]. This residual activity of viral transgenes in hiPSC-derived cells can affect their developmental potential [9]. Partially incomplete reprogramming may explain the poor differentiation capacity of hiPSC4, which was observed throughout this study.
All of the hESC and hiPSC lines differentiated efficiently into early DE progenitors. However, when the DE cells were further induced into HLCs some variability became evident. Although all the hESC lines differentiated with approximately equal efficiency, the iPSC lines were much more variable, ranging from very poor (hiPSC4) to excellent (hiPSC2). This variation was not correlated with the method used for hiPSC induction, and it is unlikely that it would be due to the different donor age (neonatal vs. adult). It has also been noted by others that there are differences in the timing of onset of expression of hepatocyte-specific genes between different cell lines [7].
The cardiomyocyte differentiation protocol used in this study produced beating areas from all the cell lines with variable efficiency. Overall, cardiac differentiation on END-2 cocultures is rather unspecific, and many other cell types besides cardiomyocytes are also induced [32]. Normally, the highest peak of Brachyury T expression is observed on day 3 in END-2 cocultures [32], and the delayed expression peak leads to poor cardiac differentiation efficiency [33, 34]. In this study, the expression peak of Brachyury T was extended up to day 6 with hiPSCs, which was associated with a lower lowest number of beating areas than in hESCs. However, high variation in cardiac differentiation efficiency was also detected between different passages/differentiation experiments within the same cell line, indicating that the cell line characteristics change over time in culture.
The neural differentiation protocol used here has been used routinely with several hESC lines previously [35, 36]. Both hESC and hiPSC lines were successfully differentiated toward neural cells regardless of their origin. Previous studies have, however, demonstrated the differences between the innate differentiation propensities within hESC and hiPSC lines [11, 16–18, 35] and between hESC and hiPSC lines [6]. In contrast, two other studies have suggested that in general, hESCs and hiPSCs have similar differentiation capacity toward neural cells, but line-specific variation can be detected in both groups [16, 17]. Our results were more compatible with the latter view.
Electrophysiological properties are an essential aspect of the characterization of neuronal cells. In the present study, all the cell lines differentiated into neuronal networks that were able to display some form of spontaneous electrical activity regarded as a feature of functional neuronal networks [27, 37]. However, obvious maturation stage-related functional variability was observed. None of the gene or protein level markers of neuronal differentiation directly correlated with the functional properties of the derived neuronal networks. Thus, it is difficult to predict the efficiency of a particular cell line to produce functional neuronal networks without electrophysiological analyses.
Lastly, we differentiated the hPSCs into another ectodermal cell type, RPE cells. During mammalian development, RPE is derived from optic neuroepithelium by approximately the seventh week of gestation [38], and RPE cell fate specification in vitro has been shown to follow a time course reminiscent of normal retinal development [39]. All the cell lines examined produced pigmented cells within 3 weeks after initiation of differentiation. On average, hiPSC lines produced pigmented cells slightly more slowly than hESC lines. This is compatible with the findings by Meyer et al., who also reported longer differentiation times with hiPSCs than hESCs [39]. hESC3 produced pigmented cells the fastest. This cell line also produced eventually mature RPE cells. Two hiPSC lines also produced mature RPE cells, suggesting that the time of pigment appearance is not a crucial factor for the later maturation of RPE cells.
Consistent reactivation of the OCT4 transgene was observed in all retrovirally induced hiPSC lines during RPE differentiation (Fig. 1B; supplemental online Fig. 2B). The reactivation was most dramatic in hiPSC1. In addition, the NANOG and LIN28 transgenes were also reactivated in hiPSC1 during the RPE differentiation. On the contrary, transgene reactivation was not observed with the Sendai virus-induced iPSC5 line (supplemental online Fig. 3). Interestingly, hiPSC1 was differentiated successfully into both HLCs and cardiomyocytes, and transgene reactivation was not seen during those experiments. During RPE differentiation, hiPSC1 appeared to produce a high number of MITF and bestrophin-1-positive cells. However, hiPSC1-derived RPE cells peeled off from the culture membranes easily, allowing only one successful experiment to be completed. The RPE differentiation protocol is much (almost 3 months) longer and more spontaneous than the other protocols, which could be one explanation for the difference. Obviously, these observations raise concerns about the safety of hiPSCs that have integrated transgenes in their genome.
Conclusion
Part of the variation in the differentiation efficiency between the individual hiPSCs could be explained by residual activity of viral transgene KLF4 in hiPSC4 and the reactivation of several transgenes during RPE differentiation. In contrast, the hiPSC line that was derived through the nonintegrating Sendai virus technology differentiated well into both HLCs and RPE cells and did not show signs of transgene expression. Our study strongly suggests that many of the “first-generation” retrovirally derived iPSC lines are hampered by potential transgene reactivation, with specific effects on their further differentiation properties. These findings highlight the need for integration-free reprogramming technologies, resulting in transgene-free iPSCs, which could also be potentially therapeutically applicable, unlike the retrovirally derived cells used in this study. Several such technologies have been established, in addition to Sendai viruses [40]: polycistronic minicircle vectors [41], PiggyBac transposons [42], and modified mRNA-based [43] or protein transduction-based methods [44]. Future studies should focus on nontransgenic iPSC lines generated through these methods.
Acknowledgments
We are grateful to the following individuals for technical assistance: Markus Haponen, Maria af Hällström, Merja Lehtinen, Henna Venäläinen, Elina Konsén, Hanna Koskenaho, Outi Melin, Jarkko Ustinov, and Eila Korhonen. We thank Juha Heikkilä and Meeri Mäkinen for assistance with MEA measurements. This study was supported by funding from Biocenter Finland for the national platform on stem cells and biomaterials. Additional grant support was received from the Academy of Finland (to H.S., S.N., K.A.-S., T.O.), the Competitive Research Funding of the Pirkanmaa Hospital District (to H.S., S.N., K.A.-S.), the Alfred Kordelin Foundation (to K.A.-S.), the Finnish Foundation of Cardiovascular Research (to K.A.-S.), the Finnish Funding Agency for Technology and Innovation (to K.A.-S.), the Päivikki and Sakari Sohlberg Foundation (to H.S.), the Sigrid Jusélius Foundation (to T.O.), and the Competitive Research Funding of the Uusimaa Hospital District (to T.O.).
Author Contributions
S.T., M.O., A.H., T.I., K.R., and R.Ä.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; M.P.-M.: conception and design, data analysis and interpretation; K.L.: conception and design, collection and assembly of data; J.P.: collection and assembly of data, data analysis and interpretation; J.W. and R.T.: provision of study material, collection and assembly of data; O.S.: conception and design, financial support; H.S.: conception and design, financial support, data analysis and interpretation, final approval of manuscript; S.N.: conception and design, financial support, provision of study material, final approval of manuscript; K.A.-S. and T.O.: conception and design, financial support, provision of study material, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Cao N, Liu Z, Chen Z, et al. Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res. 2012;22:219–236. doi: 10.1038/cr.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu BY, Weick JP, Yu J, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Si-Tayeb K, Noto FK, Nagaoka M, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaajasaari H, Ilmarinen T, Juuti-Uusitalo K, et al. Toward the defined and xeno-free differentiation of functional human pluripotent stem cell-derived retinal pigment epithelial cells. Mol Vis. 2011;17:558–575. [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Chin MH, Mason MJ, Xie W, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bock C, Kiskinis E, Verstappen G, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsui T, Leung D, Miyashita H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 13.Lister R, Pelizzola M, Kida YS, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Q, Friedrich AM, Johnson LV, et al. Memory in induced pluripotent stem cells: Reprogrammed human retinal-pigmented epithelial cells show tendency for spontaneous redifferentiation. Stem Cells. 2010;28:1981–1991. doi: 10.1002/stem.531. [DOI] [PubMed] [Google Scholar]
- 15.Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim K, Zhao R, Doi A, et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulting GL, Kiskinis E, Croft GF, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osafune K, Caron L, Borowiak M, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Wilson GF, Soerens AG, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikkola M, Olsson C, Palgi J, et al. Distinct differentiation characteristics of individual human embryonic stem cell lines. BMC Dev Biol. 2006;6:40. doi: 10.1186/1471-213X-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skottman H. Derivation and characterization of three new human embryonic stem cell lines in Finland. In Vitro Cell Dev Biol Anim. 2010;46:206–209. doi: 10.1007/s11626-010-9286-2. [DOI] [PubMed] [Google Scholar]
- 22.Hussein SM, Batada NN, Vuoristo S, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 23.Lahti AL, Kujala VJ, Chapman H, et al. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Dis Model Mech. 2012;5:220–230. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hay DC, Fletcher J, Payne C, et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires ActivinA and Wnt3a signaling. Proc Natl Acad Sci USA. 2008;105:12301–12306. doi: 10.1073/pnas.0806522105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Passier R, Oostwaard DW, Snapper J, et al. Increased cardiomyocyte differentiation from human embryonic stem cells in serum-free cultures. Stem Cells. 2005;23:772–780. doi: 10.1634/stemcells.2004-0184. [DOI] [PubMed] [Google Scholar]
- 26.Huttunen TT, Sundberg M, Pihlajamaki H, et al. An automated continuous monitoring system: A useful tool for monitoring neuronal differentiation of human embryonic stem cells. Stem Cell Studies. 2011;1:71–77. [Google Scholar]
- 27.Heikkilä TJ, Yla-Outinen L, Tanskanen JM, et al. Human embryonic stem cell-derived neuronal cells form spontaneously active neuronal networks in vitro. Exp Neurol. 2009;218:109–116. doi: 10.1016/j.expneurol.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Stadtfeld M, Maherali N, Breault DT, et al. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei H, Oh SP, Okano M, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 30.Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sridharan R, Tchieu J, Mason MJ, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beqqali A, Kloots J, Ward-van Oostwaard D, et al. Genome-wide transcriptional profiling of human embryonic stem cells differentiating to cardiomyocytes. Stem Cells. 2006;24:1956–1967. doi: 10.1634/stemcells.2006-0054. [DOI] [PubMed] [Google Scholar]
- 33.Bettiol E, Sartiani L, Chicha L, et al. Fetal bovine serum enables cardiac differentiation of human embryonic stem cells. Differentiation. 2007;75:669–681. doi: 10.1111/j.1432-0436.2007.00174.x. [DOI] [PubMed] [Google Scholar]
- 34.Pekkanen-Mattila M, Ojala M, Kerkela E, et al. The effect of human and mouse fibroblast feeder cells on cardiac differentiation of human pluripotent stem cells. Stem Cells Int. 2012;2012:875059. doi: 10.1155/2012/875059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lappalainen RS, Salomaki M, Yla-Outinen L, et al. Similarly derived and cultured hESC lines show variation in their developmental potential towards neuronal cells in long-term culture. Regen Med. 2010;5:749–762. doi: 10.2217/rme.10.58. [DOI] [PubMed] [Google Scholar]
- 36.Sundberg M, Jansson L, Ketolainen J, et al. CD marker expression profiles of human embryonic stem cells and their neural derivatives, determined using flow-cytometric analysis, reveal a novel CD marker for exclusion of pluripotent stem cells. Stem Cell Res. 2009;2:113–124. doi: 10.1016/j.scr.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Pine J. Recording action potentials from cultured neurons with extracellular microcircuit electrodes. J Neurosci Methods. 1980;2:19–31. doi: 10.1016/0165-0270(80)90042-4. [DOI] [PubMed] [Google Scholar]
- 38.Fuhrmann S. Eye morphogenesis and patterning of the optic vesicle. Curr Top Dev Biol. 2010;93:61–84. doi: 10.1016/B978-0-12-385044-7.00003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer JS, Shearer RL, Capowski EE, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci USA. 2009;106:16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fusaki N, Ban H, Nishiyama A, et al. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia F, Wilson KD, Sun N, et al. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woltjen K, Michael IP, Mohseni P, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D, Kim CH, Moon JI, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]