Cell sorting techniques for specific cell surface markers (CD24+CD29HCD49fHSca1−) have been used to prospectively isolate mammary stem cell (MaSC)-enriched populations, but these markers do not identify regenerative stem cells uniquely. This study found that MaSCs can be further enriched by size fractionation. These findings have critical implications for understanding mammary gland stem cell biology, an important requisite step for understanding the etiology of breast cancer.
Keywords: Adult stem cells, Cell surface markers, FACS, Transplantation
Abstract
Mammary gland reconstitution experiments, as well as lineage tracing experiments, have provided evidence for the existence of adult mammary stem cells (MaSCs). In addition, cell sorting techniques for specific cell surface markers (CD24+CD29HCD49fHSca1−) have been used to prospectively isolate MaSC-enriched populations. Although these markers enrich for cell subpopulations that harbor MaSCs, they do not identify regenerative stem cells uniquely. Here, we report that MaSCs can be further defined by the property of cell size. Fluorescence-activated cell sorting was used to analyze sizing beads and further separate populations of cells with varying degrees of forward scatter (FSC). Cells with a low FSC that were approximately <10 μm in size lacked outgrowth potential and failed to reconstitute the mammary gland when transplanted into the cleared fat pads of syngeneic mice. In contrast, cells >10 μm in size with a higher FSC had increased outgrowth potential as compared with lineage-negative (LIN−) control cells. Limiting dilution transplantation assays indicated that the repopulating ability of LIN−CD24+CD29H cells that were >10 μm in size was significantly increased as compared with cells marked by CD24 and CD29 alone. These results suggest that MaSCs can be further isolated by sorting based on size/FSC. These findings have critical implications for understanding mammary gland stem cell biology, an important requisite step for understanding the etiology of breast cancer.
Introduction
Mammary gland tissue homeostasis is maintained by adult, long-lived mammary epithelial cells, which include regenerative mammary stem cells (MaSCs) and lineage-restricted progenitor cells. Over the past decade, considerable progress has been made in the identification of MaSCs using fluorescence-activated cell sorting (FACS) techniques. Mammary gland reconstitution assays demonstrated that LIN−CD24+CD29HCD49fHSca1− cells (LIN− indicates lineage-negative) are enriched for MaSC repopulation activity [1–4]. However, only ∼6% of the LIN−CD24+CD29H subpopulation (six outgrowths from 102 single cells transplanted) showed regenerative capacity by single-cell transplantation [1], which is approximately half of the ∼1% stem cell frequency of unsorted epithelial cells [5]. Thus, surface markers enrich for stem cells but do not allow purification of a homogeneous population. Efforts are currently aimed at elucidating the genetic regulation of these cells; however, very little is understood about the biological properties of MaSCs. One consequence of these limitations is the inability to determine whether normal MaSCs, lineage-restricted progenitors, or differentiated cells are the targets for neoplastic transformation.
In addition to FACS analysis, complementary studies using electron microscopy [6, 7] identified two unique, relatively undifferentiated cell types in the mammary gland. Small light cells (SLCs), which represent ∼3% of all epithelial cells, were ∼8 μm in size, appeared pale under the electron microscope, and were characterized by a paucity of membrane-bound organelles and pale cytoplasmic staining. SLCs were postulated to give rise to larger (∼15–20 μm) “undifferentiated large light cells” (ULLCs). It was proposed that SLCs and ULLCs might represent two sets of regenerative stem cells in two distinct physiological states. However, the developmental potential and functional role of these electron microscopy-defined cell types has not been demonstrated.
In the present study, we sought to improve on already established methods of MaSC isolation. We report that MaSCs can be further defined by properties such as cell size and forward scatter (FSC). Whereas small epithelial cells with a low FSC had limited outgrowth potential, cells >10 μm in size with a higher FSC had the ability to reconstitute the mammary gland upon transplantation. These results provide new insight about the biological properties of MaSCs and suggest that cell size fractionation may be a novel tool for the isolation of MaSCs.
Materials and Methods
Primary Mammary Epithelial Cell Isolation
Mice were maintained in accordance with the NIH Guide for the Care and Use of Experimental Animals with approval from the Baylor College of Medicine Institutional Animal Care and Use Committee. Mammary epithelial cells (MECs) were derived from freshly dissected thoracic and inguinal (without the lymph node) mammary glands of 8–12-week old female mice (FVB.Cg-Tg(CAG-EGFP)B5Nagy/J; Jackson Laboratory, Bar Harbor, ME, http://www.jax.org). Glands were minced into fragments (<1 mm) using a razor blade and digested in Dulbecco's modified Eagle's medium (DMEM)/F12 medium containing 1 mg/ml collagenase A (Roche Applied Science, Indianapolis, IN, https://www.roche-applied-science.com), 100 U/ml hyaluronidase (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com), and 1× antibiotic-antimycotic (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) for 14 hours at 37°C with shaking at 75 rpm. Cells were washed three times with 1× phosphate-buffered saline (PBS) containing 5% fetal bovine serum (FBS) and incubated with 0.25% trypsin-EDTA at room temperature for 2–3 minutes with rocking. Trypsin was inactivated with 1× PBS containing 5% FBS; cells were centrifuged and filtered through a 40-μm cell strainer. Single cells were counted on a hemacytometer using trypan blue.
Fluorescence-Activated Cell Sorting
Freshly isolated MECs were resuspended at a concentration of 1 × 107 cells per ml in Hanks' balanced saline solution (HBSS) containing 2% FBS and 100 mM Hepes (HBSS+), and stained with specific antibodies as previously described [8]. A complete list of antibodies is provided in supplemental online Table 1. Cells were filtered through a 40-μm cell strainer, incubated with a dead cell exclusion dye (Sytox red/blue; Invitrogen), and sorted on a FACSAria II Cell Sorting Flow Cytometer (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com). Prior to sorting, sizing beads (SPERO Particle Size Standard Kit; Spherotech, Inc., Lake Forest, IL, http://www.spherotech.com) were analyzed to determine estimated sizes of MECs. For transplantation assays, cells were sorted into DMEM/F12 medium containing 5% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone, 10 ng/ml epidermal growth factor (EGF), and 1× antibiotic-antimycotic. A postsort analysis was performed to assess the purity of the sorted cell populations and was estimated (from four independent experiments) to be 97.9 ± 0.5% for LIN− cells, 81.6 ± 2.6% for cells >10 μm, 91.2 ± 0.9% for LIN−CD24+CD29H cells, and 92.4 ± 1.5% for LIN−CD24+CD29H cells >10 μm. Data were analyzed using FlowJo 2 v9.5.2 (Tree Star, Ashland, OR, http://www.treestar.com).
Mammosphere Assays
MECs were FACS-sorted based on size into DMEM/F12 media containing 20 ng/ml EGF, 20 ng/ml basic fibroblast growth factor, B27, and 1× antibiotic-antimycotic (MS media). Cells were washed, resuspended in MS media at a concentration of 15,000 cells per ml, and plated in ultralow-attachment plates (2 ml per well). Cells were fed every 4 days for 12 days, at which time they were dissociated as previously described [9]. Secondary mammospheres were cultured for an additional 14 days as previously described [8]. Twelve wells for each group were counted and the percentage of mammosphere forming cells was calculated as a measure of mammosphere efficiency.
Transplantation Assays and Whole Mount Analysis
FACS-sorted cell subpopulations were washed with 1× PBS and resuspended in a 1:1 solution of PBS and Matrigel (BD Biosciences). Cells were serially diluted for limited dilution transplantation assays. Inguinal glands of recipient female mice (FvB/NJ; Jackson Laboratory) were cleared at 3 weeks of age, and cells were transplanted 2–3 weeks later. Ten microliters of cells was injected into contralateral cleared fat pads using a 26-gauge needle and 50-μl Hamilton glass syringe [10]. Animals were sacrificed 8 weeks after transplantation, and contralateral mammary glands were removed, compressed between two glass slides, and visualized using a Leica MZ16F fluorescent stereoscope (Leica Microsystems, Buffalo Grove, IL, http://www.leica-microsystems.com). Mammary glands showing at least 5% fat pad filled were included in the analysis. For glands that displayed no outgrowth, the lack of epithelium was verified by neutral red staining, and these were included in the calculation of take rate. For neutral red staining, contralateral mammary glands were fixed in ice-cold 4% paraformaldehyde for 1 hour and stained as previously described [5].
Statistical Analysis
Data from mammosphere assays are presented as the means ± SEM. Significance values were calculated with a one-way analysis of variance model followed by Bonferroni pairwise comparisons between groups. Limiting dilution transplantation results were analyzed by Extreme Limiting Dilution Analysis software [11].
Results and Discussion
Mammary Repopulating Ability Can Be Defined by Cell Size
Previous studies suggested that SLCs may represent a population of undifferentiated epithelial cells with stem cell properties [6, 7]. To test whether small cells have functional stem cell properties, size parameters were established using a flow cytometer. Sizing beads were first analyzed for FSC and side scatter (SSC), and on the basis of the location of these beads, gates were drawn for different sizes (supplemental online Fig. 1). Doublets and larger aggregates were excluded by a series of gates using FSC and SSC, and lineage-positive cells were subsequently excluded. Once these parameters were established, mammary epithelial cells could be FACS-sorted on the basis of size (supplemental online Fig. 1; Fig. 1A). It should be noted that the gates set for specific sizes are approximations based on FSC relative to sizing beads.
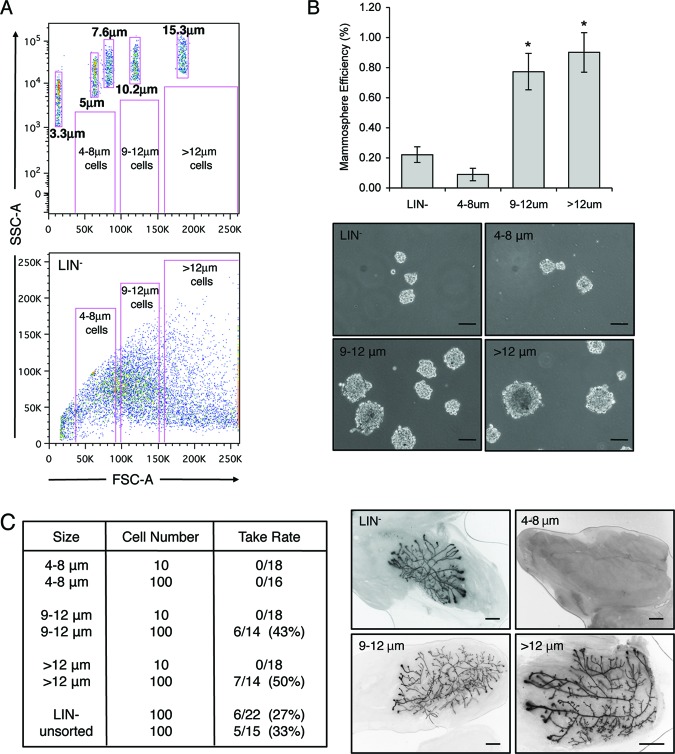
Figure 1.
Cells >8 μm in size are enriched for mammary stem cell activity. (A): Top: Exponential dot plot depicting sizing beads and theoretical gates for different sized cell populations. Bottom: Linear dot plot depicting LIN− cells, which were sorted by fluorescence-activated cell sorting on the basis of size. (B): Graph illustrates the number of secondary mammospheres formed per 5,000 cells, expressed as mammosphere efficiency of different cell sizes. The abilities of cells 9–12 μm in size and cells >12 μm in size were significantly increased as compared with LIN− cells and cells 4–8 μm in size (*, p < .0001). Data represent mean ± SEM. Representative images of mammospheres from each group are shown. Scale bars = 100 μm. (C): Table indicates repopulation activity as a function of cell size. Whereas cells 4–8 μm in size lacked outgrowth potential, cells larger than 8 μm had increased outgrowth potential as compared with LIN− and unsorted controls. Images depict representative outgrowths from 100 transplanted cells of each group. Scale bars = 5 mm. Abbreviations: FSC, forward scatter; LIN−, lineage-negative; SSC, side scatter.
To initiate these studies, in vitro assays were performed to determine whether small cells had the ability to form mammospheres. Mammary epithelial cells cultured in serum-free media under nonadherent conditions were previously shown to form spheres that are enriched for stem/progenitor cells [9, 12]. Surprisingly, only 0.09% of small cells (4–8 μm) had the ability to form secondary mammospheres, which was substantially lower than that of LIN− control cells (0.22%). In contrast, mammosphere formation was significantly increased in larger cells (0.77% for 9–12 μm, and 0.9% for >12 μm) with a higher FSC as compared with LIN− control cells (Fig. 1B). Notably, the overall diameter of mammospheres from the larger cell populations was greater than the diameter of those formed by LIN− or small cells.
Although mammospheres have been shown to be enriched for stem/progenitor cells, previous studies demonstrated that as little as 15%–30% of mammosphere cells possess the ability to repopulate the mammary gland in vivo [5]. Therefore, in vivo transplantation assays were performed to test whether larger cells were enriched for stem cell activity. Various sizes of cells were FACS-sorted and transplanted into the cleared fat pads of syngeneic mice. In support of the mammosphere assays, cells 4–8 μm in size with a low FSC failed to recapitulate the mammary gland upon transplantation, suggesting that this population lacked regenerative stem cells (Fig. 1C). On the other hand, cells 9–12 μm in size (six outgrowths per 14 transplants) or cells >12 μm (seven outgrowths per 14 transplants) were able to regenerate the mammary gland. Outgrowth abilities of these populations were dramatically increased as compared with LIN− (six per 22) or unsorted (five per 15) control cells. Notably, 10 transplanted cells of any size failed to reconstitute the mammary gland. Staining of outgrowths with the luminal cell marker keratin 8 (K8) and the myoepithelial marker K14 demonstrated that all ductal cell lineages were present. Furthermore, normal lobuloalveolar differentiation occurred in recipient mice subjected to pregnancy, as evidenced by lipid droplet formation (supplemental online Fig. 2). These results suggest that whereas small cells lacked in vivo outgrowth potential, cells >8 μm in size were enriched for regenerative MaSC activity and could recapitulate all cell lineages in the mammary gland.
To refine the cell size parameters for regenerative capacity more precisely, a series of experiments was conducted in which the size gates used for fractionation were modified incrementally. Various cell numbers were injected into the cleared fat pads of syngeneic mice, and mammary glands were harvested 8 weeks post-transplantation. The results from these experiments are summarized in supplemental online Table 2. Overall, cells that were <10 μm in size had limited outgrowth potential (1 outgrowth per 26 transplants), whereas cells >10 μm in size with a higher FSC possessed significant mammary repopulation ability (19 per 38). Although the sizes of the sorted populations are only estimations relative to the sizing beads, the approximate sizes of the cells were verified by electron microscopy (supplemental online Fig. 3). On the basis of these analyses, size gates of <10 μm and >10 μm were used for the remainder of the study.
LIN−CD24+CD29H MaSCs Can Be Enriched by Size Fractionation
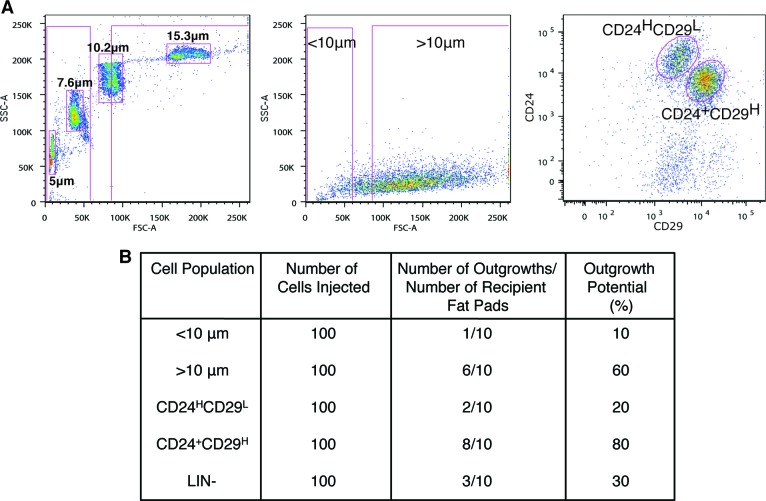
The MaSC population has been defined as a basal subpopulation of cells that are LIN−CD24+CD29H [1, 13]. Therefore, we next compared the outgrowth potential of cells expressing different levels of CD24 and CD29 to those separated by size. As expected, smaller cells (<10 μm) with a low FSC, as well as LIN−CD24HCD29L luminal cells, had limited outgrowth potential upon transplantation (Fig. 2). In contrast, cells with a high FSC that were >10 μm in size demonstrated a twofold increase in outgrowth potential (60%) as compared with LIN− cells (30%). Despite this increase, mammary repopulating activity was even further enriched in basal LIN−CD24+CD29H cells (80%) (Fig. 2B).
Figure 2.
Cells >10 μm in size are enriched for outgrowth potential. (A): Dot plot depicting sizing beads and gating strategy for cells <10 μm and >10 μm in size (left). LIN− cells <10 μm and >10 μm in size (middle), and LIN−CD24HCD29L and LIN−CD24+CD29H cells (right) were sorted by fluorescence-activated cell sorting and transplanted into cleared fat pads of FvB mice. (B): Transplantation results demonstrated that when 100 cells were transplanted, cells <10 μm in size, LIN−CD24HCD29L cells, and LIN− cells had limited repopulation ability. Repopulation of cells >10 μm in size was similar to that of mammary stem cells defined by LIN−CD24+CD29H expression. Outgrowth potential is the number of outgrowths/number of recipient fat pads expressed as a percentage. Abbreviations: FSC, forward scatter; LIN−, lineage-negative; SSC, side scatter.
To determine whether basal MaSCs could be further enriched by sorting for cell size, limiting dilution transplantation assays were performed. At clonal cell numbers, the outgrowth potential of LIN−CD24+CD29H cells was significantly increased (p = .036) as compared with cells >10 μm in size (Table 1). The mammary repopulating unit (MRU) of LIN−CD24+CD29H cells was determined to be 1 stem cell per 132 cells. Interestingly, the frequency of MaSCs was significantly increased twofold in LIN−CD24+CD29H cells that were >10 μm in size, with a calculated MRU of 1 in 66 (p = .014). These results strongly suggest that regenerative MaSCs can be further enriched by sorting for cell size without the use of additional cell surface markers.
Table 1.
LIN−CD24+CD29H cells that are >10 μm in size are enriched for mammary repopulating activity
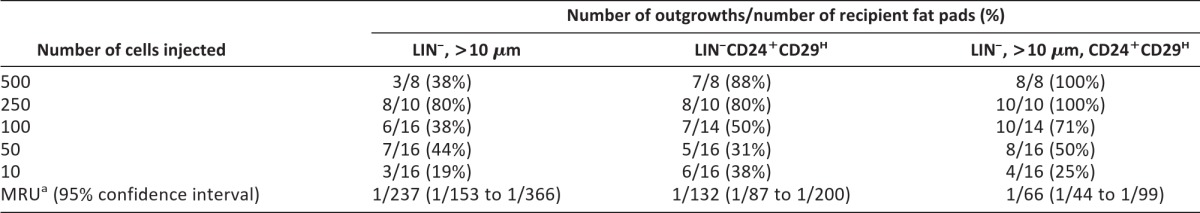
aThe frequency of stem cells is the MRU.
Abbreviations: LIN−, lineage negative; MRU, mammary repopulating unit.
Conclusion
The isolation and characterization of MaSCs is a fundamental requisite step to understanding the pathogenesis of human breast cancer. Here, we report that MaSCs can be further enriched by size fractionation. Whether the large cells identified in this study represent the previously described ULLCs remains unknown. Nevertheless, we propose that FACS based on size and cell surface markers will be an additional tool for future studies elucidating the genetic regulation of the stem cell hierarchy.
See www.StemCellsTM.com for supporting information available online.
Acknowledgments
We thank Joel Sederstrom of the Baylor College of Medicine Cytometry and Cell Sorting Core for technical assistance (NCRR S10RR024574 and NCI P30CA125123) and the Integrated Microscopy Core at Baylor College of Medicine (U54 HD-07495-39, P30 DX56338-05A2, P39 CA125123-04, S10RR027783-01A1). This work was supported by an NIH/NCI K99 (CA154605; H.L.M.), an NIH/NCI MERIT Award (CA030195-22; J.M.R.), a Breast Cancer SPORE P50 (CA58183; M.T.L), an NIH/NCI P30 (CA125123), and an NIH/NCI R01 (CA127857; M.T.L.).
Author Contributions
H.L.M.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; F.S.K.: collection and assembly of data, data interpretation, manuscript writing; D.E. and A.N.W.: collection and assembly of data, data analysis and interpretation; R.L.A.: data analysis and interpretation; J.M.R.: data analysis and interpretation, financial support; D.M. and M.T.L.: conception and design, financial support, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
M.T.L. is an uncompensated limited partner in StemMed Ltd.
References
- 1.Shackleton M, Vaillant F, Simpson KJ, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 2.Sleeman KE, Kendrick H, Robertson D, et al. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol. 2007;176:19–26. doi: 10.1083/jcb.200604065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smalley MJ, Kendrick H, Sheridan JM, et al. Isolation of mouse mammary epithelial subpopulations: A comparison of leading methods. J Mammary Gland Biol Neoplasia. 2012;17:91–97. doi: 10.1007/s10911-012-9257-1. [DOI] [PubMed] [Google Scholar]
- 4.Stingl J, Eirew P, Ricketson I, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 5.Moraes RC, Zhang X, Harrington N, et al. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134:1231–1242. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- 6.Chepko G, Smith GH. Three division-competent, structurally-distinct cell populations contribute to murine mammary epithelial renewal. Tissue Cell. 1997;29:239–253. doi: 10.1016/s0040-8166(97)80024-9. [DOI] [PubMed] [Google Scholar]
- 7.Smith GH, Chepko G. Mammary epithelial stem cells. Microsc Res Tech. 2001;52:190–203. doi: 10.1002/1097-0029(20010115)52:2<190::AID-JEMT1005>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 8.LaMarca HL, Visbal AP, Creighton CJ, et al. CCAAT/enhancer binding protein beta regulates stem cell activity and specifies luminal cell fate in the mammary gland. Stem Cells. 2010;28:535–544. doi: 10.1002/stem.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deome KB, Faulkin LJ, Jr., Bern HA, et al. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 1959;19:515–520. [PubMed] [Google Scholar]
- 11.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Dontu G, Wicha MS. Survival of mammary stem cells in suspension culture: Implications for stem cell biology and neoplasia. J Mammary Gland Biol Neoplasia. 2005;10:75–86. doi: 10.1007/s10911-005-2542-5. [DOI] [PubMed] [Google Scholar]
- 13.Asselin-Labat ML, Shackleton M, Stingl J, et al. Steroid hormone receptor status of mouse mammary stem cells. J Natl Cancer Inst. 2006;98:1011–1014. doi: 10.1093/jnci/djj267. [DOI] [PubMed] [Google Scholar]