Abstract
The mouse connexin 36 (Cx36) gene was mapped on chromosome 2 and an identical transcriptional start site was determined in brain and retina on exon I. Rabbit polyclonal antibodies to the presumptive cytoplasmic loop of the Cx36 protein recognized in immunohistochemical analyses Cx36 expression in the retina, olfactory bulb, hippocampus, inferior olive and cerebellum. In olivary neurons strong punctate labeling at dendritic cell contacts and weaker labeling in the cytoplasm of dendrites were shown by immuno electron microscopy. After expression of mouse Cx36 cDNA in human HeLa cells, neurobiotin transfer was increased 1.8-fold and electrical conductance at least 15-fold compared to untransfected HeLa cells. No Lucifer Yellow transfer was detected in either untransfected or Cx36 transfected HeLa cells. Single Cx36 channels in transfected HeLa cells showed a unitary conductance of 14.3 ± 0.8 pS. The sensitivity of Cx36 channels to transjunctional voltage was low in both HeLa-Cx36 cells and Xenopus oocytes expressing mouse Cx36. No increased transfer of neurobiotin was detected in heterotypic gap junctions formed by Cx36 and 9 other connexins expressed in HeLa cells. Our results suggest that Cx36 channels function as electrical synapses for transmission of electrical and metabolic signals between neurons in the central nervous system.
Keywords: Gap junctions, Electrical synapses, Neuronal connexin, Transcriptional start site, Cx36
Introduction
Neurons in various regions of the mammalian brain have previously been shown to be electrically coupled (reviewed in Spray and Dermietzel, 1996). The morphological structures responsible for electrical coupling between neurons are gap junctions, as demonstrated by electron microscopy and freeze fracture analyses (Schmalbruch & Jahnsen, 1981; Kosaka & Hama, 1985). Gap junctions are formed by the docking of two hemi-channels in apposed plasma membranes of adjacent cells. These channels allow passage of molecules up to 1000 Daltons, e.g., ions, metabolites, or second messengers. The protein subunits constituting gap junctional channels are termed connexins, which are encoded by a gene family of at least 15 members in rodents (Simon & Good-enough, 1998; Manthey et al., 1999). Each of these gap junction proteins is characterized by a cell type specific but overlapping expression pattern. Regulation takes place at the transcriptional and translational level, and the extent of cell-cell coupling can be dependent on the phosphorylation status of certain connexins (Bruzzone, White & Paul, 1996; Saez et al., 1998). In the nervous system, murine astrocytes express Cx43 (Dermietzel et al., 1989) and, after 3 weeks of age, Cx30 (Kunzelmann et al., 1999; Nagy et al., 1999). Oligodendrocytes express Cx32 (Dermietzel et al., 1989) and Cx45 (Kunzelmann et al., 1997; Dermietzel et al., 1997); and neurons have been reported to express Cx26, Cx32, or Cx43, depending on the type of neuron, state of differentiation and phase of cell cycle (Dermietzel et al., 1989; Bittmann & LoTurco, 1999).
Recently, the murine Cx36 gene has been described (Condorelli et al., 1998; Söhl et al., 1998). Cx36 mRNA was found in neurons of the retina and other restricted brain areas. In the present study, we further characterized the Cx36 gene by determining its genomic localization and transcriptional start site and analyzed the expression pattern of Cx36 protein in brain and retina. The latter was accomplished by analyses of immunofluorescence and immuno electron microscopy using affinity purified rabbit polyclonal antibodies directed to the presumptive cytoplasmic loop of Cx36. Furthermore, we investigated the functional properties of mouse Cx36 channels in Xenopus oocytes and in transfected human HeLa cells. Our results show that Cx36 channels are permeable to neurobiotin, have a low unitary conductance, display weak transjunctional voltage dependence and do not form heterotypic gap junction channels with other connexin channels expressed in transfected HeLa cells or microinjected Xenopus oocytes.
Materials and Methods
Genomic Mapping
The Cx36 gene was mapped by analysis of two sets of multiloci crosses: (NFS/N × M. musculus) × M. musculus (Kozak et al., 1990) and (NFS/N × M. spretus) × M. spretus or C58/J (Adamson, Silver & Kozak, 1991). Recombinational distances were calculated according to Green (1981) and gene loci were ordered by minimizing the number of recombinants.
Reverse Transcription-Polymerase Chain Reaction, “Primer Walk”
Total RNA from brain (at postnatal day 7) and retina (adult) of C57BL/6 mice was isolated with the TRIzol reagent (Life Technologies, Eggenstein, Germany). Two μg of RNA were incubated with 1 μg oligo(dT)15 primer (Promega, Madison, WI) for 10 min at 68°C in a total volume of 20 μl and briefly chilled on ice. Poly(A)+ RNA was reverse transcribed using 5 units of AMV reverse transcriptase (Promega). The RT buffer consisted of 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 6 mM MgCl2, 10 mM dithiothreitol, 40 units RNAsin (Promega) and 125 μM of each dNTP. Samples (50 μl total volume) were incubated for 60 min at 42°C and thereafter for 5 min at 95°C. Approximately 100 pg cDNA were amplified with the upstream primers P01-P7 specific for the mouse Cx36 5′ flanking region and the Cx36 exon II specific downstream primer DSP4: P01, position −554 to −534 with respect to the start codon ATG; P02, position −528 to −505; P03, position −480 to −460; P1, position −600 to −580; P2, position −445 to −424; P3, position −296 to −275; P4, position −219 to −195; P5, position −118 to −97; P6, position −58 to −36; P7, position +44 to +67; DSP4, position +1951 to +1928. Reaction mixtures (50 μl) contained 20 mM Tris-HCl (pH 8.4), 250 μM dNTPs, 1.5 mM MgCl2, 50 mM KCl, 1 μM of each primer and 5 units Ampli Taq DNA-Polymerase (Perkin Elmer, Foster City, CA). PCR was carried out for 40 cycles using a PTC-100 thermal cycler (MJ Research, Watertown, MA) with the following touch down program: denaturing at 94°C for 1 min, annealing at 65°C for 1 min and lowering by 0.5°C during each cycle until 60°C were reached, elongation at 72°C for 2 min. The PCR products were separated on a 1% agarose gel and analyzed by Southern blotting as described in Söhl et al. (1998).
Primer Extension Analysis
Poly(A)+ RNA was isolated from mouse brain RNA (see above) using an mRNA purification kit (Amersham Pharmacia Biotech, Freiburg, Germany). Cx36 specific primers Pex1 (position −458 to −478 with respect to the start codon ATG), and Pex2 (position −363 to −394) were phosphorylated with [γ-32P] ATP to a specific activity of 2 × 107 cpm/μg using an AMV reverse transcriptase primer extension system (Promega). One μg of brain derived mRNA and 10 μg of retina derived total RNA were annealed to both primers Pex1 and Pex2 with a specific activity of 2 × 105 cpm in PE buffer containing 50 mM Tris-HCl (pH 8.3), 50 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM dNTPs each and 0.5 mM spermidine at declining temperature from 78 to 58°C within 20 min and then left for 10 min at room temperature. The annealed primers were extended in 20 μl PE buffer containing additionally 2.8 mM sodium pyrophosphate, 5 units AMV reverse transcriptase (Promega) and 40 units Superscript II reverse transcriptase (Life Technologies) for 30 min at 42°C. The reactions were stopped by adding one volume loading dye containing 98% formamide, 10 mM EDTA, 0.1% xylene/cyanol and 0.1% bromphenol blue. Extended products were examined by electrophoresis in a denaturing sequencing gel.
Plasmids
Cx36 coding DNA was cloned in three steps from mouse and rat sequences into the mammalian expression vector pBEHpac18 (Horst, Harth & Hasilik, 1991). Rat Cx36 exon II was ligated as a blunted XhoI/sticky BamHI fragment (955 bp) into the pBEHpac18 vector digested with XbaI, blunted, and cut with BamHI. The rat BamHI/XbaI fragment (660 bp, corresponding to amino acids 25-245 of Cx36 [Söhl et al., 1998]) was then replaced by the respective mouse BamHI/XbaI fragment isolated from a phage containing mouse genomic Cx36 DNA (Söhl et al., 1998). Mouse Cx36 exon I was amplified by PCR from this phage clone using the primers USPCx36Kozak (5′-CGG AAT TCC GCC ATG GGG GAA TGG ACC ATC-3′) and DSP4 (see above) and the following PCR program: 94°C, 2 min; 40 cycles (95°C, 30 sec; 40°C, 30 sec; 72°C, 2 min) with 1.25 units Pfu polymerase (Stratagene, La Jolla, CA) on a PTC-100 thermal cycler (MJ Research). Of the 500 bp PCR product, only the 74 bp BamHI/EcoRI fragment was cloned in the expression vector containing Cx36 exon II cut with BamHI and EcoRI. The resulting plasmid pBEHpacCx36 confered resistence towards puromycin selection and Cx36 expression was controlled by the SV40 promoter.
Rabbit Polyclonal Antibodies to Cx36
A fifteen amino acid peptide (LNQTETTSKETEPDC) corresponding to part of the cytoplasmic loop of mouse Cx36 (amino acids 154–168) was synthetized, coupled to keyhole limpet hemocyanin and injected into rabbits at the Eurogentec company (Seraing, Belgium). Serum was affinity purified with the same peptide using a HiTrap affinity column (Amersham Pharmacia Biotech). After elution with 3 M KSCN in PBS and dialysis against PBS, the antibodies were ultrafiltrated by Centricon micro tubes 30 (Amicon, Beverly, MA) and finally stored in PBS with 0.5% BSA (Miles Diagnostics, Kankakee, IL) and 0.02% sodium azide.
Immunoprecipitation and Phosphorylation Analysis
Cells on a 35 mm dish were metabolically labeled prior to immunoprecipitation with [35S] methionine (50 μCi/ml, Amersham Pharmacia Biotech) in methionine-free medium for 4 hr. After two washes in PBS, cells were harvested in 250 μl ice-cold RIPA buffer (10 mM NaPO4, pH 7.2; 40 mM NaF, 2 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, 10 mM Na3VO4, 1× Complete™ [protease inhibitor cocktail, Roche, Mannheim, Germany]) and un-soluble material was centrifuged for 15 min at 10,000 × g and 4°C. The supernatant was precleared for 2 hr with 30 μl Sepharose CL-4B (Amersham Pharmacia Biotech) in PBS. Affinity purified rabbit antibodies (1.5 μl) to Cx36 were incubated with 10 μl protein A Sepharose (Amersham Pharmacia Biotech) on ice for 30 min. The precleared lysates were precipitated with the antibodies overnight, washed 3 times with RIPA wash (10 mM NaPO4, pH 7.2; 1 M NaCl, 40 mM NaF, 10 mM EDTA, 0.2% Triton X-100), and once with water. For dephosphorylation, the beads were incubated in 50 μl of 1 mM Tris, pH 9.7, 0.5 mM MgCl2, 0.4 μM ZnCl2, and 25 U alkaline phosphatase (Roche) at 37°C for 3 hr and centrifuged. The proteins were incubated in 15 μl sample buffer (80 mM Tris, pH 6.8, 10% glycerol, 5% SDS, 150 mM dithiothreitol, 0.005% bromophenol blue) at 90°C for 5 min and separated by SDS-PAGE on an 8% gel. After electrophoresis the gels were fixed in 10% acetic acid, enhanced in Amplify (Amersham Pharmacia Biotech), dried and exposed to Kodak BMR films.
Immunohistochemistry and Immunocytochemistry
Brains were prepared from adult decapitated C57BL/6 mice or adult rats, frozen on dry ice and cut to 25 μm sections on a cryostat. HeLa cells were seeded on coverslips, cultivated for three days and washed in PBS. Air dried sections or washed cells were fixed in ethanol or 4% paraformaldehyde in PBS and blocked with 5% low fat milk powder, 10% normal goat serum, 0.1% Tween 20 in PBS. Affinity purified Cx36 antibodies were added at a 1:1000 dilution and the incubation was carried out overnight at 4°C in a humidified chamber. After several washes, the secondary antibodies (goat anti-rabbit IgG conjugated to Alexa, Molecular Probes, Eugene, OR), diluted 1:1000 in blocking solution, were added, incubated for 2 hr at room temperature and washed extensively with PBS. The sections or cells were mounted using DAKO Fluorescent mounting medium (DAKO, Carpinteria, CA) and photographed with a Zeiss Axiophot microscope.
Electron Microscopy
Adult C57BL/6 mice were anesthetized with sodium pentobarbital (75 mg/kg) and perfused transcardially with 10 ml of Ringer's solution saturated with 95% CO2 and 5% O2 to pH 7.1, followed by 20 ml of 4% freshly depolymerized formaldehyde and 0.1% glutaraldehyde in 0.12 M sodium phospate buffer (pH 7.4, room temperature). The brains were removed 1 hr after perfusion, rinsed in ice-cold Tris-buffered saline (TBS, 290 mOsm, pH 7.5), and cut coronally as 40 μm sections on a vibratome. The sections were rinsed in TBS with 0.05% Triton X (TBST) and subsequently blocked in 5% normal donkey serum in TBST, incubated with the affinity purified Cx36 antibody diluted 1:100 in 2% normal donkey serum in TBST for 48 hr on a shaker at 4°C, rinsed in TBST, incubated for 1 hr in goat anti-rabbit IgG (Sternberger-Meyer, Mamhead, Exeter, Devon, UK) diluted 1:50 in TBST, rinsed in TBST, incubated for 1 hr in rabbit PAP (Sternberger-Meyer) diluted 1:100 in TBST, and rinsed as above. The sections were incubated for 15−20 min in 0.05% 3,3-diaminobenzidine (DAB) and 0.01% H2O2 in TBST. Finally, the sections were rinsed in 0.12 M phosphate buffer at pH 7.3 (PB), osmicated in 2% osmium tetroxide in PB, thoroughly rinsed in H2O, stained en bloc in 2% uranyl acetate in H2O overnight, dehydrated in alcohol and propylene oxide, and flat embedded in Araldite. Pyramids were made and cut to ultrathin sections on an LKB ultratome from the most superficial portions of the pyramids, collected on single-hole, Formvar-coated grids, counterstained with lead citrate, and examined with a Philips CM100 electron microscope operating at 80 kV.
Cell Culture,Transfection and Expression in Xenopus Oocytes
HeLa cells (ATCC CCL 2, human cervical carcinoma cells) were cultivated in DMEM low glucose, 2 mM glutamine, 10% FCS, 1 × Pen/Strep (all from Life Technologies) at 37°C with 10% CO2 and transfected with Tfx20 (Promega). Resistant clones were isolated after 2 weeks of selection with 1 μg/ml puromycin (Sigma, Deisenhofen, Germany), and tested for Cx36 expression by immunocytochemistry.
Cx36 cDNA from pBEHpacCx36 (see above) was cloned into the transcription vector pGEM-7zf (+) (Promega) in the T7 orientation. Cx36 RNA was made with the mMessage mMachine kit from Ambion (Austin, TX). Each oocyte was injected with 50 nl of Cx36 RNA (∼1 μg/μl) together with a phosphorothionate antisense oligonucleotide (∼0.5 pmol/nl) complemetary to XenCx38.
Dye Transfer Measurements
For dye transfer measurements, cells were grown on 35 mm dishes for 2-3 days. Glass micropipettes were pulled from capillary glass (World Precision Instruments, Berlin, Germany) with a horizontal pipette puller (Model P-97, Sutter Instruments, Novato, CA) and backfilled with dye solution (see below). Dyes were injected iontophoretically (Iontophoresis Programmer model 160; World Precision Instruments) and cell-to-cell transfer was monitored using an inverted microscope (IM35; Zeiss) equipped with fluorescent illumination. Cell culture dishes were kept on a heated block at 37°C. Lucifer Yellow CH (Molecular Probes) at 4% (w/v) in 1 M LiCl was injected by applying hyperpolarizing currents for 10 sec (I = 20 nA). Cell-to-cell transfer was evaluated 30 min after dye injection. Neurobiotin (N-2(2-aminoethyl)-biotinamide hydrochloride) (Vector Lab, Burlingame, CA) and rhodamine 3-isothiocyanate dextran 10S (Sigma) at concentrations of 6 and 0.4% (w/v), respectively, in 0.1 M Tris-Cl (pH 7.6) were injected by applying depolarizing currents for 10 sec (I = 20 nA). Thirty min after injection, cells were washed twice with phosphate-buffered saline (PBS), fixed for 10 min in 1% glutaraldehyde in PBS, washed twice with PBS, incubated in 2% Triton X-100 in PBS overnight at 4°C, washed three times with PBS, incubated with horseradish peroxidase-avidin D (Vector Lab) diluted 1:1000 in PBS for 90 min, washed three times with PBS, and incubated in 0.05% diaminobenzidine, 0.003% hydrogen peroxide solution for 30 sec. The staining reaction was stopped by washing three times with PBS. Cell-to-cell transfer was quantified by counting the number of stained neighboring cells around the microinjected cell. To identify heterotypic cell pairs, HeLa-Cx36 cells were stained with DiI as described in (Goldberg, Bechberger & Nauss, 1995) and co-cultured with a 1,000-fold excess of unstained HeLa cells expressing a different connexin gene. The cells were incubated for 18 hr before microinjection of tracers.
Electrophysiological Measurements
One to three days prior to electrophysiological recording, HeLa cells transfected with Cx36 were seeded on 22 × 22 mm No. 0 coverslips (Clay Adams). The coverslips were transferred to an experimental chamber mounted on the stage of an inverted microscope equipped with phase-contrast optics. The chamber was perfused with a modified Krebs-Ringer solution containing (in mm): NaCl, 140; KCl, 4; CaCl2, 2; MgCl2, 1; glucose, 5; pyruvate, 2; HEPES, 5 (pH 7.4). Patch pipettes were filled with a solution containing (in mm): KCl, 130; sodium aspartate, 10; MgCl2, 1; MgATP, 3; CaCl2, 0.26; EGTA, 2 ([Ca2+]i = 5 × 10−8 m); HEPES, 5 (pH 7.2). Junctional conductance was measured using the dual whole-cell patch-clamp method (Neyton & Traut-mann, 1985). After establishment of whole-cell patch clamp recordings in both cells of a pair, the cells were clamped to a common holding potential (V1 = V2). Transjunctional voltages, Vj, were applied by changing the membrane potential in one cell and keeping the other constant (Vj = V2 = V1). The resulting junctional current, Ij, is observed as a change in current in the unstepped cell. Junctional conductance, gj, is determined by gj = Ij/Vj. Voltages and currents were recorded on videotape using a data recorder, VR-100 (Instrutech, Port Washington, NY), and were subsequently digitized using a MIO-16X A/D converter (National Instruments, Austin, TX) and our own aquisition software (by EBT and VKV).
In addition, in some cell pairs we simultaneously measured gj and assessed dye transfer using Lucifer Yellow (negatively charged, −2) or DAPI (4′,6-diamino-2-phenylindole, dihydrochloride; positively charged, +2). In each experiment, dye was delivered to one cell of a pair by establishing a whole-cell recording using a pipette filled with 0.1% dye diluted in normal pipette solution. After allowing sufficient time for dye to transfer (5–10 min), a whole-cell voltage clamp recording was established in the recipient cell to measure gj. Fluorescence signals were monitored using a MERLIN imaging system equipped with a UltraPix FE250 cooled digital camera (12 bit), a xenon lamp source and a SpectraMASTER high-speed monochromator. Dye distribution was monitored by acquiring images every 5 sec (0.5 sec exposures) over a period of 0–15 min. Electronic shuttering of the digital camera allowed setting exposure times and timing intervals between fluorescence measurements. Recordings of Cx36 junctional currents between Xenopus oocyte cell pairs were obtained with a dual two-electrode voltage clamp using two GeneClamp 500 amplifiers (Axon Instruments). Approximately 24–36 hours after injection, oocytes were devittelinized and paired. The procedures and solutions used for devittelinization, pairing, filling, voltage-recording and current-passing electrodes have been previously described (Rubin et al., 1992; Verselis et al., 1994). Gj was measured as described above in dual whole cell patch clamp by dividing the current measured in the unstepped cell by the voltage difference between the cells. Vj steps were 30 sec in duration and applied over a range of ±120 mV in 10 mV increments; the cells were allowed to recover for 90–120 sec between Vj steps. Because expression over the course of some experiments increased, for each cell pair, the Vj steps were preceded by a small, brief prepulse of constant amplitude (10 mV) so that a family of currents could be normalized. Each current trace was digitized at two rates, initially at 1 kHz for 1 sec and then at 0.1 kHz for the remainder. This procedure allowed for more accurate measurements of initial, as well as steady-state currents, without collecting an excessive number of data points per trace. Initial and steady-state currents were obtained by extrapolating exponential fits of the data to t = 0 and t = ∞ as described (Verselis et al., 1994). Only cell pairs with gj values not exceeding 5 μS were used to avoid effects of series access resistance on voltage dependence (Wilders & Jongsma, 1992). Data were acquired using pClamp 6.0 software and a Digidata 1200 interface (Axon Instruments).
Results
Chromosomal Mapping of the Mouse Cx36 Gene
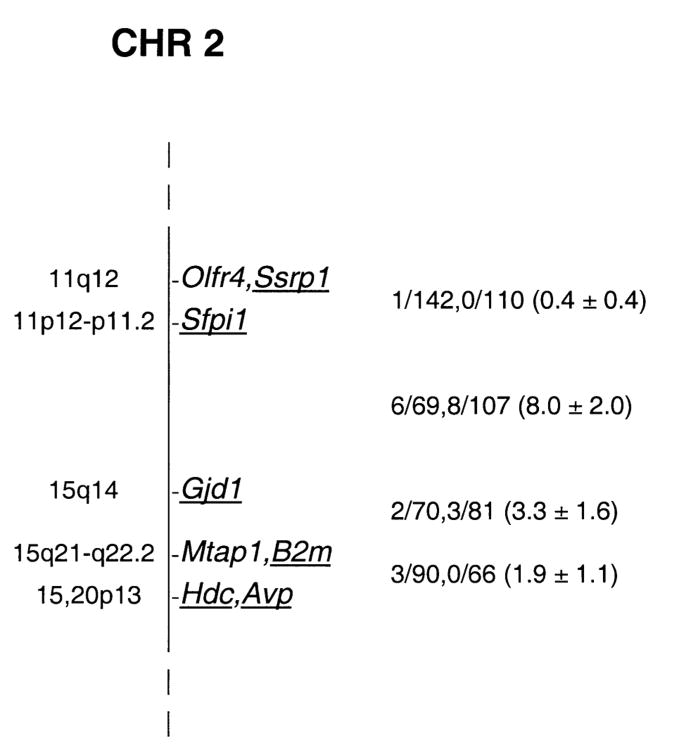
Cx36 was typed in the progeny of two sets of genetic crosses by Southern blot hybridization using an 802 bp Cx36 PCR product (Söhl et al., 1998) as probe. HpaI digestion identified fragments of 27 kb in M. musculus and 20 kb in NFS/N mice. ApaI digestion yielded fragments of 21 kb in M spretus and 15.5 kb in NFS/N. Inheritance of the variant fragments in the progeny of the two crosses revealed linkage to markers on chromosome 2 (Fig. 1).
Fig. 1.
Genetic map location of Cx36 (designated Gjd1) on mouse chromosome 2. To the right of the map, recombinational fractions between adjacent loci are listed, with the first fraction representing the M. musculus crosses and the second the M. spretus crosses. The numbers in parentheses are recombinational distances ± se. To the left of the map, chromosomal locations of the human orthologs of the underlined mouse genes are indicated.
Transcriptional Start Site of the Mouse Cx36 Gene in Brain and Retina
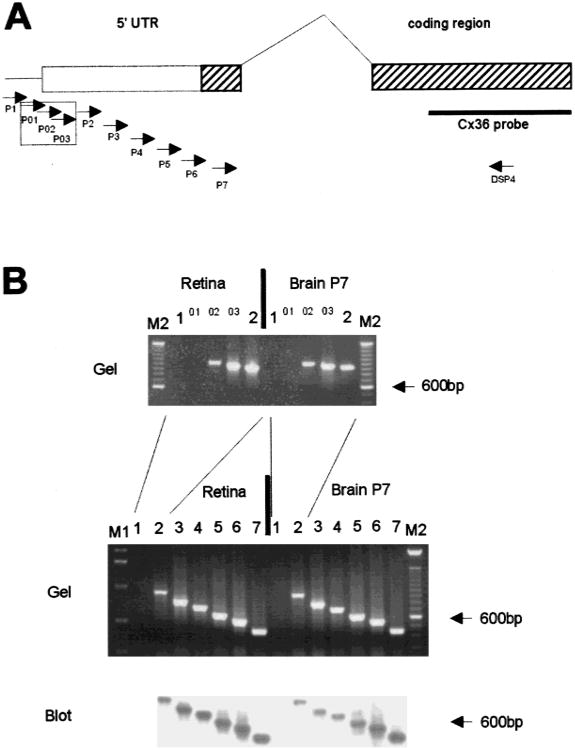
The unusual genomic structure of the murine Cx36 gene (Condorelli et al., 1998; Söhl et al., 1998) with an intron interrupting the coding sequence led us to investigate whether there are different splice variants of this gene. To determine the transcriptional start point, we sequenced mouse genomic DNA upstream of the Cx36 coding region cloned from a phage library (Söhl et al., 1998). The transcriptional start point was analyzed by applying the PCR supported method of “primer walking” as well as the conventional method of primer extension. After reverse transcription of total RNA from mouse retina and postnatal brain (P7), the cDNA templates were used in different intron-spanning PCR reactions, in which the exon II specific downstream primer DSP4 was combined with seven upstream primers P1-P7 (see Fig. 2A). As illustrated in Fig. 2B (lower part), the PCR products were further confirmed as Cx36 specific by Southern blot hybridization. Products from primers P2 through P7 were seen, but not from primer P1 thus identifying the transcriptional start site of Cx36 between upstream primers P1 and P2 (position −580 to −445 with respect to the translation start codon) in both retina and brain. To further narrow the assignment of the transcriptional start site, three additional upstream primers P01, P02 and P03 between position −580 to −445, were also combined with downstream primer DSP4 in subsequent RT-PCR reactions (Fig. 2A and B, upper part). Two additional PCR products with upstream primers P02 and P03 restricted the transcriptional start point between position −580 and −528 in both retina and brain.
Fig. 2.
PCR-mediated “primer walk” to confine the transcriptional start point of mouse Cx36 gene in retina and postnatal brain. (A) Genomic map of the mouse Cx36 gene. The positions of exons I and II are indicated as solid boxes with hatched coding regions in exons I and II. The 1424 bases of the 3 ′UTR are not shown. On the left side, the positions of the Cx36 specific upstream primers P1 to P7 are indicated by arrows. Upstream primers P01–P03 are boxed in addition. On the right side, the position of downstream primer DSP4 is shown by an arrow and the Cx36 specific hybridization probe is indicated by a solid bar. (B) Ethidium bromide stained gel and Southern blot analysis of the PCR products resulting from the intron spanning RT-PCR. Each upstream primer used is listed above the corresponding slot. The following sizes of Cx36 specific PCR products were expected: P1, 1089 bp; P01, 1040 bp, P02, 1015 bp; P03, 968 bp; P2, 934 bp; P3, 785 bp; P4, 708 bp; P5, 607 bp; P6, 548 bp; P7, 446 bp. The absence of PCR products with primers P1 and P01 indicates that the Cx36 transcript in mouse retina and postnatal brain containes at least 505 bases of 5′ untranslated region on exon I. M1: 1 kb ladder; M2: 100 bp ladder (Life Technologies).
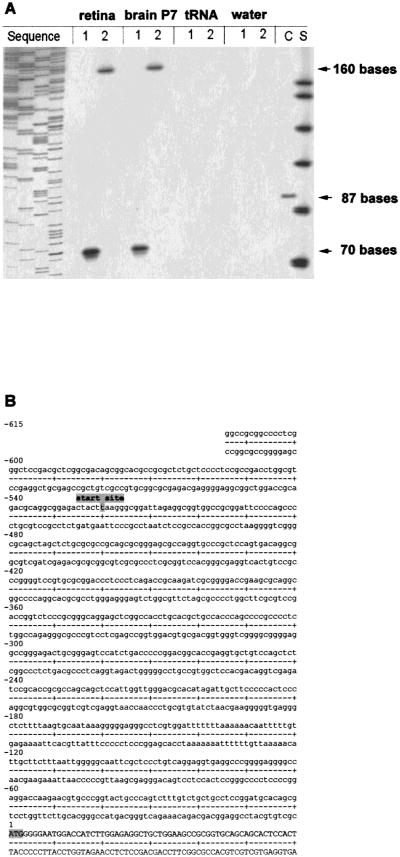
For primer extension analysis, we designed two primers Pex1 and Pex2, 50–200 bases downstream of the transcriptional start region determined by the “primer walk” method. Reverse transcription of the Cx36 mRNA with these primers yielded two distinct extension products with either total RNA from retina or poly(A)+ RNA from postnatal mouse brain. With primer Pex1, the extension products consisted of 70 bases, while primer Pex2 yielded extension products of 160 bases (Fig. 3A). Thus, considering the respective position of each primer, both extension products confined the transcriptional start point in retina and postnatal brain to position −520 (see Fig. 3B). No signals were seen in control reactions with tRNA and water. The unique transcriptional start point determined in both retina and brain corresponds to the single 2.9 kb Cx36 signal found in northern blot analyses (Condorelli et al., 1998; Söhl et al., 1998).
Fig. 3.
(A) Primer extension analysis of mouse Cx36 transcript in retina and brain and (B) Nucleotide sequence of the 5′ untranslated region of mouse Cx36. (A) Mouse retina and mouse brain RNA were annealed to Cx36 specific primers Pex1 (1) and Pex2 (2). The primers were extended using reverse transcriptase and the products were electrophoretically separated and visualized by autoradiography. Arrows indicate the size of different extension products, i.e., 70 bases for primer Pex1 and 160 bases for primer Pex2. No products were seen in control reactions using tRNA or water. Based on the length of both extension products, the transcription start point was mapped to position −520 upstream of the translation start point. The sizes of the extension products were determined with the standard S (ΦX174/HinfI DNA marker [Promega], 151, 140, 118, 100, 87, 82 and 66 bases). The sequencing reaction on the left side helped to determine the product sizes more precisely. In the control reaction (C) a synthetic mRNA coding for the kanamycin resistence gene (Promega) was extended to 87 bases. (B) The transcriptional start point is indicated at position −520. The translational start codon is at position +1. Cx36 exon I comprizes 591 bp, of which 520 bases are untranslated and 71 bases are part of the coding region.
Mouse Cx36 Protein Expression in Transfected HeLa Cells
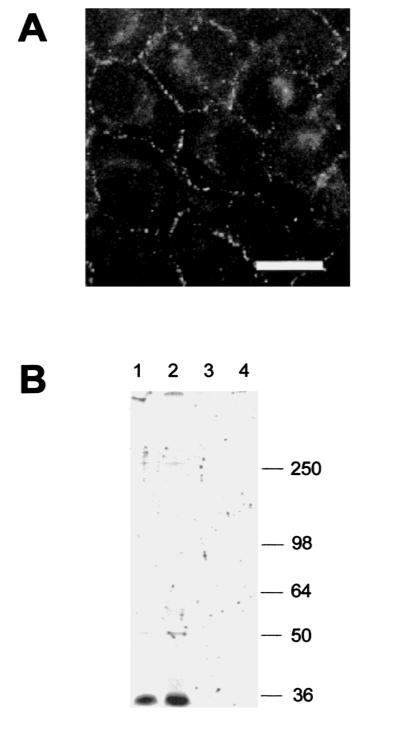
For functional expression of Cx36 channels, we transfected human HeLa cells with mouse Cx36 cDNA (see Materials and Methods). Stable Cx36 transfected clones were characterized by immunofluorescence analysis and yielded the typical punctate staining pattern of connexin expressing cultured cells on contacting membranes (Fig. 4A). The Cx36 antibodies were used to precipitate lysates from HeLa wild type or Cx36-transfected cells. One major band migrating at 36 kDa and a very faint band at ∼ 50 kDa were detected in HeLa-Cx36 cells, whereas in HeLa wild type, only the 50 kDa band was seen (after a longer exposure time; not visible on the fluorography shown in Fig. 4B). Treatment of the immunoprecipitated proteins with alkaline phosphatase did not alter the migratory pattern (Fig. 4B).
Fig. 4.
Immunofluorescence and phosphorylation analysis of HeLa cells transfected with Cx36. (A) Cx36 immunofluorescence in HeLa-Cx36 cells. Note the strong punctate labeling on contact membranes of the transfectants. There was also weak labeling in the nucleus and the cytoplasm, but no signals were visible in non transfected Hela cells (data not shown). Scale bar: 30 μm. (B). Immunoprecipitation of Cx36 in either Cx36-transfected (lanes 1 and 2) or wild type (lanes 3 and 4) HeLa cells. The beads were incubated with alkaline phosphatase (lanes 2 and 4) or without enzyme after immunoprecipitation. Molecular masses are indicated in kDa at the right.
Localization of Cx36 Protein in Rodent Retina and Brain Regions
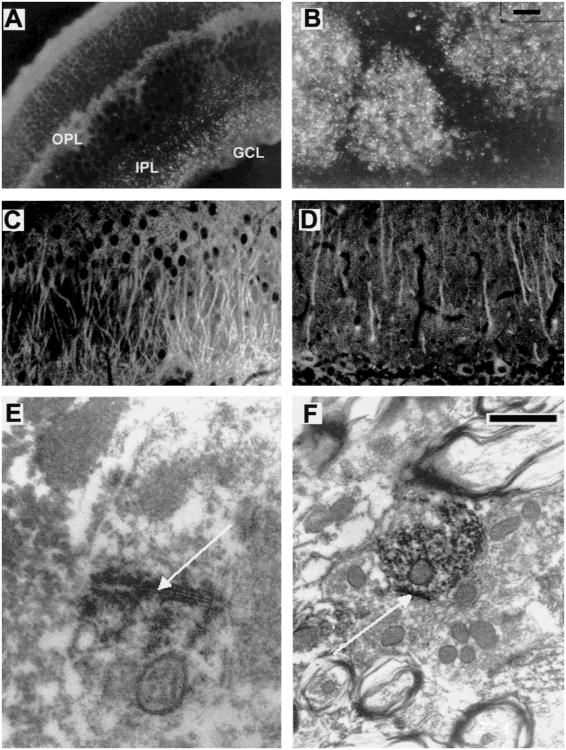
Immunofluorescence analysis using the Cx36 antibodies in both the retina and the brain also revealed strong punctate labeling. Control incubations with the preimmune serum were performed on parallel sections in order to evaluate unspecific staining, which was very low under the conditions employed (data not shown). In rat retina, punctate labeling was restricted to the the ganglion cell layer and the inner plexiform layer, with more abundance in the inner border of the inner nuclear layer where amacrine cells are localized. In the mouse retina (Fig. 5A), we observed immunolabeling within the outer plexiform layer where horizontal cell coupling has been described as well as in the inner plexiform and the ganglion cell layers. In the brain, immunoreactivity was present in the glomeruli of the olfactory bulb (Fig. 5B), the apical dendrites of the CA1 to CA3 regions of the hippocampus (Fig. 5C), the granule layer of the dentate gyrus, the primary dendrites of the cerebellar Purkinje cells (Fig. 5D), and the inferior olive. Immunoreactive cells were identified according to their position and morphology.
Fig. 5.
Cx36 immunohistochemistry in mouse and rat nervous system at light and electron microscopic levels. (A and B) Cx36 immunofluorescence analysis of ethanol-fixed mouse sections of (A) Retina (punctate staining in the outer plexiform (OPL) and the inner plexiform layers (IPL) ranging to the ganglion cell layer (GCL)) and (B) Bulbus olfactorius (punctate staining in the glomerula). (C and D) Cx36 immunofluorescence analysis of paraformaldehyde-fixed rat sections of (C) Hippocampus, CA3 region: The apical pyramidal cell dendrites in the stratum radiatum exhibit strong immunoreactivity, possibly representing intracellular transport of Cx36 protein. Staining of somata is less prominent; (D) Cerebellum: Immunoreactivity is localized in the Purkinje cells, predominantly in the dendritic processes of the molecular layer. (E and F) Cx36 immuno electron microscopy in the mouse inferior olive. Note the strong punctate labeling at a neuronal gap junction shown (arrow) in E. In F, the Cx36 antibodies labeled the cytoplasm of a dendrite. Scale bar (upper right corner) in (A–D) 30 μm; in E and F: 200 nm.
Cx36 Protein Constitutes Neuronal Gap Junctions
We confirmed the results obtained by the immunofluorescence experiments described above by immuno electron microscopic analysis of Cx36 immunolabeling in the inferior olive. Negative control experiments were done with the secondary antibodies alone or other primary antibodies, including other connexin antibodies (De Zeeuw, Hertzberg & Mugnaini, 1995). A high density of immunoreactive puncta was observed in the neuropil of the rostral medial accessory olive and principal olive, while lower densities were observed in the dorsal cap of Kooy, caudal medial accessory olive and dorsal fold of the dorsal accessory olive. Figure 5E and F shows a representative result of Cx36 immunoreactivity in the neuropil of the rostral medial accessory olive. Labeling was most prominent at the dendrodendritic gap junctions (Fig. 5E), but was also found in the cytoplasm of the dendrites (Fig. 5F). The labeled gap junctions occurred most frequently in the olivary glomeruli, which are characterized by a core of dendritic elements surrounded by several terminals and a glial sheath (see also De Zeeuw et al., 1995). No labeling was observed in the vicinity of dendritic lamellar bodies or glial gap junctions. Since there are no inhibitory neurons in the inferior olive, the Cx36 positive neurons are the projecting neurons which provide the climbing fibers to the Purkinje cells in the cerebellar cortex.
Functional Characterization of Cx36 Gap Junctions
Electrical Cell-Cell Coupling
The ability of Cx36 to induce electrical coupling was tested in HeLa-Cx36 cell pairs and pairs of Xenopus oocytes injected with Cx36 cRNA. We tested 42 spontaneously preformed HeLa-Cx36 cell pairs and found all of them to be electrically coupled. Coupling in all but three of these cell pairs exhibited voltage dependence and full electrical uncoupling with arachidonic acid and CO2, indicating that coupling in the majority of cases was mediated by gap junctions and not cytoplasmic bridges. In the cell pairs connected by gap junctions, gj varied between 0.04 and 6 nS with a mean value of 1.6 nS. Similarly, all tested pairs of Xenopus oocytes injected with Cx36 cRNA together with antisense oligonucleotides to XenCx38 were coupled (n = 14) with a mean gj of 5.5 ± 2.9 μS. Control oocytes injected only with antisense oligonucleotides showed no detectable coupling (n = 7).
Voltage and Chemical Gating
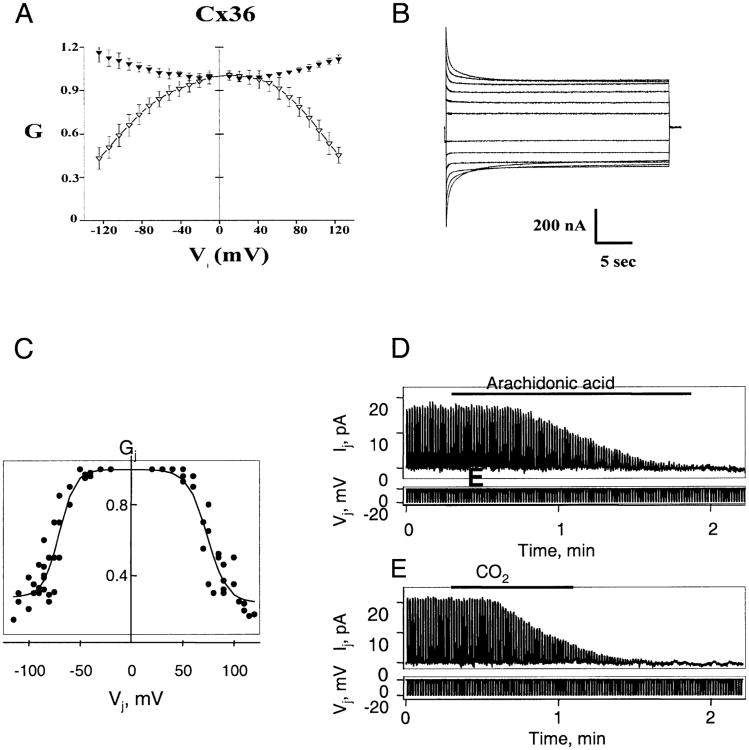
The dependence of gj on Vj was examined in both HeLa cells andXenopus oocytes expressing Cx36. Normalized initial and steady-state gj-Vj relations obtained from Xenopus oocyte pairs are plotted in Fig. 6A. Initial Gj (filled triangles) increased with Vjs of either polarity, reaching values ∼20% higher at Vj = ±120 mV than at Vj = 0. Steady-state Gj (open triangles) decreased about Vj = 0, but did not reach a plateau Gj within the range examined; Vj steps larger than ±120 mV compromised the integrity of the cells and were not used. Although the steady-state Gj – Vj relation could not be reliably fit to a Boltzmann equation, the data clearly illustrate the weak Vj dependence associated with Cx36. Gj is nearly insensitive to Vjs up to ±30 mV and declines gradually ∼60% over a 90 mV range. Representative junctional currents are shown in Fig. 6B. The time course of the decline in junctional current is typical of gap junction channels, requiring hundreds milliseconds to seconds to reach steady-state over the Vj range examined. Equal and simultaneous hyperpolarization and depolarization of both cells showed no measurable effect on gj indicating a lack of sensitivity to membrane potential, Vm.
A similar weak dependence of gj on Vj was observed in Cx36 transfected HeLa cells, but differed from the data in Xenopus oocytes in that g reached a plateau value at large Vjs. Data summarizing the steady-state gj-Vj dependence obtained from HeLa cells are presented in Fig. 6C The solid lines are fits of the data to the Boltzmann equation of the form Gj = {(1 − Gmin)/[1 + exp[A(Vj−V0)]}, where for each polarity of Vj, V0 corresponds to the Vj at which Gj is half maximum, A characterizes the steepness of Vj dependence and Gmin is the residual plateau Gj at large Vjs. Fitting parameters were as follows: V0+ = 74 mV, V0− = −71 mV, A+ = 0.1, A− = 0.11, Gmin+ = 0.23 and Gmin− = 0.24, where subscript + or − corresponds to depolarization and hyperpolarization of one cell of the cell pair, respectively. In three tested cell pairs there was no significant change in gj in response to simultaneous depolarization or hyperpolarization of both cells (data not shown).
Fig. 6.
Voltage and chemical gating of Cx36 gap junctions. (A–C) Voltage dependence of Cx36 homotypic junctions expressed in Xenopus oocytes and transfected HeLa cells. (A) Graph of initial and steady-state Gj (filled and open symbols, respectively) as a function of Vj. Gj is gj normalized to its value at Vj = 0. Data represent mean values obtained from 9 Xenopus oocyte cell pairs with maximum gs less than 5 μS. Both initial and steady-state Gj- Vj relations are nearly symmetric about Vj = 0 with initial Gj showing an increase and steady-state Gj a decrease with increasing Vjs of either polarity. (B) Representative junctional currents for Vj steps up to ±120 mV in 20 mV increments. A +20 mV Vj step, 200 msec in duration, preceded each long-duration (30 sec) Vj step. Upward and downward currents are elicited by negative and positive Vj 's, respectively. Current calibration, 200 nA. Time calibration, 5 sec. (C) Graph of steady-state Gj as a function of Vj in HeLa-Cx36 cell pairs. Conductance is normalized as described in A. Solid lines are fits of the experimental data with the Boltzmann equation (see Results for details). (D and E) Examples of chemical gating of Cx36 homotypic junctions expressed in transfected HeLa cells. Junctional current, Ij, was measured by applying repeated negative pulses (Vj) of 20 mV to one cell of a pair. Holding potentials of −20 mV were maintained for both cells in each case. (D) Application of arachidonic acid (see horizontal bar) to a cell pair in which gj≈ 0.8 nS produced full uncoupling. (E) Application of 100% CO2 (horizontal bar) to a cell pair with gj ≈ 1 nS produced full uncoupling within ∼1 min.
Sensitivity of g to chemical agents was examined using arachidonic acid and CO2. Application of arachidonic acid (10−5 m) to four cell pairs in which gj ranged from 0.5 to 1 nS produced full electrical uncoupling (Fig. 6D). Upon washout we observed very slow and only partial recovery (data not shown). Application of 100% CO2 to five cell pairs in which gj ranged from 0.4 to 2 nS also produced full electrical uncoupling within 1–2 min (Fig. 6E). Electrical coupling recovered slowly after washout, often taking 10–20 min to reach 50% of the control g(data not shown). In one experiment, where recording was stable for >30 min we observed full recovery of coupling. Thus, Cx36 exhibits similar properties as other members of the connexin family in response to these uncoupling agents.
Conductance and Permeability of Cx36 Channels
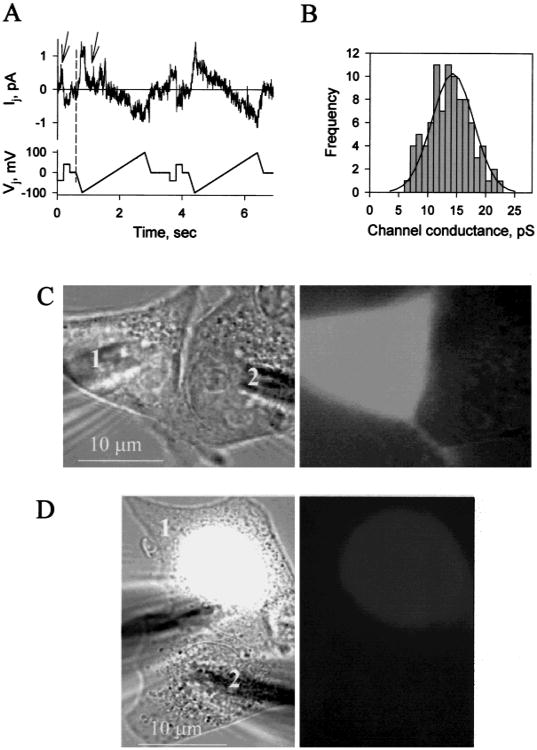
Currents of single Cx36 channels were examined in weakly coupled HeLa-Cx36 cell pairs. Figure 7A shows an I-V relation of single open channel obtained by applying a voltage ramp protocol from −100 to +100 mV to one cell of a pair. The open channel I-V relation remained linear over the entire voltage range. The frequency histogram in Fig. 7B summarizes single channel conductance data obtained from currents measured at constant Vjs. Mean value of single channel conductance is 14.3 ± 0.8 pS (n = 92).
Fig. 7.
Single channel conductance and dye permeability of Cx36 gap junctions. (A) Example of single HeLa-Cx36 channel currents from a cell pair elicited by repeated segments of ±40 mV, 200 msec Vj steps followed by ± 100 mV, 2 sec Vj ramps. Several opening and closing gating transitions occurred during the first Vj step and ramp (indicated by arrows). Both cells were initially clamped to −20 mV. Single channels were obtained during the early phase of recovery from full uncoupling with 10 μM arachidonic acid. A single channel conductance in the range of 12–15 pS was approximated from the slope of the I-Vj relation. (B) Frequency histogram of single open channel conductance. Data were pooled from different cell pairs with Vj steps varying in amplitude from ±50 to ±100 mV. Solid line shows a fit of the data to a Gaussian distribution and gives a mean single channel conductance of 14.3 ± 0.8 pS (n = 92). (C and D) Lack of dye transfer between electrically coupled cell pairs expressing Cx36. Panels on the left and on the right show phase contrast and fluorescent images, respectively. (C) Lucifer yellow was loaded in cell 1. A fluorescent image taken 5 min after patch opening in cell 1 demonstrates no Lucifer Yellow transfer from cell 1 to cell 2; gj = 3 nS. (D) DAPI was loaded in cell 1. DAPI preferentially stains the nucleus. A fluorescent image taken 12 min after patch opening shows no DAPI transfer from cell 1 to cell 2; gj = 4 nS.
To examine whether larger molecules would pass through the small conductance Cx36 channels, permeability was examined by injection of neurobiotin (Mr 287, net charge +1), and Lucifer Yellow (Mr 448, net charge -2) into one cell of a cluster. Thirty minutes after neurobiotin injection, 44% (±5.25 sem, n = 31) of first order neighboring cells surrounding the injected cell were stained in untransfected HeLa cells, whereas in HeLa-Cx36 cells, 77% (±3.8 sem, n = 44) of first order neighboring cells were coupled. Dye transfer to higher order surrounding cells was very rarely observed. The neurobiotin spread in untransfected HeLa cells was due to the long incubation time after injection. Neither untransfected nor Cx36 transfected HeLa cells showed dye transfer after injection with Lucifer Yellow.
Dye transfer to cells paired heterotypically with Cx36 was tested in co-cultures of HeLa-Cx36 cells with one each of the following murine Cx HeLa transfectants: HeLa-Cx26, -30, -31, -32, -37, -40, -43, -45, -50. None of these co-cultures demonstrated transfer above background of either neurobiotin or Lucifer Yellow (data not shown).
To better assess dye permeability, we simultaneously measured dye transfer and gj in HeLa-Cx36 cell pairs. We used Lucifer Yellow and DAPI in these experiments. In four cell pairs in which gj ranged from 2–5 nS, no Lucifer Yellow transfer was detected (Fig. 7C). Similarly, in three cell pairs in which gj ranged from 2–4 nS, no DAPI transfer was detected (Fig. 7D). Coupling by Cx36 gap junctions and not cytoplasmic bridges or endogenous channels was assessed by voltage dependence and sensitivity to uncoupling agents.
Discussion
In order to ascertain whether the mouse Cx36 gene (Con-dorelli et al., 1998; Söhl et al., 1998) is located in a chromosomal region to which neuronal disorders have been assigned, we determined its chromosomal location by recombination after interspecies crosses (Kozak et al., 1990). We mapped the mouse Cx36 gene on mouse chromosome 2 to a region where a mutation leading to juvenile myoclonic epilepsy had been assigned (Rise et al., 1991). This region corresponds to the human chromosome 15. Very recently, Belluardo et al. (1999) reported that the human Cx36 gene is located on the human chromosome 15q14, to which a juvenile form of myoclonic epilepsy has been mapped. It will be of interest to establish whether mutation(s) in the Cx36 gene cause this form of epilepsy.
Furthermore, we mapped the transcriptional start point of the Cx36 gene 520 bases upstream of the translational start codon ATG. The next 71 bases of exon I are translated. The coding region is interrupted by an intron, and exon II consists of 894 bases of the coding region (Condorelli et al., 1998; Söhl et al., 1998) and 1424 bases of 3′ UTR. Thus, the transcribed region of the Cx36 gene comprises 2909 bases which corresponds to the signal at 2.9 kb found by Northern blot analyses (Condorelli et al., 1998; Söhl et al., 1998). The “primer walk” confirmed not only the length, but also the sequence identity of the transcripts in retina and brain. We conclude that there are no alternative variants of exon I in the two tissues tested. The possibility of additional variants of exon I was checked, since in this connexin, in contrast to the other 14 murine connexin genes, the coding region is interrupted by an intron. The splice junction is located close to the presumptive transition from the N-terminus to the first transmembrane domain of the protein, and this region has been shown to form part of the transjunctional voltage sensor in other connexins (Verselis, Ginter & Bargiello, 1994). For the mouse Cx32 gene, two alternative variants of exon I were described: one expressed in liver (Hennemann et al., 1992) and another in sciatic nerve (Neuhaus, Dahl & Werner, 1995; Söhl et al., 1996). In the case of Cx32, both transcripts lead to the same protein, since the intron does not interrupt the coding region but only the 5′UTR. An alternate splicing mechanism of Cx36 expression could lead to expression of different forms of Cx36 protein.
We have demonstrated the specificity of the Cx36 antibodies by immunoprecipitation of lysates from Cx36- and untransfected HeLa cells. The band at 36 kDa corresponds to the theoretical molecular mass (36084 Da; Söhl et al., 1998) of this protein. A band similar to the weak signal at ∼50 kDa was very recently also described with antibodies to skate Cx35 in liver at 42 kDa (Srinivas et al., 1999) and might have been due to cross-reaction of each of the antibodies with an unknown antigen. In the primary sequence of Cx36, we found several putative phosphorylation sites for different protein kinases (Söhl et al., 1998; see also O'Brien et al., 1998), but no shift of the apparent molecular mass of Cx36 expresssed in HeLa cells was seen after phosphatase treatment.
Our findings on the immunoreactivity of Cx36 in the retina are consistent with the published in situ localization of Cx36 mRNA described by Condorelli et al. (1998). We found strong punctate staining in the ganglion cell layer and the inner plexiform layer. Exclusively in mice, weaker discrete labeling was observed at the border of the inner nuclear layer and the outer plexiform layer, where the terminals of horizontal cell axons are located. Definite identification of the neuronal cell types expressing Cx36 in the retina will require further experiments with immuno electron microscopy.
The immunolocalization of Cx36 in the hippocampus is also in accordance with in situ hybridization data (Condorelli et al., 1998; Belluardo et al., 1999) and immunolabeling with antibodies to skate Cx35 (Srinivas et al., 1999). Staining of dendrites was prevalent in the pyramidal cells of the entire hippocampus, but no definite pattern occurred. Granule cells of the dentate gyrus were less extensively stained, but showed slight labeling in the stratum moleculare. Interestingly, we found consistent staining of Purkinje cells in the cerebellar cortex, although no functional data on gap junction coupling of this neuronal cell type have been reported thus far. However, inhibitory interneurons in the molecular layer of the guinea pig cerebellar cortex have been reported to be electrotonically coupled (Mann-Metzer & Yarom, 1999). We have found Cx36 immunoreactivity, both in inter-neurons and pyramidal neurons, of the adult mouse cortex. Cx36 channels could mediate electrotonic coupling in these cells (Strata et al., 1997; Draguhn et al., 1998). Our electron microscopic analysis of the inferior olive demonstrated that Cx36 is present in gap junctions between olivary neurons. This is in accordance with the light microscopic demonstration of Cx36 mRNA in olivary neurons by Condorelli et al. (1998) and Belluardo et al. (1999), and it provides direct evidence that Cx36 protein is in fact involved in the formation of neuronal gap junctions. Other connexins that have been suggested to be part of neuronal gap junctions include Cx26, Cx32 and Cx43 (Dermietzel et al., 1989; Shiosaka et al., 1989; Yamamoto et al., 1990). Cx36 is the only connexin, thus far, that may be specifically associated with neuronal gap junctions in adults, since it has not been found in glial gap junctions where Cx43 and Cx32 have been located. Previously, we have observed that dendritic lamellar bodies can be associated with dendrodendritic gap junctions (De Zeeuw et al., 1995). Since Cx36 was not present in dendritic lamellar bodies, it cannot be responsible for this association.
The most prominent features of the mouse Cx36 channel, functionally expressed in human HeLa cells and Xenopus oocytes, are its low voltage sensitivity, small single channel conductance and inability to form hetero-typic gap junction channels with many other connexins. The latter was assessed by transfer of neurobiotin and will require complementary electrophysiological studies. The low voltage sensitivity of Cx36 channels in HeLa cells (V0 is ∼ 75 mV) is similar to that measured for rat Cx26, skate Cx35 (White et al., 1999) and Cx36 expressed in N2A or PC12 cells (Srinivas et al., 1999). The low sensitivity of Cx36 channels to transjunctional voltage as well as their insensitivity to absolute membrane voltage suggests that Cx36 channels can maintain electrical coupling between neurons undergoing substantial fluctuations in membrane potential. The conductance between HeLa Cx36 transfected cells was low in spite of a relatively high level of Cx36 protein expressed at the cell membranes (Fig. 4A), suggesting that either only a small fraction of channels were functional or that these channels have a low open probability. Possibly, Cx36 channels when expressed in neuronal cells need to be modified in order to reach full activity. Alternatively, Cx36 channels when exogenously expressed in HeLa cells, may be partially inactivated, leading to the relatively low level of functional Cx36 channels found in HeLa-Cx36 transfectants. Marked reduction of gap junctional conductance has been shown to occur in cultured horizontal cells from fish retina (Lasater, 1987), mediated by dopamine and cAMP. Although the molecular identity of the corresponding gap junction protein is not yet known, it appears possible that Cx36 channels could be downregulated or closed when expressed in an ectopic environment. Here we demonstrate a low single channel conductance, ∼ 15 pS (Fig. 7) which is in accordance with the data published by Srinivas et al. (1999). Thus low levels of coupling in Cx36 transfected cells may be due, in part, to a small single channel conductance. Despite the low single channel conductance Srinivas et al., 1999 reported that Cx36-transfected N2A and PC12 cells transferred Lucifer Yellow. This contrasts our results showing no detectable transfer of Lucifer Yellow between HeLa-Cx36 cell pairs. Although junctional conductance in our HeLa-Cx36 transfectants was lower, on average, than that reported by Srinivas et al., 1999, we did not detect Lucifer Yellow transfer in Cx36-HeLa cell pairs displaying gj as high as 5 nS, a level comparable to that reported for the N2A cells. We have no explanation for this discrepancy. In addition, we did not detect transfer of DAPI in HeLa-Cx36 cell pairs with gj as high as 4 nS. The observation that Cx36 did not form functional heterotypic channels with nine other connexin transfected Hela cells may be indicative of a specialized role of this neuronal connexin or explained by one of the arguments discussed above. The biological function of Cx36 containing synapses between neuronal cells may be revealed when Cx36-deleted “knock-out” mice can be investigated. Alternatively, due to the now known chromosomal location in humans (Belluardo et al., 1999) and mouse (this paper), it may be possible to characterize phenotypically affected individuals exhibiting neuronal disorders caused by or associated with a defective Cx36 gene.
During revision of this manuscript, Al-Ubaidi et al. (2000) published further results on the characterization of Cx36. Whereas the chromosomal localization of the mouse gene and the electrophysiological properties of Cx36 channels expressed in oocytes are consistent with our results, we found Cx36 immunolabeling of the mouse retina in the inner plexiform and not the inner nuclear layer. This discrepancy might be due to the use of different antibodies and fixation protocols. Al-Ubaidi et al. employed antibodies to skate Cx35, the antibodies used in our study were directed against mouse Cx36. The cells of the inner nuclear layer (most likely amacrine cells) that were positive in the insitu hybridization (Con-dorelli et al., 1998) are connected via gap junctions in the inner plexiform layer (Vaney 1994), where Cx36 immuno signals should then be expected.
We would like to thank Meike Weigel and Angele Bukauskiene for their excellent technical assistance. Drs. Ute Preuss, Gerald Seifert and Otto Traub helped with advice for immunochemical analyses. This work was supported by grants of the Deutsche Forschungsgemeinschaft (through SFB 400 and direct grant), by the Fonds der Chemischen Industrie to K.W., by NIH Grant NS367060 to F.F.B. and by grants of the Human Frontiers Science Programme and NWO-MW to C.I.D.Z.
References
- Adamson MC, Silver J, Kozak CA. The mouse homolog of the Gibbon ape leukemia virus receptor: genetic mapping and a possible receptor function in rodents. Virology. 1991;183:778–781. doi: 10.1016/0042-6822(91)91010-e. [DOI] [PubMed] [Google Scholar]
- Al-Ubaidi MR, White TW, Ripps H, Poras I, Avner P, Gomes D, Bruzzone R. Functional properties, developmental regulation, and chromosomal localization of murine connexin36, a gap-junctional protein expressed preferentially in retina and brain. J Neurosci Res. 2000;59:813–826. doi: 10.1002/(SICI)1097-4547(20000315)59:6<813::AID-JNR14>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Belluardo N, Trovato-Salinaro A, Mudo G, Hurd YL, Condorelli DF. Structure, chromosomal localization, and brain expression of human Cx36 gene. J Neurosci Res. 1999;57:740–752. [PubMed] [Google Scholar]
- Bittmann KS, LoTurco JJ. Differential regulation of connexin 26 and 43 in murine cortical precursors. Cerebral Cortex. 1999;9:188–195. doi: 10.1093/cercor/9.2.188. [DOI] [PubMed] [Google Scholar]
- Bruzzone R, White TW, Paul DL. Connections with con-nexins: the molecular basis of direct intercellular signaling. Eur J Biochem. 1996;238:1–27. doi: 10.1111/j.1432-1033.1996.0001q.x. [DOI] [PubMed] [Google Scholar]
- Condorelli DF, Parenti R, Spinella F, Trovato-Salinaro A, Belluardo N, Cardile V, Cicirata F. Cloning of a new gap junction gene (Cx36) highly expressed in mammalian brain neurons. Eur J Neurosci. 1998;10:1202–1208. doi: 10.1046/j.1460-9568.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- Dermietzel R, Traub O, Hwang TK, Beyer E, Bennett MVL, Spray DC, Willecke K. Differential expression of three gap junction proteins in developing and mature brain tissues. PNAS USA. 1989;86:10148–10152. doi: 10.1073/pnas.86.24.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermietzel R, Farooq M, Kessler JA, Althaus H, Hertzberg EL, Spray DC. Oligodendrocytes express gap junction proteins connexin32 and connexin45. Glia. 1997;20:101–114. [PubMed] [Google Scholar]
- De Zeeuw CI, Hertzberg EL, Mugnaini E. The dendritic lamellar body: a new neuronal organelle putatively associated with dendrodendritic gap junctions. J Neurosci. 1995;15:1587–1604. doi: 10.1523/JNEUROSCI.15-02-01587.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draguhn A, Traub RD, Schmitz D, Jefferys JGR. Electrical coupling underlies high-frequency oscillations in the hippocampus in vitro. Nature. 1998;394:189–192. doi: 10.1038/28184. [DOI] [PubMed] [Google Scholar]
- Goldberg GS, Bechberger JF, Nauss CCG. A pre-loading method of evaluating gap junctional communication by fluorescent dye transfer. Biotechniques. 1995;18:490–497. [PubMed] [Google Scholar]
- Green EL. Genetics and Probability in Animal Breeding Experiments. Oxford University Press; New York: 1981. [Google Scholar]
- Hennemann H, Kozjek G, Dahl E, Nicholson BJ, Willecke K. Molecular cloning of mouse connexins26 and -32: similar genomic organization but distinct promoter sequences of two gap junction genes. Eur J Cell Biol. 1992;58:81–89. [PubMed] [Google Scholar]
- Horst M, Harth N, Hasilik A. Biosynthesis of glycosylated human lysozyme mutants. J Biol Chem. 1991;266:13914–13919. [PubMed] [Google Scholar]
- Kosaka T, Hama K. Gap junctions between non-pyramidal cell dendrites in the rat hippocampus (CA1 and CA3 regions): a combined Golgi-electron microscopy study. J Comp Neurol. 1985;231:150–161. doi: 10.1002/cne.902310203. [DOI] [PubMed] [Google Scholar]
- Kozak CA, Peyser M, Krall M, Mariano TM, Kumar CS, Pestka S, Mock BA. Molecular genetic markers spanning mouse chromosome 10. Genomics. 1990;8:519–524. doi: 10.1016/0888-7543(90)90039-w. [DOI] [PubMed] [Google Scholar]
- Kunzelmann P, Blümcke I, Traub O, Dermietzel R, Willecke K. Coexpression of connexin45 and -32 in oligodendrocytes of rat brain. J Neurocytol. 1997;26:17–22. doi: 10.1023/a:1018555207379. [DOI] [PubMed] [Google Scholar]
- Kunzelmann P, Schröder W, Traub O, Steinhäuser C, Dermietzel R, Willecke K. Late onset and increasing expression of the gap junction protein connexin30 in adult murine brain and long-term cultured astrocytes. Glia. 1999;25:111–119. doi: 10.1002/(sici)1098-1136(19990115)25:2<111::aid-glia2>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Lasater EM. Retinal horizontal cell gap junctional conductance is modulated by dopamine through a cyclic AMP-dependent protein kinase. PNAS USA. 1987;84:7319–7323. doi: 10.1073/pnas.84.20.7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann-Metzer P, Yarom Y. Electrotonic coupling interacts with intrinsic properties to generate synchronized activity in cerebellar networks of inhibitory interneurons. J Neurosci. 1999;19:3298–306. doi: 10.1523/JNEUROSCI.19-09-03298.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey D, Bukauskas FF, Lee CG, Kozak CA, Willecke K. Molecular cloning and functional expression of the mouse gap junction gene connexin-57 in human HeLa cells. J Biol Chem. 1999;274:14716–14723. doi: 10.1074/jbc.274.21.14716. [DOI] [PubMed] [Google Scholar]
- Nagy JI, Patel D, Ochalski PAY, Stelmack GL. Con-nexin30 in rodent, cat and human brain: Selective expression in gray matter astrocytes, colocalization with connexin43 at gap junctions and late developmental appearance. Neuroscience. 1999;88:447–468. doi: 10.1016/s0306-4522(98)00191-2. [DOI] [PubMed] [Google Scholar]
- Neuhaus IM, Dahl G, Werner R. Use of alternate promoters for tissue-specific expression of the gene coding for connexin32. Gene. 1995;158:257–262. doi: 10.1016/0378-1119(94)00899-4. [DOI] [PubMed] [Google Scholar]
- Neyton A, Trautmann A. Single-channel currents of an intercellular junction. Nature. 1985;317:331–335. doi: 10.1038/317331a0. [DOI] [PubMed] [Google Scholar]
- O'Brien J, Bruzzone R, White TW, Al-Ubaidi MR, Ripps H. Cloning and expression of two related connexins from the perch retina define a distinct subgroup of the connexin family. J Neurosci. 1998;18:7625–7637. doi: 10.1523/JNEUROSCI.18-19-07625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rise ML, Frankel WN, Coffin JM, Seyfried TN. Genes for epilepsy mapped in the mouse. Science. 1991;253:669–673. doi: 10.1126/science.1871601. [DOI] [PubMed] [Google Scholar]
- Rubin JB, Verselis VK, Bennett MV, Bargiello TA. Molecular analysis of voltage dependence of heterotypic gap junctions formed by connexins 26 and 32. Biophys J. 1992;62:183–193. doi: 10.1016/S0006-3495(92)81804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez JC, Martinez AD, Branes MC, Gonzalez HE. Regulation of gap junctions by protein phosphorylation. Braz J Med Biol Res. 1998;31:593–600. doi: 10.1590/s0100-879x1998000500001. [DOI] [PubMed] [Google Scholar]
- Schmalbruch H, Jahnsen H. Gap junctions on CA3 pyramidal cells of guinea pig hippocampus shown by freeze-fracture. Brain Res. 1981;217:175–178. doi: 10.1016/0006-8993(81)90196-7. [DOI] [PubMed] [Google Scholar]
- Shiosaka S, Yamamoto T, Hertzberg EL, Nagy JI. Gap junction protein in rat hippocampus: correlative light and electron microscope immunohistochemical localization. J Comp Neurol. 1989;281:282–297. doi: 10.1002/cne.902810210. [DOI] [PubMed] [Google Scholar]
- Simon AM, Goodenough DA. Diverse functions of vertebrate gap junctions. Trends Cell Biol. 1998;8:477–483. doi: 10.1016/s0962-8924(98)01372-5. [DOI] [PubMed] [Google Scholar]
- Söhl G, Gillen C, Bosse F, Gleichmann M, Müller HW, Willecke K. A second alternative transcript of the gap junction gene connexin32 is expressed in murine Schwann cells and modulated in injured sciatic nerve. Eur J Cell Biol. 1996;69:267–75. [PubMed] [Google Scholar]
- Söhl G, Degen J, Teubner B, Willecke K. The murine gap junction gene connexin36 is highly expressed in mouse retina and regulated during brain development. FEBS Lett. 1998;428:27–31. doi: 10.1016/s0014-5793(98)00479-7. [DOI] [PubMed] [Google Scholar]
- Spray DC, Dermietzel R. Gap Junctions in the Nervous System. Springer; New York: 1996. pp. 1–311. [Google Scholar]
- Srinivas M, Rozental R, Kojima T, Dermietzel R, Mehler M, Condorelli DF, Kessler JA, Spray DC. Functional properties of channels formed by the neuronal gap junction protein Connexin36. J Neurosci. 1999;19:9848–9855. doi: 10.1523/JNEUROSCI.19-22-09848.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strata F, Atzori M, Molnar M, Ugolini G, Tempia F, Cherubini E. A pacemaker current in dye-coupled hilar interneurones contributes to the generation of giant GABAergic potentials in developing hippocampus. J Neurosci. 1997;17:1435–1446. doi: 10.1523/JNEUROSCI.17-04-01435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaney DJ. Patterns of neuronal coupling in the retina. Prog Ret Eye Res. 1994;13:301–355. [Google Scholar]
- Verselis VK, Ginter CS, Bargiello TA. Opposite voltage gating polarities of two closely related connexins. Nature. 1994;368:348–51. doi: 10.1038/368348a0. [DOI] [PubMed] [Google Scholar]
- White TW, Deans MR, O'Brien J, Al-Ubaidi MR, Goodenough DA, Ripps H, Bruzzone R. Functional characteristics of skate connexin35, a member of the gamma subfamily of connexins expressed in the vertebrate retina. Eur J Neurosci. 1999;11:1883–1890. doi: 10.1046/j.1460-9568.1999.00607.x. [DOI] [PubMed] [Google Scholar]
- Wilders R, Jongsma HJ. Limitations of the dual voltage clamp method in assaying conductance and kinetics of gap junction channels. Biophys J. 1992;63:942–953. doi: 10.1016/S0006-3495(92)81664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Ochalski A, Hertzberg EL, Nagy JI. LM and EM immunolocalization of the gap junctional protein connexin 43 in rat brain. Brain Res. 1990;508:313–319. doi: 10.1016/0006-8993(90)90415-8. [DOI] [PubMed] [Google Scholar]