Abstract
An automated method is reported for segmenting 3D fluid and fluid-associated abnormalities in the retina, so-called Symptomatic Exudate-Associated Derangements (SEAD), from 3D OCT retinal images of subjects suffering from exudative age-related macular degeneration. In the first stage of a two-stage approach, retinal layers are segmented, candidate SEAD regions identified, and the retinal OCT image is flattened using a candidate-SEAD aware approach. In the second stage, a probability constrained combined graph search – graph cut method refines the candidate SEADs by integrating the candidate volumes into the graph cut cost function as probability constraints. The proposed method was evaluated on 15 spectral domain OCT images from 15 subjects undergoing intravitreal anti-VEGF injection treatment. Leave-one-out evaluation resulted in a true positive volume fraction (TPVF), false positive volume fraction (FPVF) and relative volume difference ratio (RVDR) of 86.5%, 1.7% and 12.8%, respectively. The new graph cut – graph search method significantly outperformed both the traditional graph cut and traditional graph search approaches (p<0.01, p<0.04) and has the potential to improve clinical management of patients with choroidal neovascularization due to exudative age-related macular degeneration.
Index Terms: Age-related Macular Degeneration, Symptomatic Exudate-Associated Derangement (SEAD), Retinal Layer Segmentation, graph cut, graph search
I. Introduction
Age -related macular degeneration (AMD) is the primary cause of blindness and vision loss among the adults (>50year-old) [1].Exudative AMD or neovascular AMD is an advanced form of AMD, due to the growth of abnormal blood vessels from the choroidal vasculature, leading to subretinal and intra-retinal leakage of vascular fluid. Recently, treatment of exudative AMD has become available in the form of anti-vascular endothelial growth factor agents (including Ranibizumab and be vacizumab) [2, 3, 4], through intra vitreal injection, leading to a regression of the neovascularization and resulting resoroption of fluid. The frequency of the injections is primarily guided by the amount of intra-retinal fluid, which is clinically estimated subjectively from a limited number of spectral domain optical coherence tomography (SD-OCT)slices [5, 6, 7, 8]. The intra- and inter-observer variability is high potentially leading to substantial inconsistency in treatment, and automated fluid segmentation has the potential to improve this[9, 10]. In the following text, we use the term symptomatic exudate-associated derangement (SEAD) for the main retinal manifestations of AMD, including intraretinal fluid, subretinal fluid, and pigment epithelial detachment (as shown in Fig. 1). The segmentation of SEADs is a challenging task due to their relatively low signal to noise ratio (SNR) in SD-OCT scans and considerable shape variability.
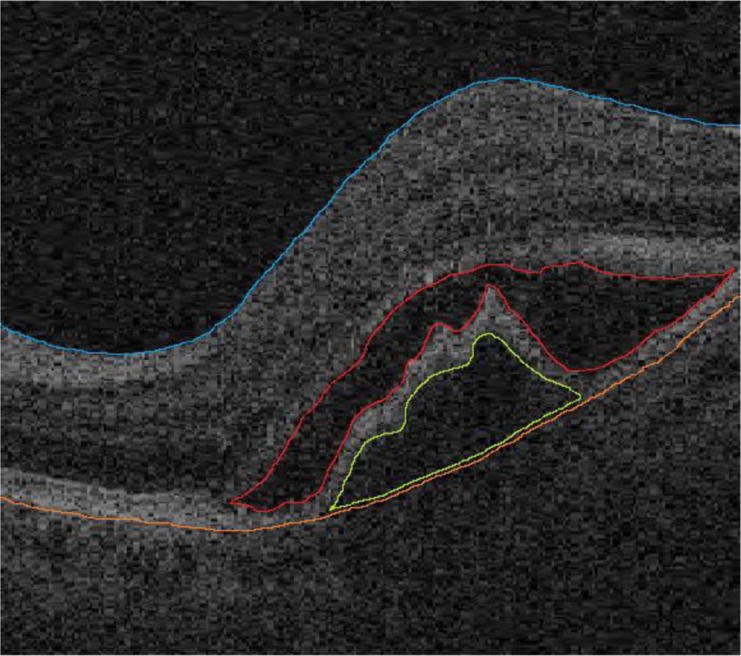
Fig. 1.
Examples of SEADs. The red curve is a manual segmentation of the SEAD consisting by the intraretinal fluid and the green curve outlines a SEAD resulting from a pigment epithelial detachment.
Prior to the work reported here, only semi-automated methods have been proposed, relying on manual initialization in 2D OCT slices to roughly quantify the SEAD volume [11]. We have previously reported a method for the identification of 2D SEAD footprints [12]. This method [12] was initiated by a fully automated retinal layer segmentation [13, 14]. Twenty-three features were then extracted in each intra-retinal layer to characterize its texture and thickness across the imaged portion of the macula. Abnormalities caused by the presence of SEADs were detected by classifying the local differences between the properties of the normal retinas and the diseased AMD under analysis. The method was successful in identifying the SEAD footprint (2D region in the OCT-imaged retinal domain). However, full segmentation of the 3D SEAD volumes is more challenging and only a few approaches have been reported in the past with a limited level of success. An automatic segmentation tool was reported in [15] but it failed to segment up to 30% of the analyzed scans. We have previously reported an early two-step attempt to 3D SEAD segmentation [16]. In the first step, our optimal surface approach was employed to segment the intra-retinal layers [13, 14] followed by a [ 17]. However, initialization (foreground and background voxels) required manual interaction and was not automated. The method was also directly dependent on the result of layer segmentation which worked well in certain types of SEADs but may have resulted in mis-segmentation of layers in some pigment epithelial detachments and intra-retinal fluid cases, which reduced the resulting accuracy of this semi-automated 3D SEAD segmentation.
In this paper, we report a fully 3D and fully automated graph-theoretic method for SEAD segmentation. As can be seen in Fig.1, the top and bottom surfaces provide natural constraints for the SEAD segmentation. As previously shown in our paper [18], a multi-object strategy may be useful for segmenting the SEADs. This is due to the identifiable constraints between objects that cause the search space to become substantially smaller thus yielding a more accurate object (SEAD) segmentation. As shown in [18], even when considering only one object of interest, including other objects as the contextual targets may be a good strategy. As hinted earlier, for segmenting SEADs, two layers (one above the potentially SEAD containing regions and another below) are identified as the related target objects that serve as constrains. Graph search (GS) methods were successfully applied to segment the retinal layers [28, 29]. For the segmentation of the region object, the GC methods [17, 19, 20] have been used widely. In this paper, we aim to effectively combine the GS and GC methods for segmenting the SEADS and layers simultaneously. An automatic voxel classification-based initialization is utilized that is based on the layer-specific texture features following the success of our previous work [12, 19].
II. Related Works
A. Conventional graph-cut algorithm
GC methods have been very popular in the studies of image segmentation in recent years [17, 20, 21, 22, 23, 24, 25, 26, 27]. A conventional graph-cut framework [17, 20] was thought to be feasible to solve SEAD segmentation in 3D OCT[16]. By introducing both a regional term and a boundary term into the graph-cut energy function, the method computed a minimum cost s/t cut on an appropriately constructed graph [21, 22]. For multiple object-region segmentation, an interaction term can be introduced to the energy function as a hard geometric constraint [28]. The overall problem can also be solved by computing an s/t cut with a maximum-flow algorithm. The conventional graph-cut framework can be applied to objects with different topological shapes, but it cannot avoid segmentation leaks in lower-resolution images.
B. Optimal surface approach – graph-search approach
Optimal surface approach (GS methods) [29, 30, 31, 32] is important for the analysis of multiple intra-retinal layers in 3D OCT images [13, 14]. For SEAD cases, most of the lesions – intra-retinal fluid, sub-retinal fluid and the pigment epithelial detachments are all associated with surrounding retinal layers. The optimal surface approach modeled the boundaries in-between layers as terrain-like surfaces and suggested representing the terrain-like surface as a related closed set. By finding an optimal closed set, the approach was able to segment the terrain-like surface. For the multiple-surface case, the approach constructed a corresponding sub-graph for each terrain-like surface [30]. Weighted inter-graph arcs were added, which enforced geometry constraints between sub-graphs. The multiple optimal surfaces could be solved simultaneously as a single s/t cut problem by using a maximum-flow algorithm. The method worked well in finding stable results of globally optimal terrain-like surfaces. However, it was limited by the prior shape requirement. The multiple-SEAD case, in a single OCT image, can be modeled as a problem with multiple regions interacting with multiple surfaces. A surface-region graph-based method was proposed to segment multiple regions and multiple surfaces simultaneously [ 33 ]. Like the optimal surface approach, surface-region method constructed a sub-graph for each target region and surface. However, shape priors describing the geometric relations among the regions and surfaces were needed.
III. Probability-Constrained Graph-Search-Graph-Cut
A. Method Overview
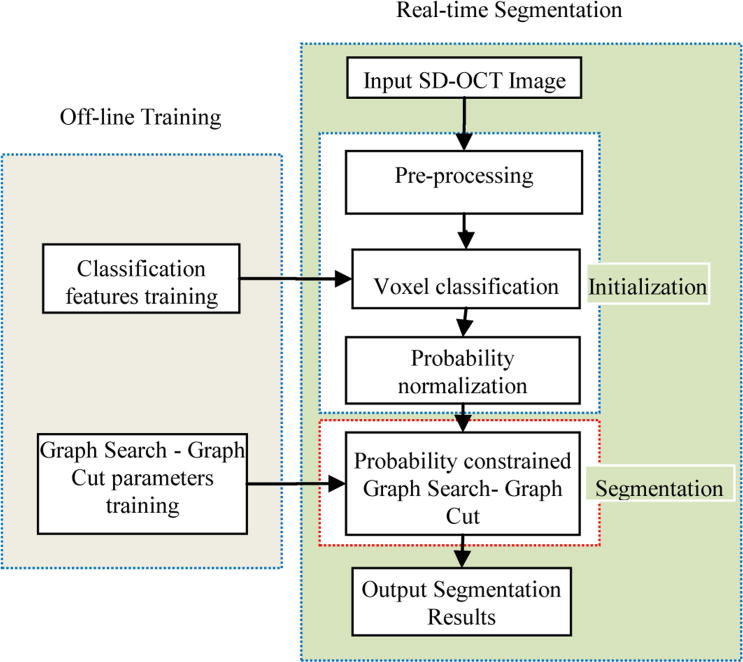
The proposed method consists of two main steps: Initialization and segmentation (Fig. 2). In the initialization step, several pre-processing steps are first applied to the input OCT image which include: segmenting the layers, fitting a surface to the bottom (retinal pigment epithelium (RPE)) layer, determining SEAD footprints [12], ignoring points within the SEAD footprints, and flattening the scan images; a texture classification based method is employed producing the initialization results. After initialization, probability normalization refines the initialization results. In the segmentation phase, the GS-GC method synergistically integrates the results from the initialization as described in the following sub-sections.
Fig. 2.
Flowchart of the proposed system.
B. Initialization
To initialize the graph based SEAD segmentation algorithm, an initial segmentation of the SEAD regions is needed. To find voxels that are likely inside of a SEAD region, a statistical voxel classification approach is applied directly to the pre-processed input image. The classifier assigns a likelihood to each voxel that it belongs to a SEAD. This likelihood map serves as constraints for the graph based segmentation algorithm.
B.1 Pre-processing
First, the upper and lower surface of the retina is determined in the scan using the output of our 11-surface segmentation [14]. For these 11 surfaces, the top retinal surface corresponds to the inner limiting membrane and bottom layer corresponds to the RPE. For more details, see figure 2 in [12]. While the top retinal surface is usually segmented successfully even in OCT scans with SEADs, the bottom surface - surface 11 - can be problematic, especially with SEADs located under the retinal pigment epithelium (RPE, see Fig. 3). In these cases, the layer segmentation may follow the top of the SEAD instead of identifying the bottom of the retina.
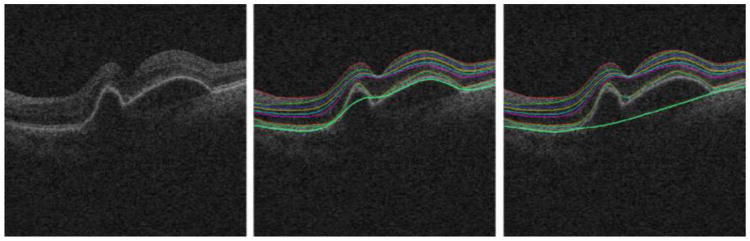
Fig. 3.
Illustration of retinal layer correction. (a) One slice from the original OCT image. (b) Segmentation of all surfaces, with surface 11 at the bottom (cyan). (c) Surface 11 after thin-plate spline fitting.
As mentioned above, we have previously presented a method for detection of SEAD locations in the XY-plane [12]. This method finds a 2D SEAD “footprint” by analyzing the textural and thickness properties of individual layers in groups of A-scans. The likelihood that an A-scan belongs to a SEAD footprint is calculated from the number of standard deviations from the normal atlas value. The binary footprint is generated by thresholding the likelihood map and the binary footprint is used to enhance the bottom layer segmentation result so that it is approximately located at the position in the scan where the bottom of the retina would have been located had the SEAD not been present. This is accomplished by fitting a thin plate spline to a set of 1000 randomly sampled points from the bottom surface 11, located outside of the 2D SEAD footprint map. A representative example of the bottom surface before and after thin plate spline fitting is shown in Fig. 3. The retinal images are subsequently flattened according to the identified thin-plate spline surface.
B.2 Voxel classification
To generate an initial segmentation of the fluid-filled SEAD areas we apply a supervised voxel classification approach trained on the voxels between the previously segmented top and bottom surface of the retina. To speed-up feature extraction and subsequent voxel classification, the training images are first sub-sampled by a factor of 2 in the X and Y directions and a factor of 4 in depth, the Z direction yielding images sized 100×100×256 voxels.
1) Features
For each voxel, many of textural, structural and positional features are calculated (see Table 1). Structural features (1-15) describe the local image structure while textural features (16-45) describe local texture. The location (height) of the voxel in the retina is encoded in three location features (46-48), the L2 distance in voxels from previously segmented surfaces 1, 7 and 11. Finally, 4 features (49-52) are included that were determined in our previous work [12] as relevant to SEAD detection and description.
Table 1.
Used classification features.
| Feature Nr | Feature Description |
| 1-5 | First eigenvalues of the Hessian matrices at scales σ=1, 3, 6, 9 and 14 |
| 6-10 | Second eigenvalues of the Hessian matrices at scales σ=1, 3, 6, 9 and 14 |
| 11-15 | Third eigenvalues of the Hessian matrices at scales σ=1, 3, 6, 9 and 14 |
| 16-45 | Output of a Gaussian filter bank up to and including second order derivatives at scales σ=2, 4, 8 |
| 46-48 | Voxel distances from surfaces 1,7 and 11 |
| 49-52 | Layer texture features as described in [12]: mean intensity, co-occurrence matrix entropy and inertia, wavelet analysis standard deviation (level 1) |
2) Training Phase
In the training phase, the pre-processed training images are randomly sampled to collect voxels that are either inside or outside of the SEADs. Due to differences in the number of SEAD voxels in individual OCT images both the normal and the SEAD voxels in a scan are sampled separately to ensure that a sufficient number of positive training samples are obtained in each scan. For each training image, 50,000 negative samples and 10,000 positive samples (i.e., two classes) were randomly collected. If there were less than 10,000 positive voxels in any training image, all available positive voxels were included in the training set [34]. Two-class classification was used because the SEADs in our data are fluid filled, voxels inside the SEADs correspond to fluid while voxels outside of the SEADs do not.
A k-Nearest Neighbor classifier (k=21) was chosen based on its performance in comparative preliminary experiments on a small, independent set of images. The employed k-NN implementation [ 35 ] allows approximate Nearest Neighbor classification and the maximum error parameter epsilon was set to 2 for this algorithm. Training time for this classifier is low, not taking more than 20 seconds. The training phase only needs to be completed once, after this, the trained classifier can be used to classify unseen voxels [36].
3) Testing Phase
In the testing phase, the previously described trained classifier was applied to test images. After pre-processing and feature extraction, each voxel between the top and the bottom surfaces was assigned a likelihood between 0 and 1 that the voxel is inside of a SEAD region.
B.3 Initialization Post-Processing by Probability Normalization
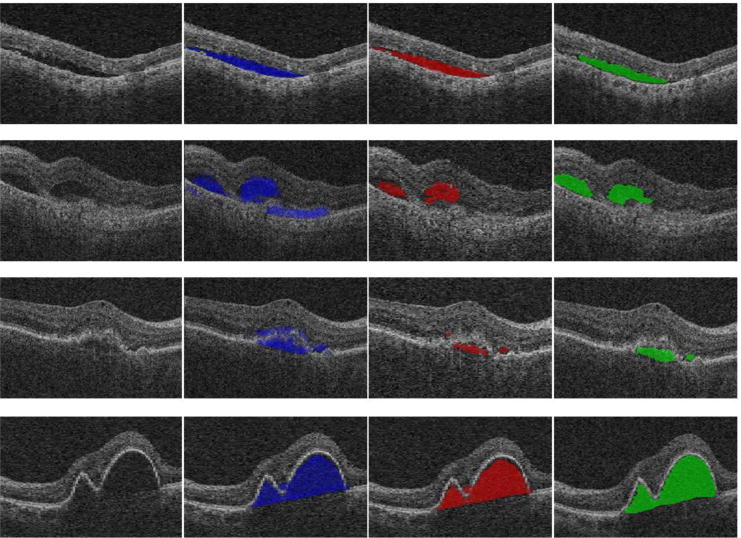
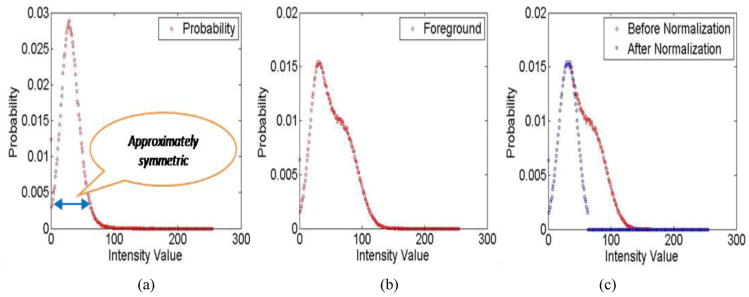
The above described initialization is not always successful (see 2nd and 3rd rows of Fig. 7). To cope with the high image noise in these cases, a post-processing method was proposed. The ground truth identified in the training data set revealed that the intensity distribution of the SEAD regions closely follows the Gaussian distribution in the low intensity range. This knowledge was used to post-process the initialization results as shown in Fig. 4: (1) Find the largest intensity value on the original curve. (2) Using this value, flip-duplicate the left part of the curve. (3) Set the probability of those intensity values outside the symmetric part to zero. After the post-processing, the resulting likelihood map is used to constraint the subsequent graph based segmentation.
Fig. 7.
Experimental results for four examples of SEAD initialization. The first column shows the original image, the 2nd column shows the initialization results, the 3rd column shows the final initialization results after probability normalization, and the last column shows the ground truth. Note the improvements obtained by probability normalization in columns 3 and 4.
Fig. 4.
Initialization post-processing: Probability normalization. (a) Intensity distribution of SEAD regions in the reference standard. (b) Intensity distribution in a specific initialization result. (c) Probability normalization resulting from the flip-duplicate step (see text).
C. Graph Search-Graph Cut SEAD Segmentation
The GS and GC methods were synergistically combined to segment the SEADs. Two layers (one layer above the SEAD region and another below the SEAD region) are included as the auxiliary target objects to constraint the SEAD segmentation.
C.1 Cost Function Design
The segmentation problem can be formulated as an energy minimization problem such that for a set of pixels P and a set of labels L, the goal is to find a labeling f: P → L that minimizes the energy function En(f). Our cost function is designed as follows:
| (1) |
where E(Surfaces) represents the cost associated with the segmentation of all surfaces, E(Regions) represents the cost associated with the segmented regions, and E(Interactions) represents the cost of constraints between the surfaces and regions. More details are given below.
(1) Surface cost function
For the terrain-like multiple-surface segmentation, the graph search method [29] is utilized. Similar to [29], the cost function is designed as
| (2) |
where S is the desired surface, cv is an edge-based cost which is inversely related to the likelihood that S contains the voxel v. (p, q) is a pair of neighboring columns N. hp,q is a convex function penalizing the surface S shape change on p and q.
(2) Region cost function
The graph cut method [17] has been successfully applied to regional segmentation. The typical graph cut energy function is defined as,
| (3) |
where Np is the set of pixels in the neighborhood of p. Rp(fp) is the cost of assigning label fp ∊ L to p which is usually defined based on the image intensity and can be considered as a log likelihood of the image intensity for the target object, and Bp,q(fp, fq) is the cost of assigning labels fp,fp ∊ L to p and q that could be based on the gradient of the image intensity.
Importantly, the results of the initialization step are effectively integrated into the whole framework: 1) The high likelihood voxels (over 0.8, followed by morphologic erosion) were used as source seeds. Voxels with low probability (here 0) were used as sink seeds. 2) The proposed probability-constrained energy function was defined as follows:
| (4) |
where α,β,γ are the weights for the data term, probability constrained term, and boundary term, respectively, satisfying α+β+γ=1. These components are defined as follows:
| (5) |
| (6) |
and
| (7) |
where Ip is the intensity of pixel p, object label is the label of the object (foreground). P(Ip|O) and P(Ip|B) are the probabilities of intensity of pixel p belonging to object and background, respectively, which are estimated from object and background intensity histograms during the separate training phase (details given below). d(p, q) is the Euclidian distance between pixels p and q, and σ is the standard deviation of the intensity differences of neighboring voxels along the boundary,
| (8) |
where InitP(p) is the probability of p which is the initialization result, λ is a constant (here λ =1).
During the training stage, the intensity histogram of each object is estimated from the training images. Based on this, P(Ip|O) and P(Ip |B) can be computed. As for the parameters α, β and γ in Eqn. (9), since α + β + γ = 1, we only estimate α and β by optimizing the accuracy as a function of α and β and set γ= 1-α-β. We use the gradient descent method [37] for the optimization.
(3) Interaction between the surfaces and regions
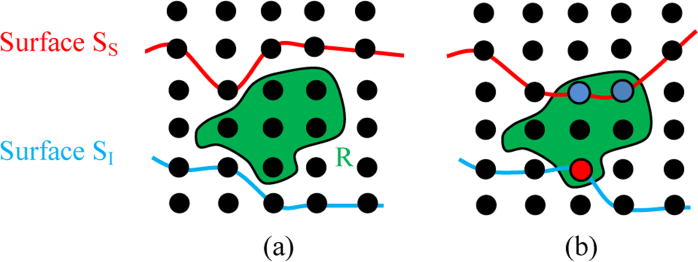
The E(Interactions) represents the interactions between the surfaces and regions. We included two surfaces: SI and SS to constrain the regions, as shown in Fig. 5. If the voxel in the region is located higher than surface SS, then a penalty is given. Similarly if the voxel in the region is located lower than surface SI, then a penalty is given. The proposed interaction term is designed as follows,
Fig.5.
Illustration of surface-region interactions on a 2D example. (a) The two terrain-like surfaces: SS and SI, and the region R in green. (b) Incorporation of the constraints between the region and surfaces. If the voxel in the region is superior to surface SS, then a penalty is given (as illustrated in blue). And if the voxel in the region is inferior than surface SI, then a penalty is given (as illustrated in red).
| (9) |
where z(v) represents the z coordinate of voxel v, p is a column which contains v, SS(p) and SI(p) are the z values for the surfaces SS and SI on the column p, respectively, d is a pre-defined distance threshold (here, d=1), wv is a penalty weight for v and fv = 1 if v ε region R.
C.2 Graph Construction
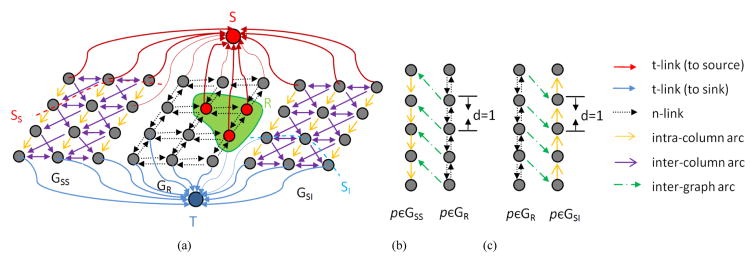
Three sub-graphs are constructed for superior surface SS, inferior surface SI and region R. These three sub-graphs are merged together to form as a single s-t graph G which can be solved by a min-cut/max-flow technique [17].
For the surface SS, a sub-graph GSS(VSS, ASS) is constructed by following the method in [29]. Each node in VSS corresponds to exactly one voxel in the image. Two types of arcs are added to the graph: (1) The intra-column arcs with +∞ weight, which enforces the monotonicity of the target surface; and (2) the inter-column arcs incorporating the penalties hp,q between the neighboring columns p and q. Each node is assigned a weight wn such that the total weight of a closed set in the graph GSS equals to the edge-cost term of E(surface). Following the method in [38], each node is connected to either the source S with the weight –wn if wn < 0 or the sink T with the weight wn if wn > 0.
For the surface SI, the same graph construction method is applied creating another sub-graph GSI(VSI, ASI).
For the region term cost function, the third sub-graph GR(VR, AR) is constructed following the graph cut method in [17]. Here, each node in VR is also corresponding to exactly one voxel in the image. The two terminal nodes: source S and sink T are the same nodes already used in GSS and GSI. Each node has t-links to the source and sink, which encode the data term. Each pair of neighboring nodes is connected by an n-link, which encodes the boundary term. Fig. 6 shows the graph construction. The nodes in VR, VSS and VSI are all corresponding, so these three sub-graphs can be merged into a single graph G.
Fig.6.
Illustration of graph construction on a 2D example. (a) Final constructed graph G which consists of three sub-graphs GSS, GR and GSI. (b) Geometric constraints between surfaces GSS and GR. (c) Geometric constraints between surfaces GR and GSI.
Additional inter-graph arcs are added between GSS and GR, as well as between GR and GSI to incorporate geometric interaction constraints. For GSS and GR, if a node (x, y, z) in the sub-graph GR is labeled as “source” and the node (x, y, z+d) in the sub-graph GSS is labeled as “sink”, i.e. z-SSS(x,y) > d, then a directed arc with a penalty weight from each node GR(x, y, z) to GSS (x, y, z + d) will be added, as shown in Fig. 6(b). For GR and G SI, the same approach is employed.
IV. Experimental Methods
A. Subject Data and Independent Standard
Macula-centered 3D OCT volumes (200 × 200 × 1024voxels, 6 × 6 × 2 mm3, voxel size 30 × 30 × 1.95 μm3) were obtained from 15 eyes of 15 patients with exudative AMD. The study protocol was approved by the institutional review board of the University of Iowa. For the validation purpose, a leave-one-out strategy was employed during voxel classification and segmentation.
For the reference standard, a retinal specialist (MDA) manually segmented the intra- and sub-retinal fluid in each slice of each eye using Truthmarker software [39] on iPad.
B. Assessment of Initialization and Segmentation Performance
The accuracy in terms of true positive volume fraction (TPVF), false positive volume fraction (FPVF) [35], and relative volume difference ratio (RVDR) were used as performance indices. TPVF indicates the fraction of the total amount of fluid in the reference standard delineation; FPVF denotes the amount of fluid falsely identified; RVDR measures the volume difference ratio comparing to the reference standard volume; which are defined as follows,
| (10) |
| (11) |
| (12) |
where, Ud is assumed to be a binary scene with all voxels in the scene domain set to have a value 1, and Ctd is the set of voxels in the true delineation, |·| denotes volume. |VM| is the segmented volume by method M, and |VR| is the volume of the reference standard. More details can be seen in [40].
For the initialization, an experiment was performed to show the efficacy of the probability normalization by comparing the performance before and after probability normalization.
For the segmentation, three methods of the traditional GC in [17], the traditional GS method in [14] and the proposed probability constraints GS-GC were compared. A multivariate analysis of variance (MANOVA) test [41] was based on the three performance measures: TPVF, FPVF and RVDR to show the statistical significance of performance differences.
C. Statistical Correlation Analysis and Reproducibility Analysis
For statistical correlation analysis, linear regression analysis [42] and Bland-Altman plots [43] were used to evaluate the relationship and agreement between the manual and automatic segmentations.
For the reproducibility analysis, the retinal specialist was asked to manually segment the intra- and sub-retinal fluid at the onset of the project and again after more than 3 months. The manual segmenting of all the slices for one eye required more than 2 hours of expert tracing, so the re-tracing was performed on 5 randomly selected eyes from the entire data set.
V. Results
A. Assessment of Initialization Performance
Fig. 7 shows four examples of initialization and its post-processing by probability normalization. We can see that for the 3rd and 4th images, reflective of probability normalization, the numbers of falsely detected voxels have decreased substantially. The 1st and 2nd rows of Table 2 show the initialization performance of TPVF, FPVF and RVD before and after probability normalization. We can see that after the probability normalization, the FPVF decreased noticeably (from 5.2% to 3.0%).
Table 2.
Mean ± standard deviation (median) of TPVF, FPVF and RVDR for initialization, initialization after probability normalization, traditional GC in [17], traditional GS [14] and the proposed probability constrained GS-GC.
| TPVF (%) | FPVF (%) | RVDR % | |
| Initialization | 72.3 ±17.6 (77.5) | 4.5 ±3.7 (3.6) | 21.1±41.2 (16.4) |
| Initialization after probability normalization | 72.5 ±17.5 (77.5) | 3.0 ±3.2 (2.5) | 20.8±40.5 (16.2) |
| Traditional GC [17] | 77.9±23.9 (81.4) | 3.6 ±3.3 (3.2) | 20.2±37.6 (6.5) |
| Traditional GS [14] | 82.8±10.5 (86.0) | 3.2 ±4.5 (2.6) | 22.8±45.6 (12.5) |
| The proposed probability constrained GS-GC | 86.5±9.5 (90.2) | 1.7 ±2.3 (0.5) | 12.8±32.1 (4.5) |
B. Assessment of Segmentation Performance
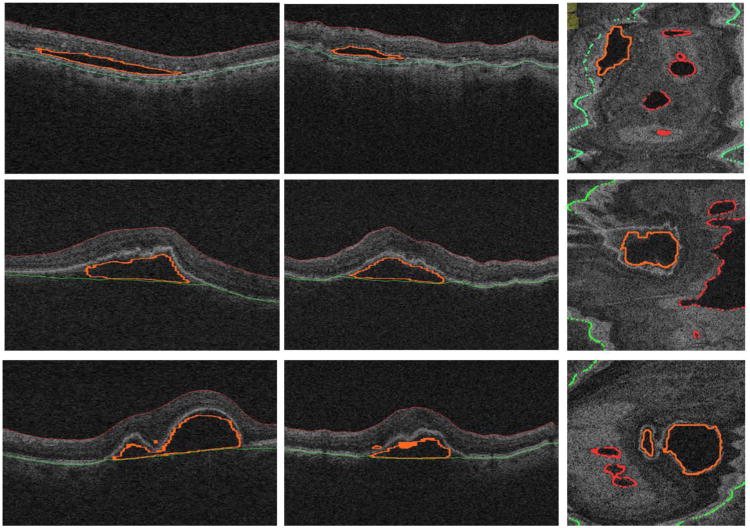
Fig. 8 shows three examples of the obtained segmentation results. The quantitative assessment of the segmentation performance achieved by the proposed method expressed in TPVF, FPVF and RVDR is summarized in Table 2. Compared to the traditional GC[17] and GS[14], the proposed probability constrained GS-GC method achieved a better performance. . The p-value of the MANOVA test for the proposed method vs. the traditional GC [17] and the proposed method vs. the traditional GS [14] is p<0.01 and p<0.04, respectively, i.e., both of the performance improvements are statistically significant. The average TPVF, FPVF and RVDR for the proposed method are about 86.5%, 1.7% and 12.8%, respectively. Fig. 9 shows a 3D visualization of the typical SEAD segmentation results.
Fig. 8.
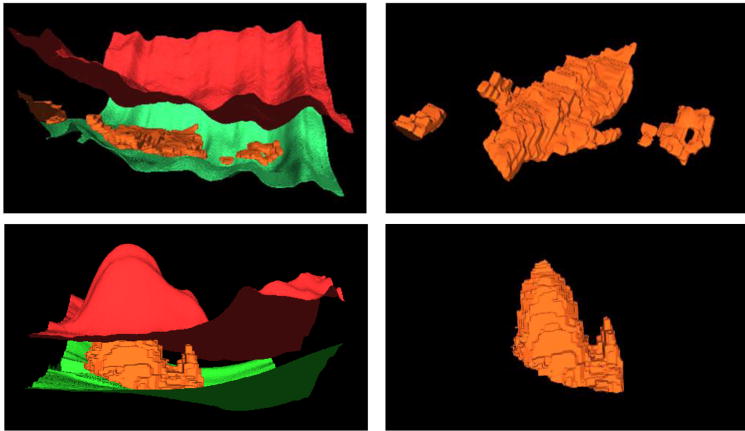
Experimental results for three examples of SEAD segmentation. The 1st, 2nd and 3rd columns correspond to the axial, sagittal and coronal views, respectively. Red color represents the upper retinal surface, green color represents the lower retinal surface, and yellow color depicts the surface of the segmented SEAD.
Fig. 9.
3D visualization of SEAD segmentation on two examples (the 1st and 3rd cases in Fig. 8). Red color represents the upper retinal surface, green color the lower retinal surface, and orange color depicts the surface of the segmented SEAD.
In terms of efficiency, the proposed method was tested on an HP Z400 workstation with 3.33GHz CPU, 24 GB of RAM. The computation times for the initialization and segmentation were 15 and 10 minutes, respectively.
C. Statistical Correlation Analysis and Reproducibility Analysis
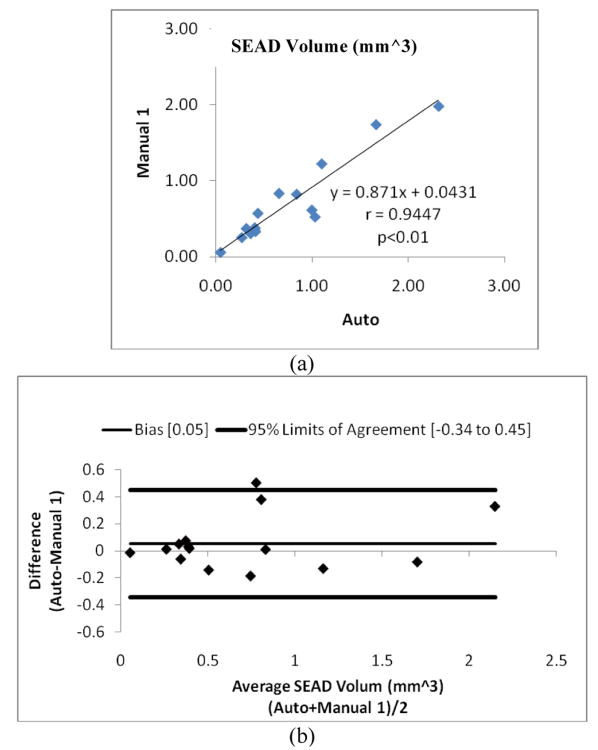
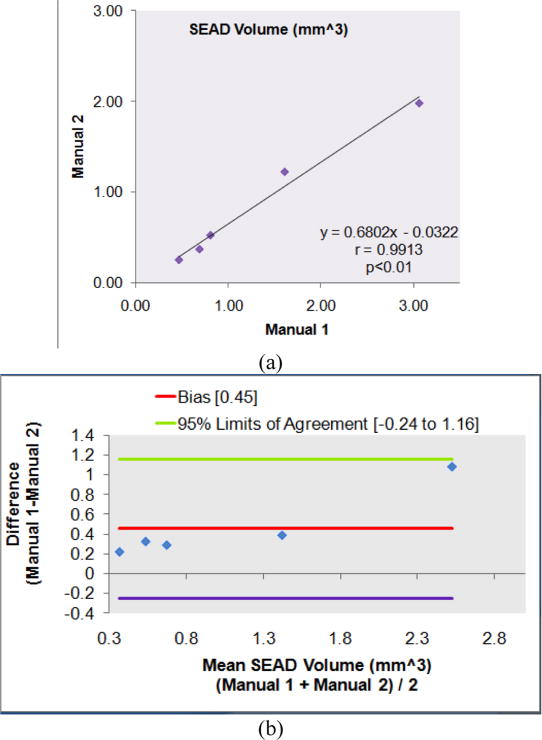
Fig. 10 shows the linear regression analysis comparing SEAD volumes and Bland-Altman plots for the fully automated probability constrained GS-GC method vs. Manual 1. Fig. 11 gives the reproducibility assesssment of manual tracing Manual 1 vs. Manual 2. The figures demonstrate that: 1) the intra-observer reproducibility has the highest correlation with r=0.991. In comparison, the automated analysis achieves a high correlation with the Manual 1 segmentation (r=0.945). 2) Analyzing the Bland-Altman plotsreveals that the 95% limits of agreement were [-0.34, 0.45] and [-0.24, 1.16] for the Automated method vs. Manual 1, and Manual 1 vs. Manual 2, respectively. The Automated vs. Manual 1 showed a much lower bias compared to the Manual 1 vs. Manual 2.
Fig. 10.
Statistical correlation analysis between the Automated method GS-GC and manual tracings Manual 1. (a) Linear regression analysis results comparing SEAD volumes. (b) Bland-Altman plots.
Fig. 11.
Statistical correlation analysis of reproducibility comparing manual tracings Manual 1 and Manual 2. (a) Linear regression analysis results comparing SEAD volumes. (b) Bland-Altman plots.
VI. Discussion
The results show that our new probability constrained graph cut – graph search method significantly outperforms both the traditional graph cut and traditional graph search approaches, and its performance to segment intra- and subretinal fluid in SD-OCT images of patients with exudative AMD is comparable to that of a clinician expert.
A. Importance of SEAD segmentation
As mentioned in the Introduction, current treatment is entirely based on subjective evaluation of intra- and subretinal fluid amounts from SD-OCT by the treating clinician. Though never confirmed in studies, anecdotal evidence and experience in other fields show that the resulting intra- and interobserver variability will lead to considerable variation in treatment and therefore, under- and overtreatment. Though each treatment, based on regular and frequent intravitreal injections of anti-VEGF, has less than a 1:2000 risk of potentially devastating endophthalmitis and visual loss, because of the high number of lifetime treatments, the cumulative risk is still considerable. In addition, the cost of each injection is high millions of patients are being treated every month so that the total burden on health care systems is in billions of US$ (year 2012). The potential of our approach to avoid overtreatment is therefore double attractive, because both lowering of the risk to patients and cost-savings can be achieved. However, validation in larger studies are required before our approach can be translated to the clinic.
B. Advantages of the reported method
We have reported a graph-theoretic based method for SEAD segmentation. The multi-object strategy was employed for segmenting the SEADs, during which two retinal surfaces (one above the SEAD region and another below the SEAD region) were included as auxiliary target objects for helping the SEAD segmentation. The two auxiliary surfaces provide natural constraints for the SEAD segmentation and make the search space become substantially smaller, thus yielding a more accurate segmentation result. The similar idea has also been proved in [18]. The proposed graph-theoretic based method effectively combined the GS and GC methods for segmenting the SEADs and layers simultaneously. An automatic voxel classification-based method was used for initialization which was based on the layer-specific texture features following the success of our previous work [12, 19]. The probability constraints from the initialization were effectively integrated into the later GS-GC method which further improved the segmentation accuracy.
The contributions of the presented work can be summarized as follows:
To the best of our knowledge, this is the first approach for fully automated 3D SEAD segmentation.
The improved flattening method can deal with sub RPE structures based on spline fitting over the structure footprint.
Texture-based voxel classification is used for automated initialization, additionally providing probability constraints inherently integrated in the later graph based segmentation method.
Our probability-constrained GS-GC method integrates the initialization results in two ways: The initialization a) defines the source and sink seeds for the graph search; and 2) modifies the cost function via local probability constraints.
C. Limitations of the reported method
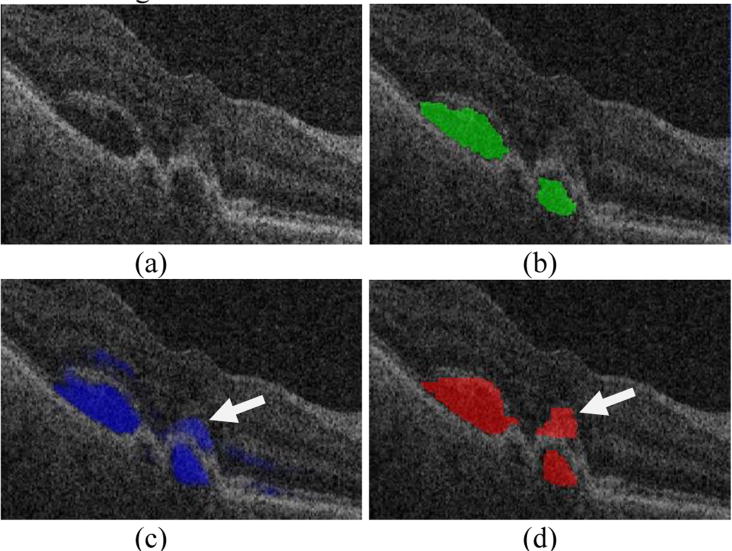
Our approach has some limitations. The first limitation is that it largely relies on the initialization results. If the probability constraints from the initialization step are incorrect, the final segmentation results may fail. Fig. 12 shows one example that mis-detects the SEAD due to the inaccurate initialization.
Fig. 12.
One example of erroneous segmentation of a SEAD due to inaccurate initialization. (a) Original OCT slice. (b) Ground truth. (c) Initialization. (d) SEAD segmentation result. Arrow points to the mis-initialized and therefore mis-segmented SEAD.
The proposed method shows high correlation with manual segmentation and if validated in a larger study, may be applicable to clinical use. From Figs. 10 and 11, it can be seen that the Automated vs. Manual 1 showed a much lower bias compared to the Manual 1 vs. Manual 2, which may be caused by the Manual 2 analysis being available for a subset of only 5OCT images - because of the laboriousness of expert tracing, even when accelerated with Truth marker. We plan to expand our labelling efforts in the future.
D. Segmentation of abnormal retinal layers
Several methods were proposed for the retinal surface and layer segmentation [13, 14, 28, 44, 45, 46]. However, all these methods have been evaluated on datasets from non-AMD subjects, where the retinal layers and other structures are intact. When the retinal layers are disrupted, and additional structures are present that transgress layer boundaries, as in exudative AMD or Diabetic Macular Edema, segmentation becomes exponentially more challenging. This paper has provided an idea for the abnormal layer segmentation. The main task, the SEAD segmentation, has been tackled by our innovative approach of combining two auxiliary surfaces. In this process, the normal (surface) provides constraints for the abnormal (SEAD) segmentation, and as a return, the abnormal help refine the segmentation of normal. As shown by our results (see Fig. 8), whenever a successful SEAD segmentation is achieved, the bottom surface is also correctly segmented. This idea may also be applied to segment other targets in abnormal data set, such as liver tumor segmentation in liver CT scans.
VII. Conclusion
In summary, a fully automated framework for 3D SEAD segmentation was reported. The proposed framework effectively combined the GS and GC methods, and employed a multi-object strategy during which two retinal layers were included as auxiliary target objects for helping the SEAD segmentation. An automatic voxel classification based on the texture features was used for initialization. Probability constraints further improved the graph-based segmentation. The method was tested on SD-OCT data from 15 eyes of 15 patients with AMD. The experimental results yielded an overall segmentation accuracy of TPVF > 86.5%, FPVF < 1.7%, and RVDR < 12.8%.
Acknowledgments
This work was supported in part by the National Institutes of Health under Grant EY017066, Grant EB004640, Research to Prevent Blindness and the Veterans Administration Center of Excellence for Prevention and Treatment of Visual Loss.
Contributor Information
Meindert Niemeijer, Email: mniemeij@healthcare.uiowa.edu, M. Niemeijer is with the Department of Electrical and Computer Engineering and the Department of Ophthalmology and Visual Sciences, the University of Iowa, Iowa City, IA 52242 USA.
Li Zhang, Email: li-zhang-1@uiowa.edu, L. Zhang is with the Department of Electrical and Computer Engineering, The University of Iowa, Iowa City, IA 52242 USA.
Kyungmoo Lee, Email: kyungmle@engineering.uiowa.edu, K. Lee is with the Department of Electrical and Computer Engineering , the University of Iowa, Iowa City, IA 52242 USA.
Michael D. Abràmoff, Email: michael-abramoff@uiowa.edu, M. D. Abràmoff is with the Department of Ophthalmology and Visual Sciences, the Department of Electrical and Computer Engineering, the Department of Biomedical Engineering, the University of Iowa, Iowa City, IA 52242 USA, and also with the VA Medical Center, Iowa City, IA 52246 USA.
Milan Sonka, Email: milan-sonka@uiowa.edu, M. Sonka is with the Department of Electrical and Computer Engineering, the Department of Ophthalmology and Visual Sciences, and the Department of Radiation Oncology, the University of Iowa, Iowa City, IA 52242 USA.
References
- 1.Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 2.Rosenfeld PJ, Brown DM, Heier JS, Boyer DS, Kaiser PK, Chung CY, Kim RY. Ranibizumab for neovascular Age-Related Macular Degeneration. N Engl J Med. 2006;355:1419–1431. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 3.Ferrara N, Hilan KJ, Gerber H, Novetny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nature Reviews Drug Discovery. 2004 May;3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 4.Algvere PV, Steén B, Seregard S, Kvanta A. A prospective study on intravitreal bevacizumab (Avastin®) for neovascular age-related macular degeneration of different durations. Acta Ophthalmologica. 2008 Aug;86:482–489. doi: 10.1111/j.1600-0420.2007.01113.x. [DOI] [PubMed] [Google Scholar]
- 5.Cukras C, Wang YD, Meyerle CB, Forooghian F, Chew EY, Wong WT. Optical coherence tomography-based decision making in exudative age-related macular degeneration: comparison of time- vs spectral-domain devices. Eye. 2010;24:775–783. doi: 10.1038/eye.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lalwani GA, Rosenfeld PJ, Fung AE, Dubovy SR, Michels S, Feuer W, Davis JL, Flynn HW, Jr, Esquiabro M. A variable-dosing regimen with intravitreal ranibizumab for neovascular age-related macular degeneration: year 2 of the PrONTO study. Am J Ophthalmol. 2009 Jul;148:43–58. doi: 10.1016/j.ajo.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 7.Coscas F, Coscas G, Souied E, Tick S, Soubrane G. Optical coherence tomography identification of occult choroidal neovascularization in age-related macular degeneration. Am J Ophthalmol. 2007 Oct;144:592–599. doi: 10.1016/j.ajo.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Kashani AH, Keane PA, Dustin L, Walsh AC, Sadda SR. Quantitative subanalysis of cystoid spaces and outer nuclear layer using optical coherence tomography in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50:3366–3373. doi: 10.1167/iovs.08-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung AE, Lalwani GA, Rosenfeld PJ, Dubovy SR, Michels S, Feuer WJ, Puliafito CA, Davis JL, Flynn HW, Jr, Esquiabro M. An optical coherence tomography-guided, Variable dosing regimen with intravitreal ranibizumab (Lucentis) for neovascular age-related macular degeneration. Am J Ophthalmol. 2007 Aprl;143:566–583. doi: 10.1016/j.ajo.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 10.Dadgostar H, Ventura AA, Chung JY, Sharma S, Kaiser PK. Evaluation of injection frequency and visual acuity outcomes for ranibizumab monotherapy in exudative age-related macular degeneration. Am J Ophthalmol. 2009 Sep;116:1740–1747. doi: 10.1016/j.ophtha.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 11.Fernández DC. Delineating fluid-filled region boundaries in optical coherence tomography images of the retina. IEEE Trans Med Imag. 2005 Aug;24:929–945. doi: 10.1109/TMI.2005.848655. [DOI] [PubMed] [Google Scholar]
- 12.Quellec G, Lee K, Dolejsi M, Garvin MK, Abramoff MD, Sonka M. Three-dimensional analysis of retinal layer texture: identification of fluid-filled regions in SD-OCT of the macula. IEEE Trans Med Imag. 2010 Jun;29:1321–1330. doi: 10.1109/TMI.2010.2047023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garvin MK, Abramoff MD, Kardon R, Russell SR, Wu X, Sonka M. Intraretinal layer segmentation of macular optical coherence tomography images using optimal 3-D graph search. IEEE Trans Med Imag. 2008 Oct;27:1495–1505. doi: 10.1109/TMI.2008.923966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garvin MK, Abramoff MD, Wu X, Russell SR, Burns TL, Sonka M. Automated 3-D intraretinal layer segmentation of macular spectral-domain optical coherence tomography images. IEEE Trans Med Imag. 2009 Sep;28:1436–1447. doi: 10.1109/TMI.2009.2016958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahlers C, Simader C, Geitzenauer W, Stock G, Stetson P, Dastmalchi S, Schnidt-Erfurth U. Automatic segmentation in three-dimensional analysis of fibrovascular pigment epithelial detachment using high-definition optical coherence tomography. Br J Ophthalmol. 2007 Oct;92:197–203. doi: 10.1136/bjo.2007.120956. [DOI] [PubMed] [Google Scholar]
- 16.Dolejší M, Abràmoff MD, Sonka M, Kybic J. Semi-Automated Segmentation of Symptomatic Exudate-Associated Derangements (SEADs) in 3D OCT Using Layer Segmentation. Biosignal. 2010 [Google Scholar]
- 17.Boykov Y, Funka-Lea G. Graph cuts and efficient N-D image segmentation. International Journal of Computer Vision (IJCV) 2006;70:109–131. [Google Scholar]
- 18.Chen Xinjian, Jayaram K Udupa, Abass Alavi, Drew A Torigian. Automatic Anatomy Recognition via MultiObject Oriented Active Shape Models. Medical Physics. 2010;37(12):6390–6401. doi: 10.1118/1.3515751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meindert Niemeijer, Kyungmoo Lee, Xinjian Chen, Li Zhang, Milan Sonka, Michael D Abràmoff. Automated Estimation of Fluid Volume in 3D OCT Scans of Patients with CNV Due to AMD, Submitted to ARVO. 2012 [Google Scholar]
- 20.Boykov Y, Kolmogorov V. An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision. IEEE Trans Pattern Anal Mach Intell. 2004 Sep;26:1124–1137. doi: 10.1109/TPAMI.2004.60. [DOI] [PubMed] [Google Scholar]
- 21.Boykov Y, Jolly MP. Interactive graph cuts for optimal boundary & region segmentation of objects in ND images. IEEE International Conference on Computer Vision (ICCV) 2001;I:105–112. [Google Scholar]
- 22.Kolmogorov V, Zabih R. What energy function can be minimized via graph cuts? IEEE Trans Pattern Anal Mach Intell. 2004;26:147–159. doi: 10.1109/TPAMI.2004.1262177. [DOI] [PubMed] [Google Scholar]
- 23.Veksler O. Star shape prior for graph-cut image segmentation. 10th European Conference on Computer Vision (ECCV) 2008;5304:454–467. [Google Scholar]
- 24.Freedman D, Zhang T. Interactive graph cut based segmentation with shape priors. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2005:755–762. [Google Scholar]
- 25.Malcolm J, Rathi Y, Tannenbaum A. Graph Cut Segmentation with Nonlinear Shape Priors. IEEE International Conference on Image Processing. 2007:365–368. [Google Scholar]
- 26.Vu N, Manjunath BS. Shape prior segmentation of multiple objects with graph cuts. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2008:1–8. [Google Scholar]
- 27.Chen X, Bagci U. 3D automatic anatomy segmentation based on iterative graph-cut-ASM. Med Phys. 2011;38(8):4610–4622. doi: 10.1118/1.3602070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delong A, Boykov Y. Globally optimal segmentation of multi-region objects. IEEE International Conference on Computer Vision (ICCV) 2009 [Google Scholar]
- 29.Li K, Wu X, Chen DZ, Sonka M. Efficient optimal surface detection: theory, implementation and experimental validation. Proc SPIE Int'l Symp Medical Imaging: Image Processing. 2004 May;5370:620–627. [Google Scholar]
- 30.Li K, Wu X, Chen DZ, Sonka M. Optimal surface segmentation in volumetric images - a graph-theoretic approach. IEEE Trans Pattern Anal Mach Intell. 2006 Jan;28:119–134. doi: 10.1109/TPAMI.2006.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin Y, Zhang X, Williams R, Wu X, Anderson DD, Sonka M. LOGISMOS—Layered Optimal Graph Image Segmentation of Multiple Objects and Surfaces: Cartilage Segmentation in the Knee Joint. IEEE Transactions on Medical Imaging. 2010;29(12):2023–2037. doi: 10.1109/TMI.2010.2058861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Appleton B, Talbot H. Globally minimal surfaces by continuous maximal flows. IEEE transactions on Pattern Analysis and Pattern Recognition (PAMI) 2006;28(1):106–118. doi: 10.1109/TPAMI.2006.12. [DOI] [PubMed] [Google Scholar]
- 33.Song Q, Chen M, Bai J, Sonka M, Wu X. Surface-region context in optimal multi-object graph-based segmentation: Robust delineation of pulmonary tumors. Proc of the 22nd Biennial International Conference on Information Processing in Medical Imaging (IPMI); 2011; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abramoff Michael D, Alward Wallace LM, Greenlee Emily C, Shuba Lesya, Kim Chan Y, Fingert John H, Kwon Young H. Automated Segmentation of the Optic Disc from Stereo Color Photographs Using Physiologically Plausible Features. Investigative Ophthalmology & Visual Science. 2007 Apr;48(4):1665–1673. doi: 10.1167/iovs.06-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arya S, Mount DM, Netanyahu NS, Silverman R, Wu AY. An optimal algorithm for approximate nearest neighbor searching. Journal of the ACM. 1998;45:891–923. [Google Scholar]
- 36.Niemeijer Meindert, Ginneken Bram van, Staal Joes, Suttorp-Schulten Maria SA, Abràmoff Michael D. Automatic detection of red lesions in digital color fundus photographs. IEEE Trans Med Imaging. 2005;24(5):584–592. doi: 10.1109/TMI.2005.843738. [DOI] [PubMed] [Google Scholar]
- 37.Snyman JA. Practical Mathematical Optimization: An Introduction to Basic Optimization Theory and Classical and New Gradient-Based Algorithms. Springer Publishing; 2005. ISBN 0-387-24348-8. [Google Scholar]
- 38.Wu X, Chen DZ. Optimal net surface problems with applications. ICALP. 2002:1029–1042. [Google Scholar]
- 39.Christopher M, Moga DC, Russell SR, Folk JC, Scheetz T, Abràmoff MD. Validation of tablet-based evaluation of color fundus images. Retina. 2012 doi: 10.1097/IAE.0b013e3182483361. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Udupa JK, Leblanc VR, Zhuge Y, Imielinska C, Schmidt H, Currie LM, Hirsch BE, Woodburn J. A framework for evaluating image segmentation algorithms. Computerized Medical Imaging and Graphics. 2006;30(2):75–87. doi: 10.1016/j.compmedimag.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Krzanowski WJ. Principles of Multivariate Analysis. Oxford University Press; 1988. [Google Scholar]
- 42.Cox DR, Hinkley DV. Theoretical Statistics, Chapman & Hall (Appendix 3) 1974 ISBN 0412124203. [Google Scholar]
- 43.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 44.Fernandez DC, Salinas HM, Puliafito CA. Automated detection of retinal layer structures on optical coherence tomography images. Optics Express. 2005;13(25):10200–10216. doi: 10.1364/opex.13.010200. [DOI] [PubMed] [Google Scholar]
- 45.Mayer Markus, Hornegger Joachim, Mardin Christian Y, Tornow Ralf P. Retinal Nerve Fiber Layer Segmentation on FD-OCT Scans of Normal Subjects and Glaucoma Patients Biomedical. Optics Express. 2010;1(5):1358–1383. doi: 10.1364/BOE.1.001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiu SJ, Li XT, Nicholas P, Toth CA, Izatt JA, Farsiu S. Automatic segmentation of seven retinal layers in SDOCT images congruent with expert manual segmentation. Opt Express. 2010;18(18):19413–19428. doi: 10.1364/OE.18.019413. [DOI] [PMC free article] [PubMed] [Google Scholar]